Measurement and Characterization of Yeast Cell Size Using a Digital Optical Microscope
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Yeast Preparation
2.3. Measurement of Cell Area and Length
2.4. Statistical Analysis
3. Results and Discussion
3.1. Validation of the Proposed Method in Comparison with Conventional Methods
3.2. Measurement of Cell Area and Length
3.3. Results of Cell Size Obtained Using Conventional Automated Methods
3.4. Discussion of the Results and Future Applications
4. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turner, J.J.; Ewald, J.C.; Skotheim, J.M. Cell size control in yeast. Curr. Biol. 2012, 22, R350–R359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, J.M.; Garcia-Diego, F.J. Dielectrophoretic force measurements in yeast cells by the Stokes method. In Proceedings of the 1997 IEEE Industry Applications Conference Thirty-Second IAS Annual Meeting, New Orleans, LA, USA, 5–9 October 1997; Volume 2012–2018. [Google Scholar] [CrossRef]
- ImageJ. Wo Tsukatta Saibou Su No Keisoku. Available online: https://slideshowjp.com/doc/1559964/imagej-%E3%82%92%E4%BD%BF%E3%81%A3%E3%81%9F%E7%B4%B0%E8%83%9E%E6%95%B0%E3%81%AE%E8%A8%88%E6%B8%AC (accessed on 14 October 2022).
- Yoda, T.; Ogura, A.; Saito, T. Influence of ethyl caproate on the size of lipid vesicles and yeast cells. Biomimetics 2020, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Katakura, Y.; Ohmasa, T.; Naganuma, T.; Ono, H. Yuyo-Biseibutsu-No-Iroha [ABC of Useful Microorganisms]; NTS Ltd.: Tokyo, Japan, 2016. (In Japanese) [Google Scholar]
- Chan, L.L.; Lyettefi, E.J.; Pirani, A.; Smith, T.; Qiu, J.; Lin, B. Direct concentration and viability measurement of yeast in corn mash using a novel imaging cytometry method. J. Ind. Microbiol. Biotechnol. 2011, 3, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Yoda, T. Quality evaluation of drinks based on liposome shape changes induced by flavor molecules. ACS Omega 2022, 7, 5679–5686. [Google Scholar] [CrossRef]
- Yoda, T.; Saito, T. Size of cells and physicochemical properties of membranes are related to flavor production during sake brewing in the yeast Saccharomyces cerevisiae. Membrane 2020, 10, 440. [Google Scholar] [CrossRef]
- Kaewprasit, C.; Hequet, E.; Abidi, N.; Gourlot, J.P. Application of methylene blue adsorption to cotton fiber specific surface area measurement: Part I. Methodology. J. Cotton Sci. 1998, 2, 164–173. Available online: https://www.cotton.org/journal/1998-02/4/upload/jcs02-164.pdf (accessed on 14 October 2022).
- Yoda, T.; Shibuya, K.; Miura, K.; Myoubuddani, H. Characterization of the adsorption ability of silk-derived activated carbon fibers using X-ray analysis and camera imaging methods. Measurement 2017, 101, 103–110. [Google Scholar] [CrossRef]
- Yoda, T.; Shibuya, K.; Myoubudani, H. Preparation of activated carbon fibers from mixtures of cotton and polyester fibers. Measurement 2018, 110, 572–576. [Google Scholar] [CrossRef]
- Yoda, T.; Shibuya, K.; Myoubudani, H. Preparation of activated carbon fibers from fiber mixtures. J. Text. Sci. Fash. Technol. 2018, 1, JTSFT.MS.ID.000510. [Google Scholar] [CrossRef]
- Yoda, T.; Shibuya, K.; Miura, K.; Myoubudani, H. Effects of two-step heat treatment on the structure of cotton-derived activated carbon fibres. Int. J. Mater. Struct. Integr. 2020, 14, 33–43. [Google Scholar] [CrossRef]
- Chen, W.; Qian, J.; Zhang, M.; Lu, W.; Zhang, S.; Xu, H. Recycle of cotton waste by hard templating with magnesium acetate as MgO precursor. Environ. Sci. Pollut. Res. 2019, 26, 29908–29916. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.L.Y.; Rice, W.L.; Qiu, J. Observation and quantification of the morphological effect of trypan blue rupturing dead or dying cells. PLoS ONE 2020, 15, e0227950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ImageJ for Image Resolution by Marco Language. Available online: https://lp-tech.net/articles/paDGX (accessed on 5 February 2022).
