Bayesian Analysis Used to Identify Clinical and Laboratory Variables Capable of Predicting Progression to Severe Dengue among Infected Pediatric Patients
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. General Description of the Cases and Controls
3.2. Comparison between Groups of the Clinical Conditions and Laboratory Parameters
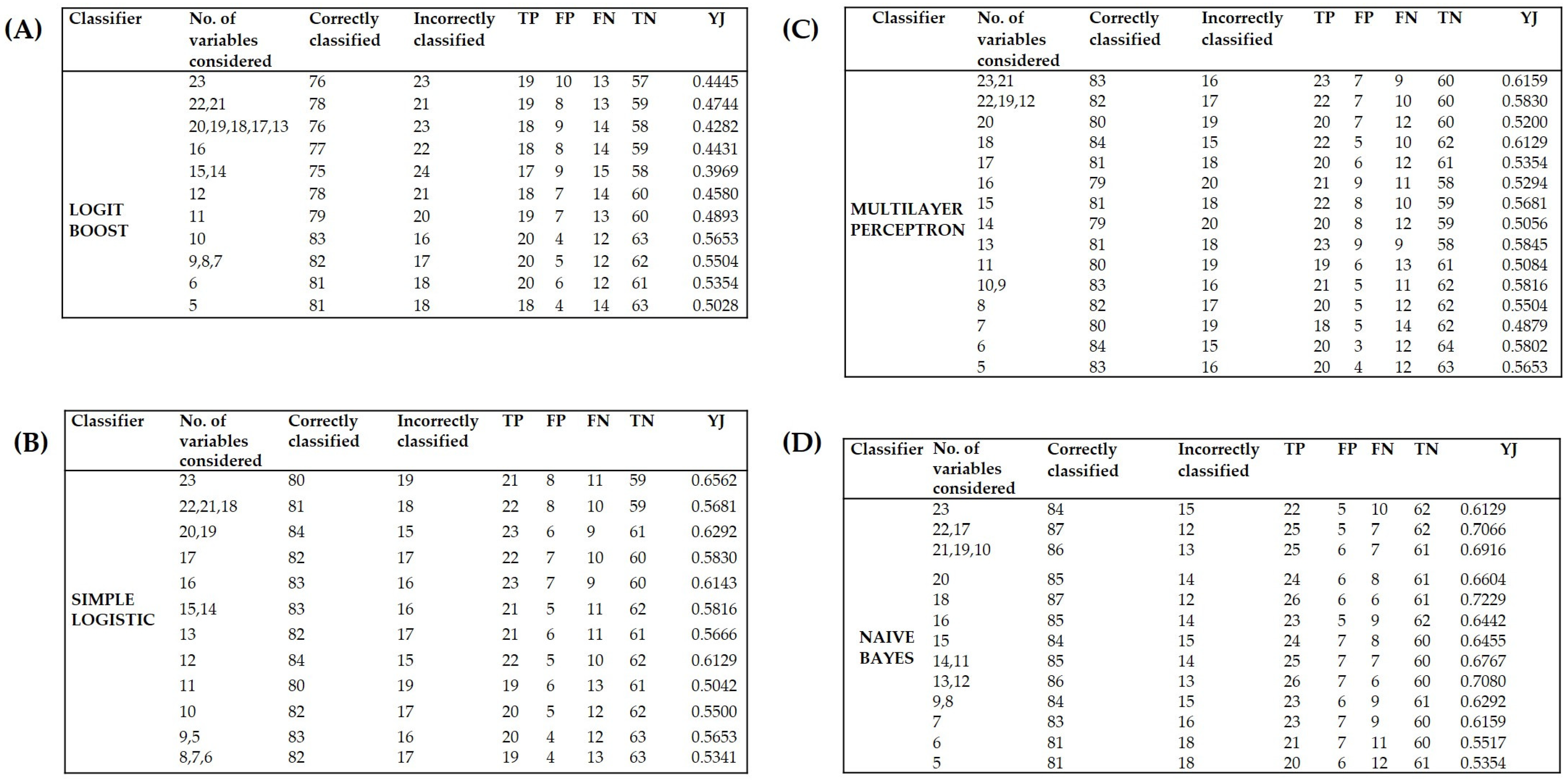
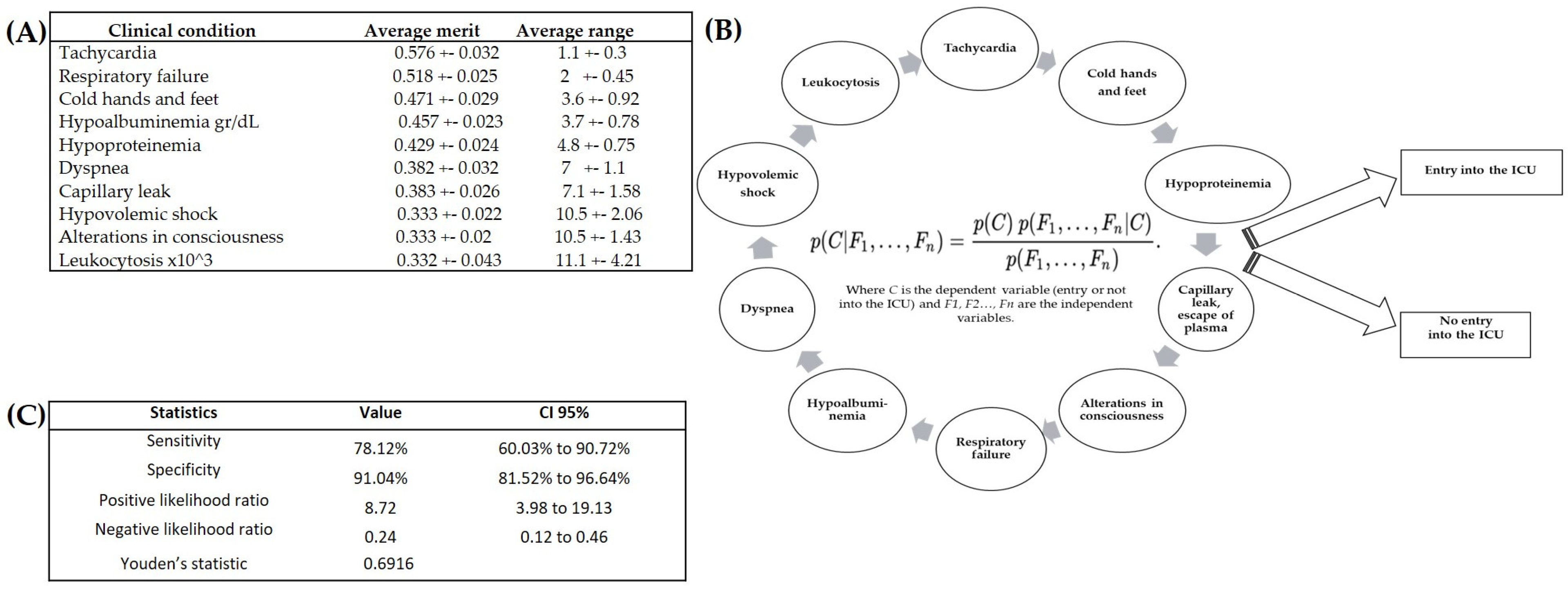
3.3. Identification of the Variables Capable of Prognosis of Severe Dengue
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- San Martín, J.L.; Brathwaite, O.; Zambrano, B.; Solórzano, J.O.; Bouckenooghe, A.; Dayan, G.H.; Guzmán, M.G. The Epidemiology of Dengue in the Americas over the Last Three Decades: A Worrisome Reality. Am. J. Trop. Med. Hyg. 2010, 82, 128–135. [Google Scholar] [CrossRef]
- Messina, J.P.; Brady, O.J.; Golding, N.; Kraemer, M.U.G.; Wint, G.R.W.; Ray, S.E.; Pigott, D.M.; Shearer, F.M.; Johnson, K.; Earl, L.; et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019, 4, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Dengue. Available online: https://www.paho.org/data/index.php/en/mnu-topics/indicadoresdengue-en.html (accessed on 12 October 2021).
- Dos Santos, T.H.; Martin, J.L.S.; Castellanos, L.G.; Espinal, M.A. Dengue in the Americas: Honduras’ worst outbreak. Lancet 2019, 394, 2149. [Google Scholar] [CrossRef] [PubMed]
- SINAVE 2018. Panorama Epidemiológico de Dengue 2018. Availaboratory le Online: Panorama Epidemiológico de Dengue 2018—Semana Epidemiológica 52|Secretaría de Salud|Gobierno|gob.mx. Available online: www.gob.mx (accessed on 15 January 2023).
