Prenatal Identification of a Novel Mutation in the MCPH1 Gene Associated with Autosomal Recessive Primary Microcephaly (MCPH) Using Next Generation Sequencing (NGS): A Case Report and Review of the Literature
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Report
2.2. Molecular Karyotyping
2.3. Next Generation Sequencing (NGS)
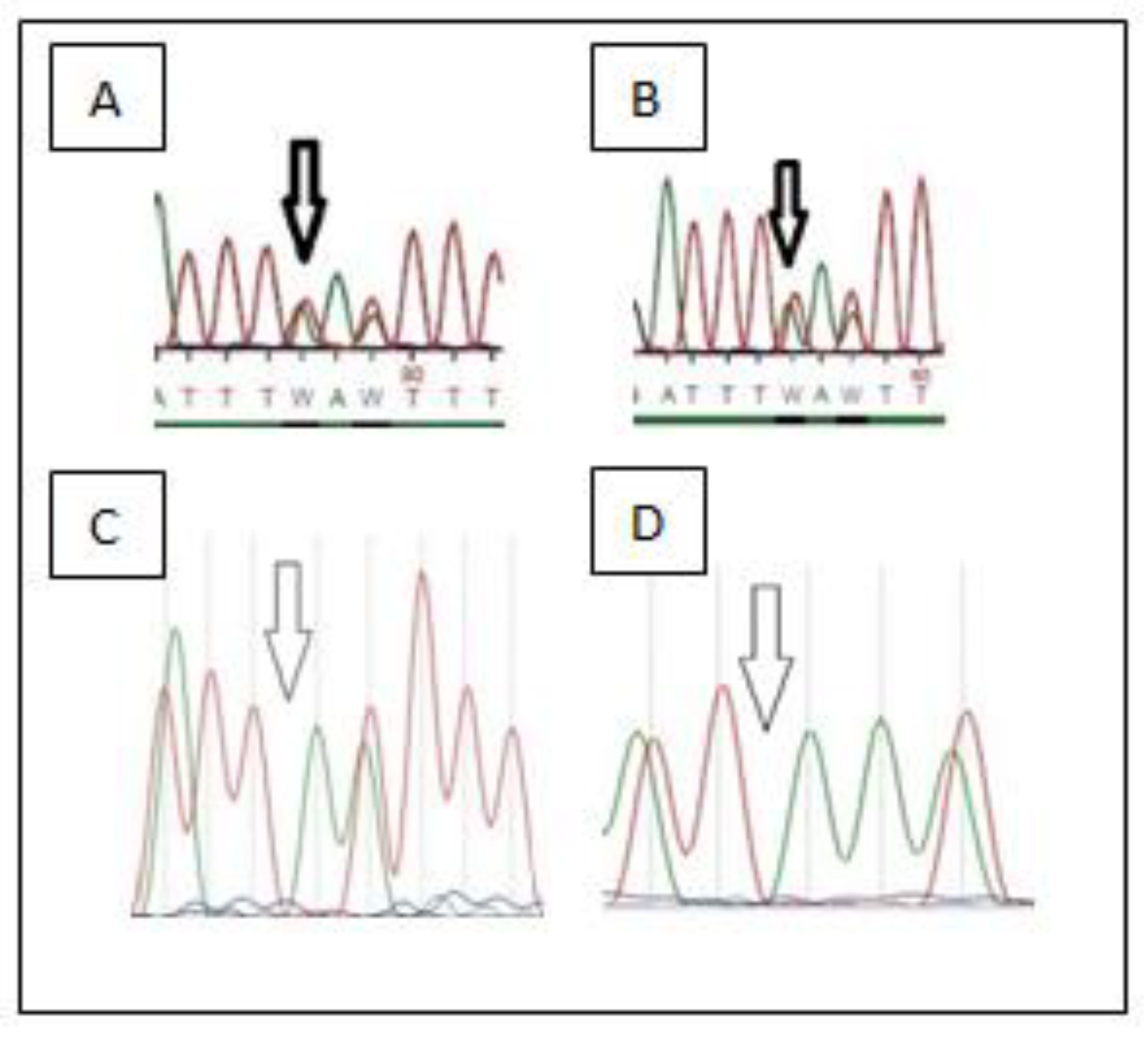
3. Results
Pathologic Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morris-Rosendahl, D.J.; Kaindl, A.M. What next-generation sequencing (NGS) technology has enabled us to learn about primary autosomal recessive microcephaly (MCPH). Mol. Cell. Probes 2015, 29, 271–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jean, F.; Stuart, A.; Tarailo-Graovac, M. Dissecting the Genetic and Etiological Causes of Primary Microcephaly. Front. Neurol. 2020, 11, 570830. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.P.; McHale, D.P.; Campbell, D.A.; Jafri, H.; Rashid, Y.; Mannan, J.; Karbani, G.; Corry, P.; Levene, M.I.; Mueller, R.F.; et al. Primary Autosomal Recessive Microcephaly (MCPH1) Maps to Chromosome 8p22-pter. Am. J. Hum. Genet. 1998, 63, 541–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, A.; Eastwood, H.; Bell, S.M.; Adu, J.; Toomes, C.; Carr, I.M.; Roberts, E.; Hampshire, D.; Crow, Y.; Mighell, A.J.; et al. Identification of Microcephalin, a Protein Implicated in Determining the Size of the Human Brain. Am. J. Hum. Genet. 2002, 71, 136–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbelanne, M.; Tsang, W.Y. Molecular and cellular basis of autosomal recessive primary microcephaly. BioMed Res. Int. 2014, 2014, 547986. [Google Scholar] [CrossRef] [Green Version]
- Cox, J.; Jackson, A.P.; Bond, J.; Woods, C.G. What primary microcephaly can tell us about brain growth? Trends Mol. Med. 2006, 12, 358–366. [Google Scholar] [CrossRef]
- Gruber, R.; Zhou, Z.; Sukchev, M.; Joerss, T.; Frappart, P.-O.; Wang, Z.-Q. MCPH1 regulates the neuroprogenitor division mode by coupling the centrosomal cycle with mitotic entry through the Chk1–Cdc25 pathway. Nat. Cell Biol. 2011, 13, 1325–1334. [Google Scholar] [CrossRef]
- Passemard, S.; Titomanlio, L.; Elmaleh, M.; Afenjar, A.; Alessandri, J.-L.; Andria, G.; de Villemeur, T.B.; Boespflug-Tanguy, O.; Burglen, L.; Del Giudice, E.; et al. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology 2009, 73, 962–969. [Google Scholar] [CrossRef]
- Kaindl, A.M.; Passemard, S.; Kumar, P.; Kraemer, N.; Issa, L.; Zwirner, A.; Gerard, B.; Verloes, A.; Mani, S.; Gressens, P. Many roads lead to primary autosomal recessive microcephaly. Prog. Neurobiol. 2010, 90, 363–383. [Google Scholar] [CrossRef]
- Zaqout, S.; Morris-Rosendahl, D.; Kaindl, A.M.; Zaqout, S. Autosomal Recessive Primary Microcephaly (MCPH): An Update. Neuropediatrics 2017, 48, 135–142. [Google Scholar] [CrossRef]