- De Antonio, R.; Amaya-Tapia, G.; Ibarra-Nieto, G.; Huerta, G.; Damaso, S. Incidence of dengue illness in Mexican people aged 6 months to 50 years old: A prospective cohort study conducted in Jalisco. PLoS ONE 2021, 16, e0250253. [Google Scholar] [CrossRef]
- World Health Organization. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control; New Edition; World Health Organization: Geneva, Switzerland, 2009; Available online: https://www.ncbi.nlm.nih.gov/books/NBK143157/ (accessed on 12 December 2021).
- Schaefer, T.J.; Panda, P.K.; Wolford, R.W. Dengue Fever. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023; Available online: https://www.ncbi.nlm.nih.gov/books/NBK430732/ (accessed on 14 November 2022).
- Rodenhuis-Zybert, I.A.; Wilschut, J.; Smit, J.M. Dengue virus life cycle: Viral and host factors modulating infectivity. Cell. Mol. Life Sci. CMLS 2010, 67, 2773–2786. [Google Scholar] [CrossRef]
- Pan American Health Organization. Guidelines for the Clinical Diagnosis and Treatment of Dengue, Chikungunya, and Zika. Available online: https://iris.paho.org/handle/10665.2/55867 (accessed on 13 July 2023).
- Toledo, J.; George, L.; Martinez, E.; Lazaro, A.; Han, W.W.; Coelho, G.E.; Runge Ranzinger, S.; Horstick, O. Relevance of Non-communicable Comorbidities for the Development of the Severe Forms of Dengue: A Systematic Literature Review. PLoS Neglected Trop. Dis. 2016, 10, e0004284. [Google Scholar] [CrossRef]
- Macias, A.E.; Werneck, G.L.; Castro, R.; Mascareñas, C.; Coudeville, L.; Morley, D.; Recamier, V.; Guergova-Kuras, M.; Etcheto, A.; Puentes-Rosas, E.; et al. Mortality among Hospitalized Dengue Patients with Comorbidities in Mexico, Brazil, and Colombia. Am. J. Trop. Med. Hyg. 2021, 105, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Fonseca-Portilla, R.; Martínez-Gil, M.; Morgenstern-Kaplan, D. Risk factors for hospitalization and mortality due to dengue fever in a Mexican population: A retrospective cohort study. Int. J. Infect. Dis. 2021, 110, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.K.; Ngoc, T.V.; Thu Thuy, T.T.; Hong Van, N.T.; Nhu Thuy, T.T.; Hoai Tam, D.T.; Dung, N.M.; Tien, N.T.H.; Kieu, N.T.T.; Simmons, C.P.; et al. The value of daily platelet counts for predicting dengue shock syndrome: Results from a prospective observational study of 2301 Vietnamese children with dengue. PLoS Neglected Trop. Dis. 2017, 11, e0005498. [Google Scholar] [CrossRef]
- Dey, S.K.; Rahman, M.M.; Howlader, A.; Siddiqi, U.R.; Uddin, K.M.M.; Borhan, R.; Rahman, E.U. Prediction of dengue incidents using hospitalized patients, metrological and socio-economic data in Bangladesh: A machine learning approach. PLoS ONE 2022, 17, e0270933. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Ho, T.N.; Nguyen, V.V.; Nguyen, T.H.; Ha, M.T.; Ta, V.T.; Nguyen, L.D.; Phan, L.; Han, K.Q.; Duong, T.H.; et al. An Evidence-Based Algorithm for Early Prognosis of Severe Dengue in the Outpatient Setting. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2017, 64, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Huy, B.V.; Toàn, N.V. Prognostic indicators associated with progresses of severe dengue. PLoS ONE 2022, 17, e0262096. [Google Scholar] [CrossRef] [PubMed]
- Thach, T.Q.; Eisa, H.G.; Hmeda, A.B.; Faraj, H.; Thuan, T.M.; Abdelrahman, M.M.; Awadallah, M.G.; Ha, N.X.; Noeske, M.; Abdul Aziz, J.M.; et al. Predictive markers for the early prognosis of dengue severity: A systematic review and meta-analysis. PLoS Neglected Trop. Dis. 2021, 15, e0009808. [Google Scholar] [CrossRef] [PubMed]
- Djossou, F.; Vesin, G.; Elenga, N.; Demar, M.; Epelboin, L.; Walter, G.; Abboud, P.; Le-Guen, T.; Rousset, D.; Moreau, B.; et al. A predictive score for hypotension in patients with confirmed dengue fever in Cayenne Hospital, French Guiana. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 705–713. [Google Scholar] [CrossRef]
- Van de Schoot, R.; Depaoli, S.; King, R.; Kramer, B.; Märtens, K.; Tadesse, M.G.; Vannucci, M.; Gelman, A.; Veen, D.; Willemsen, J.; et al. Bayesian statistics and modelling. Nat. Rev. Methods Primers 2021, 1, 1. [Google Scholar] [CrossRef]
- Chen, D.G.; Fraser, M.W. A Bayesian Approach to Sample Size Estimation and the Decision to Continue Program Development in Intervention Research. J. Soc. Soc. Work. Res. 2017, 8, 457–470. [Google Scholar] [CrossRef]
- Van Eeden, W.A.; Luo, C.; Van Hemert, A.M.; Carlier, I.V.E.; Penninx, B.W.; Wardenaar, K.J.; Hoos, H.; Giltay, E.J. Predicting the 9-year course of mood and anxiety disorders with automated machine learning: A comparison between auto-sklearn, naïve Bayes classifier, and traditional logistic regression. Psychiatry Res. 2021, 299, 113823. [Google Scholar] [CrossRef]
- Bone, C.; Simmonds-Buckley, M.; Thwaites, R.; Sandford, D.; Merzhvynska, M.; Rubel, J.; Deisenhofer, A.K.; Lutz, W.; Delgadillo, J. Dynamic prediction of psychological treatment outcomes: Development and validation of a prediction model using routinely collected symptom data. Lancet Digit. Health 2021, 3, e231–e240. [Google Scholar] [CrossRef]
- Tomasiuk, R.; Dabrowski, J.; Smykiewicz, J.; Wiacek, M. Predictors of COVID-19 Hospital Treatment Outcome. Int. J. Gen. Med. 2021, 14, 10247–10256. [Google Scholar] [CrossRef]
- Aswi, A.; Cramb, S.M.; Moraga, P.; Mengersen, K. Bayesian spatial and spatio-temporal approaches to modelling dengue fever: A systematic review. Epidemiol. Infect. 2018, 147, e33. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Gómez, M.; Danglot-Banck, C.; Huerta Alvarado, S.G.; García de la Torre, G. El estudio de casos y controles: Su diseño, análisis e interpretación, en investigación clínica. Rev. Mex. Pediatr. 2003, 70, 257–263. [Google Scholar]
- Martínez, D.; Papuzinski, C.; Stojanova, J.; Arancibia, M. General concepts in biostatistics and clinical epidemiology: Observational studies with case-control design. Medwave 2019, 19, e7716. [Google Scholar] [CrossRef]
- Schapire, R.E. The Boosting Approach to Machine Learning: An Overview. In Nonlinear Estimation and Classification; Denison, D.D., Hansen, M.H., Holmes, C.C., Mallick, B., Yu, B., Eds.; Lecture Notes in Statistics; Springer: Berlin/Heidelberg, Germany, 2003; Volume 171. [Google Scholar] [CrossRef]
- Walley, W.J.; Džeroski, S. Biological Monitoring: A Comparison between Bayesian, Neural and Machine Learning Methods of Water Quality Classification. In Environmental Software Systems; Denzer, R., Schimak, G., Russell, D., Eds.; IFIP—The International Federation for Information Processing; Springer: Boston, MA, USA, 1996. [Google Scholar] [CrossRef]
- Kamarudin, M.H.; Maple, C.; Watson, T.; Safa, N.S. A LogitBoost-Based Algorithm for Detecting Known and Unknown Web Attacks. IEEE Access 2017, 5, 26190–26200. [Google Scholar] [CrossRef]
- Bandeira, A.P. Aplicação de Rede Neural Artificial para o Reconhecimento do Diabetes Mellitus Gestacional com Marcadores Não-Glicêmicos. Master’s Thesis, Universidade Federal do Paraná, Curitiba, Braazil, 2015. [Google Scholar]
- Aler, R. Tutorial Weka 3.6.0 Contenidos 2009. Available online: https://1library.co/document/y6964ony-tutorial-weka-ricardo-aler-2009.html (accessed on 15 June 2020).
- Hernández Rosales, D.E. Modelado de la Capacidad Funcional Articular de la Mano Usando Algoritmos de Inteligencia Artificial en Pacientes con Artritis Reumatoide. Master’s Thesis, Universidad Autónoma de Guerrero, Facultad de Ingeniería, Chilpancingo, Mexico, 2017. Available online: http://ri.uagro.mx/handle/uagro/294?locale-attribute=en (accessed on 15 June 2023).