- Muhammad, F.; Baig, S.M.; Hansen, L.; Hussain, M.S.; Inayat, I.A.; Aslam, M.; Qureshi, J.A.; Toilat, M.; Kirst, E.; Wajid, M.; et al. Compound heterozygous ASPM mutations in Pakistani MCPH families. Am. J. Med. Genet. Part A 2009, 149A, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, T.; Nagashri, M.N.; Swamy, S.S.; Mohiyuddin, S.M.A.; Gopinath, K.S.; Kumar, A. Primary Microcephaly Gene MCPH1 Shows Signatures of Tumor Suppressors and Is Regulated by miR-27a in Oral Squamous Cell Carcinoma. PLoS ONE 2013, 8, e54643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulvers, J.N.; Journiac, N.; Arai, Y.; Nardelli, J. MCPH1: A window into brain development and evolution. Front. Cell. Neurosci. 2015, 9, 92. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, S.H.; Mundy, N.I. Microcephaly genes evolved adaptively throughout the evolution of eutherian mammals. BMC Evol. Biol. 2014, 14, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woods, C.G.; Bond, J.; Enard, W. Autosomal Recessive Primary Microcephaly (MCPH): A Review of Clinical, Molecular, and Evolutionary Findings. Am. J. Hum. Genet. 2005, 76, 717–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trimborn, M.; Bell, S.M.; Felix, C.; Rashid, Y.; Jafri, H.; Griffiths, P.D.; Neumann, L.M.; Krebs, A.; Reis, A.; Sperling, K.; et al. Mutations in Microcephalin Cause Aberrant Regulation of Chromosome Condensation. Am. J. Hum. Genet. 2004, 75, 261–266. [Google Scholar] [CrossRef] [Green Version]
- Garshasbi, M.; Motazacker, M.M.; Kahrizi, K.; Behjati, F.; Abedini, S.S.; Nieh, S.E.; Firouzabadi, S.G.; Becker, C.; Rüschendorf, F.; Nürn-berg, P.; et al. SNP ar-ray-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Hum. Genet. 2006, 118, 708–715. [Google Scholar] [CrossRef]
- Pfau, R.B.; Thrush, D.L.; Hamelberg, E.; Bartholomew, D.; Botes, S.; Pastore, M.; Tan, C.; Del Gaudio, D.; Gastier-Foster, J.M.; Astbury, C. MCPH1 deletion in a newborn with severe microcephaly and premature chromosome condensation. Eur. J. Med. Genet. 2013, 56, 609–613. [Google Scholar] [CrossRef]
- Darvish, H.; Esmaeeli-Nieh, S.; Monajemi, G.B.; Mohseni, M.; Ghasemi-Firouzabadi, S.; Abedini, S.S.; Bahman, I.; Jamali, P.; Azimi, S.; Mojahedi, F.; et al. A clinical and molecular genetic study of 112 Iranian families with primary microcephaly. J. Med. Genet. 2010, 47, 823–828, Erratum in J. Med. Genet. 2014, 51, 70. [Google Scholar] [CrossRef] [Green Version]
- Cowie, V. The genetics and sub-classification of microcephaly. J. Ment. Defic. Res. 1960, 4, 42–47. [Google Scholar] [CrossRef]
- Naseer, M.I.; Rasool, M.; Abdulkareem, A.A.; Bassiouni, R.I.; Algahtani, H.; Chaudhary, A.G.; Al-Qahtani, M.H. Novel compound heterozygous mutations in MCPH1 gene causes primary microcephaly in Saudi family. Neurosciences 2018, 23, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Pavone, P.; Pappalardo, X.G.; Praticò, A.D.; Polizzi, A.; Ruggieri, M.; Piccione, M.; Corsello, G.; Falsaperla, R. Primary Microcephaly with Novel Variant of MCPH1 Gene in Twins: Both Manifesting in Childhood at the Same Time with Hashimoto’s Thyroiditis. J. Pediatr. Genet. 2020, 9, 177–182. [Google Scholar] [CrossRef] [PubMed]