- Cifuentes, L.; Cerda, J. Clinical use of diagnostic tests (Part 2). Clinical application and usefulness of a diagnostic test. Rev. Chil. Infectol. 2010, 27, 316–319. (In Spanish) [Google Scholar] [CrossRef]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques, 3rd ed.; Morgan Kaufmann: San Francisco, CA, USA, 2011; Consultado el 2011-01-19. [Google Scholar]
- Pone, S.M.; Hökerberg, Y.H.; de Oliveira, R.d.V.; Daumas, R.P.; Pone, T.M.; Pone, M.V.; Brasil, P. Clinical and laboratory oratory signs associated to serious dengue disease in hospitalized children. J. Pediatr. 2016, 92, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Cerda, J.; Cifuentes, L. Uso de curvas ROC en investigación clínica: Aspectos teórico-prácticos. Rev. Chil. Infectol. 2012, 29, 138–141. [Google Scholar] [CrossRef]
- Phakhounthong, K.; Chaovalit, P.; Jittamala, P.; Blacksell, S.D.; Carter, M.J.; Turner, P.; Chheng, K.; Sona, S.; Kumar, V.; Day, N.P.J.; et al. Predicting the severity of dengue fever in children on admission based on clinical features and laboratory oratory indicators: Application of classification tree analysis. BMC Pediatr. 2018, 18, 109. [Google Scholar] [CrossRef]
- Sangkaew, S.; Ming, D.; Boonyasiri, A.; Honeyford, K.; Kalayanarooj, S.; Yacoub, S.; Dorigatti, I.; Holmes, A. Risk predictors of progression to severe disease during the febrile phase of dengue: A systematic review and meta-analysis. Lancet Infect. Dis. 2021, 21, 1014–1026. [Google Scholar] [CrossRef]
- Arora, S.K.; Nandan, D.; Sharma, A.; Benerjee, P.; Singh, D.P. Predictors of severe dengue amongst children as per the revised WHO classification. J. Vector Borne Dis. 2021, 58, 329–334. [Google Scholar] [CrossRef]
- Tsheten, T.; Clements, A.C.A.; Gray, D.J.; Adhikary, R.K.; Furuya-Kanamori, L.; Wangdi, K. Clinical predictors of severe dengue: A systematic review and meta-analysis. Infect. Dis. Poverty 2021, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Tamibmaniam, J.; Hussin, N.; Cheah, W.K.; Ng, K.S.; Muninathan, P. Proposal of a Clinical Decision Tree Algorithm Using Factors Associated with Severe Dengue Infection. PLoS ONE 2016, 11, e0161696. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Liu, S.F.; Liao, S.C.; Lee, K.; Liu, J.W.; Lin, A.S.; Lin, M.C. Acute respiratory failure in adult patients with dengue virus infection. Am. J. Trop. Med. Hyg. 2007, 77, 151–158. [Google Scholar] [CrossRef]
- Abhay, A.S.; Kattoor, S.; Paul, D.; Antony, T.P. Clinical course and outcome of dengue fever patients admitted with respiratory manifestations. Pulmon 2020, 22, 217–223. [Google Scholar]
- Gelman, A.; Simpson, D.; Betancourt, M. The prior can often only be understood in the context of the likelihood. Entropy 2017, 19, 555. [Google Scholar] [CrossRef]
- Nuzzo, R.L. An Introduction to Bayesian Data Analysis for Correlations. PM R J. Inj. Funct. Rehabil. 2017, 9, 1278–1282. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Shen, L.; Lehman, L.W.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III 2016, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Gutierrez, G. Artificial Intelligence in the Intensive Care Unit. Crit. Care 2020, 24, 101. [Google Scholar] [CrossRef]
- Greco, M.; Caruso, P.F.; Cecconi, M. Artificial Intelligence in the Intensive Care Unit. Semin. Respir. Crit. Care Med. 2021, 42, 2–9. [Google Scholar] [CrossRef]
- Bayesian statistics and modelling. Nat. Rev. Methods Primers 2021, 1, 3. [CrossRef]