| AP3B2, AP4B1, AP4E1, AP4M1, APEX1, ARFGEF2, ASNS, ASPM, ATR, ATRX, BMPR1A, BRWD3, C2, CDK5RAP2, CDON, CDT1, CENPJ, CEP152, COL2A1, CYP21A2, CZ1P-ASNS, DICER1, DPP6, EFTUD2, EHMT2, EMG1, F10, F2, F5, FAM20A, FAM20C, FLNA, FLT1, FN3K, GLI2, GNAS, HUWE1, HYMAI, IGF1R, KARS, KNL1, LIG4, LMNB2, LOC100287042, LOC102724058, MCM5, MCPH1, MCPH1-AS1, MED17, MMP2, MRE11, MTHFR, MTNR1A, MYH11, NBN, NCAPD2, NDE1, NELFE, NPC1, ORC4, ORC6, OSGEP, PACERR, PCNT, PEX2, PHB2, PLAGL1, PRKAR1A, PRKDC, PTGS1, PTGS2, PTPRJ, REV3L, RPTOR, SCAP, SCARNA10, SCN10A, SCN1A, SERPINA3, SKIV2L, SLC16A2, SLC25A19, STIL, STK4, TBCD, TCOF1, TERT, TP53, TUBA1A, TUBGCP6, VPS13B, VPS35, VRK1, WDR62, WDR81, XRN1, ZNF335, ZNF592, ZNF750 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papoulidis, I.; Eleftheriades, M.; Manolakos, E.; Petersen, M.B.; Liappi, S.M.; Konstantinidou, A.; Papamichail, M.; Papadopoulos, V.; Garas, A.; Sotiriou, S.; et al. Prenatal Identification of a Novel Mutation in the MCPH1 Gene Associated with Autosomal Recessive Primary Microcephaly (MCPH) Using Next Generation Sequencing (NGS): A Case Report and Review of the Literature. Children 2022, 9, 1879. https://doi.org/10.3390/children9121879
Papoulidis I, Eleftheriades M, Manolakos E, Petersen MB, Liappi SM, Konstantinidou A, Papamichail M, Papadopoulos V, Garas A, Sotiriou S, et al. Prenatal Identification of a Novel Mutation in the MCPH1 Gene Associated with Autosomal Recessive Primary Microcephaly (MCPH) Using Next Generation Sequencing (NGS): A Case Report and Review of the Literature. Children. 2022; 9(12):1879. https://doi.org/10.3390/children9121879
Chicago/Turabian StylePapoulidis, Ioannis, Makarios Eleftheriades, Emmanouil Manolakos, Michael B. Petersen, Simoni Marina Liappi, Anastasia Konstantinidou, Maria Papamichail, Vassilios Papadopoulos, Antonios Garas, Sotirios Sotiriou, and et al. 2022. "Prenatal Identification of a Novel Mutation in the MCPH1 Gene Associated with Autosomal Recessive Primary Microcephaly (MCPH) Using Next Generation Sequencing (NGS): A Case Report and Review of the Literature" Children 9, no. 12: 1879. https://doi.org/10.3390/children9121879
APA StylePapoulidis, I., Eleftheriades, M., Manolakos, E., Petersen, M. B., Liappi, S. M., Konstantinidou, A., Papamichail, M., Papadopoulos, V., Garas, A., Sotiriou, S., Papastefanou, I., Daskalakis, G., & Ristic, A. (2022). Prenatal Identification of a Novel Mutation in the MCPH1 Gene Associated with Autosomal Recessive Primary Microcephaly (MCPH) Using Next Generation Sequencing (NGS): A Case Report and Review of the Literature. Children, 9(12), 1879. https://doi.org/10.3390/children9121879