- Sehlabana, M.A.; Maposa, D.; Boateng, A. Modelling Malaria Incidence in the Limpopo Province, South Africa: Comparison of Classical and Bayesian Methods of Estimation. Int. J. Environ. Res. Public Health 2020, 17, 5016. [Google Scholar] [CrossRef] [PubMed]
| Clinical Variables | Cases (n = 32) | Controls (n = 67) | Fisher’s Exact Test p-Value |
|---|---|---|---|
| Mucosal bleeding | 14 | 13 | 0.0157 |
| Substantial escape of blood plasma with a weak or undetectable pulse | 8 | 1 | 0.0004 |
| Tachycardia | 15 | 1 | <0.0001 *** |
| Cold hands and feet | 11 | 1 | <0.0001 *** |
| Respiratory failure | 15 | 2 | <0.0001 *** |
| Hematemesis | 8 | 3 | 0.0045 |
| Alterations in consciousness | 5 | 0 | 0.0028 |
| Diaphoresis | 5 | 2 | 0.0342 |
| Fainting | 5 | 2 | 0.0342 |
| Conjunctivitis | 3 | 0 | 0.0316 |
| Cough | 7 | 4 | 0.0352 |
| Dyspnea | 8 | 1 | 0.0004 |
| Stupor | 4 | 0 | 0.0096 |
| Disorientation | 4 | 0 | 0.0096 |
| Shock | 5 | 0 | 0.0028 |
| Pleural effusion | 7 | 3 | 0.0120 |
| Mottled skin | 3 | 0 | 0.0316 |
| Comorbidities | 7 | 2 | 0.0047 |
| Hemorrhage | 16 | 13 | 0.0040 |
| Laboratory Variables | Cases (n = 32) x (SD) | Controls (n = 67) x (SD) | Student’s t-Test p-Value |
|---|---|---|---|
| Hematocrit | 38.58 (8.77) | 39.76 (6.07) | 0.4377 |
| Hb gr/100 mL | 13.34 (3.09) | 13.86 (2.10) | 0.3277 |
| Platelet × 103 | 39.63 (58.00) | 62.54 (55.92) | 0.0626 |
| Albumin gr/Dl | 2.95 (0.67) | 3.61 (0.50) | <0.0001 *** |
| Leukocytes × 103 | 7.77 (4.49) | 5.22 (2.70) | 0.0007 * |
| Lymphocytes × 103 | 1.94 (1.36) | 1.72 (1.13) | 0.3989 |
| ALT | 215.71 (818.36) | 53.65 (42.93) | 0.1073 |
| AST | 757.02 (2841.69) | 138.19 (95.34) | 0.0765 |
| DB | 0.26 (0.40) | 0.12 (0.07) | 0.0063 ** |
| TB | 0.80 (0.53) | 0.60 (0.24) | 0.0110 * |
| Alkaline phosphatase u/L | 133.73 (62.94) | 141.48 (40.86) | 0.4636 |
| C-reactive protein mg/Dl | 4.67 (7.57) | 1.11 (1.53) | 0.0003 ** |
| Neutrophils × 103 | 4.13 (3.26) | 2.75 (2.00) | 0.0109 * |
| Total proteins | 5.22 (1.05) | 6.15 (0.71) | <0.0001 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corzo-Gómez, J.; Guzmán-Aquino, S.; Vargas-De-León, C.; Megchún-Hernández, M.; Briones-Aranda, A. Bayesian Analysis Used to Identify Clinical and Laboratory Variables Capable of Predicting Progression to Severe Dengue among Infected Pediatric Patients. Children 2023, 10, 1508. https://doi.org/10.3390/children10091508
Corzo-Gómez J, Guzmán-Aquino S, Vargas-De-León C, Megchún-Hernández M, Briones-Aranda A. Bayesian Analysis Used to Identify Clinical and Laboratory Variables Capable of Predicting Progression to Severe Dengue among Infected Pediatric Patients. Children. 2023; 10(9):1508. https://doi.org/10.3390/children10091508
Chicago/Turabian StyleCorzo-Gómez, Josselin, Susana Guzmán-Aquino, Cruz Vargas-De-León, Mauricio Megchún-Hernández, and Alfredo Briones-Aranda. 2023. "Bayesian Analysis Used to Identify Clinical and Laboratory Variables Capable of Predicting Progression to Severe Dengue among Infected Pediatric Patients" Children 10, no. 9: 1508. https://doi.org/10.3390/children10091508
APA StyleCorzo-Gómez, J., Guzmán-Aquino, S., Vargas-De-León, C., Megchún-Hernández, M., & Briones-Aranda, A. (2023). Bayesian Analysis Used to Identify Clinical and Laboratory Variables Capable of Predicting Progression to Severe Dengue among Infected Pediatric Patients. Children, 10(9), 1508. https://doi.org/10.3390/children10091508