- Ohnuki, S.; Okada, H.; Friedrich, A.; Kanno, Y.; Goshima, T.; Hasuda, H.; Inahashi, M.; Okazaki, N.; Tamura, H.; Nakamura, R.; et al. Phenotypic diagnosis of lineage and differentiation during sake yeast breeding. G3 2017, 7, 2807–2820. [Google Scholar] [CrossRef] [Green Version]
- Biggemann, J.; Diepold, B.; Pezoldt, M.; Stumpf, M.; Greil, P.; Feyet, T. Automated 3D assembly of periodic alumina-epoxy composite structures. J. Am. Ceram. Soc. 2018, 101, 3864–3873. [Google Scholar] [CrossRef]
- Leksycki, K.; Królczyk, J.B. Comparative assessment of the surface topography for different optical profilometry techniques after dry turning of Ti6Al4V titanium alloy. Measurement 2020, 169, 108378. [Google Scholar] [CrossRef]
- Rappaz, B.; Cano, E.; Colomb, T.; Kuhn, J.; Depeursinge, C.D.; Simanis, V.; Magistretti, P.J.; Marquet, P.P. Noninvasive characterization of the fission yeast cell cycle by monitoring dry mass with digital holographic microscopy. J. Biomed. Opt. 2009, 14, 034049. [Google Scholar] [CrossRef]
- Mir, M.; Wang, Z.; Shen, Z.; Bednarz, M.; Bashir, R.; Golding, I.; Prasanth, S.G.; Popescu, G. Optical measurement of cycle-dependent cell growth. Proc. Natl. Acad. Sci. USA 2011, 108, 13124–13129. [Google Scholar] [CrossRef] [Green Version]
- Kastl, L.; Isbach, M.; Dirksen, D.; Schnekenburger, J.; Kemper, B. Quantitative phase imaging for cell culture quality control. Cytom. Part A 2017, 91A, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Jo, M.C.; Liu, W.; Gu, L.; Dang, W.; Qin, L. High-throughput analysis of yeast replicative aging using a microfluidic system. Proc. Natl. Acad. Sci. USA 2015, 112, 9364–9369. [Google Scholar] [CrossRef] [Green Version]
- Isozaki, A.; Mikami, H.; Tezuka, H.; Matsumura, H.; Huang, K.; Akamine, M.; Hiramatsu, K.; Iino, T.; Ito, T.; Karakawa, H.; et al. Intelligent image-activated cell sorting 2.0. Lab Chip 2020, 20, 2263–2273. [Google Scholar] [CrossRef]
- Chadani, T.; Ohnuki, S.; Isogai, A.; Goshima, T.; Kashima, M.; Ghanegolmohammadiet, F.; Nishi, T.; Hirata, D.; Watanabe, D.; Kitamoto, K.; et al. Genome editing to generate sake yeast strains with eight mutations that confer excellent brewing characteristics. Cells 2021, 10, 1299. [Google Scholar] [CrossRef] [PubMed]

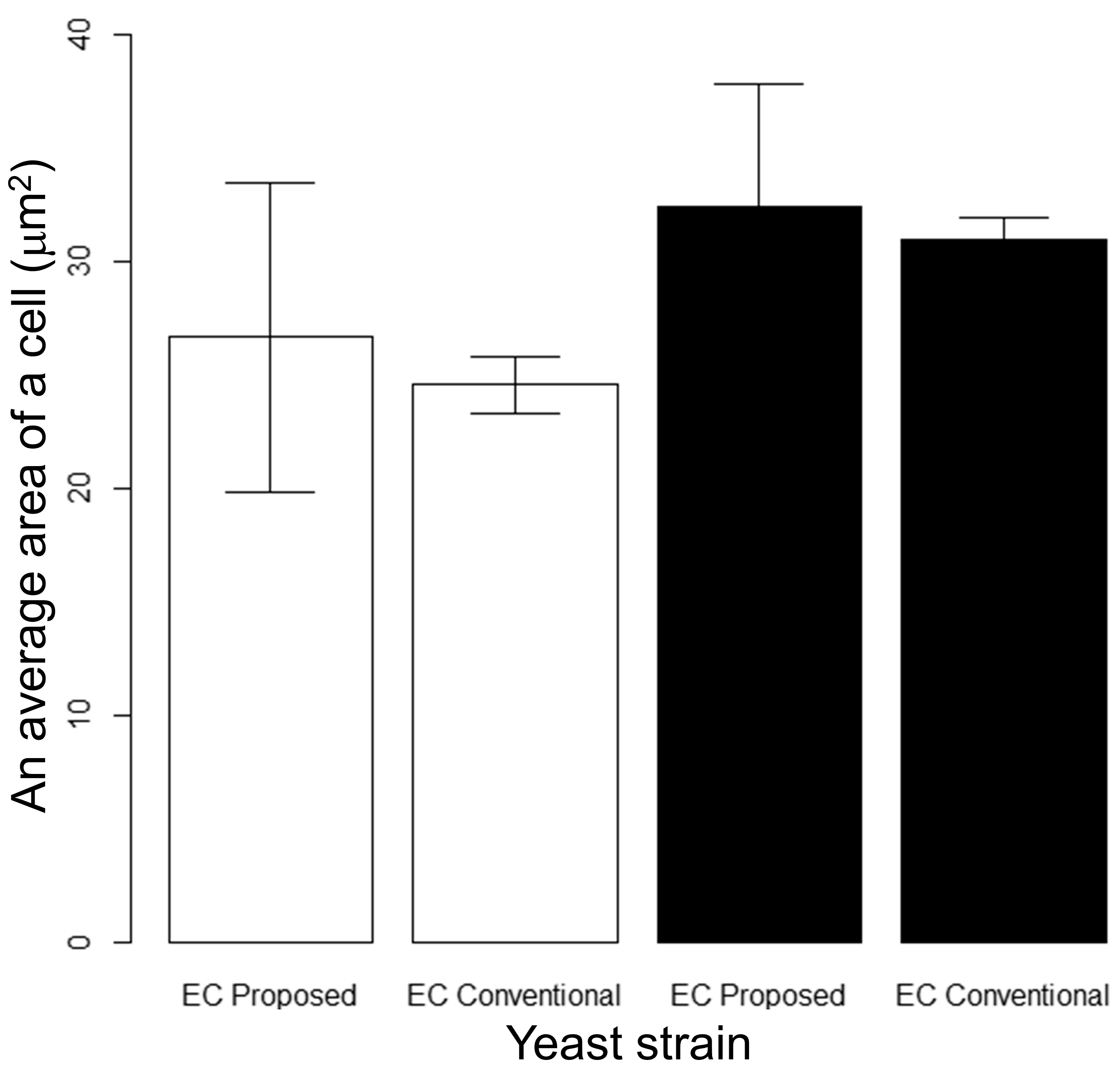
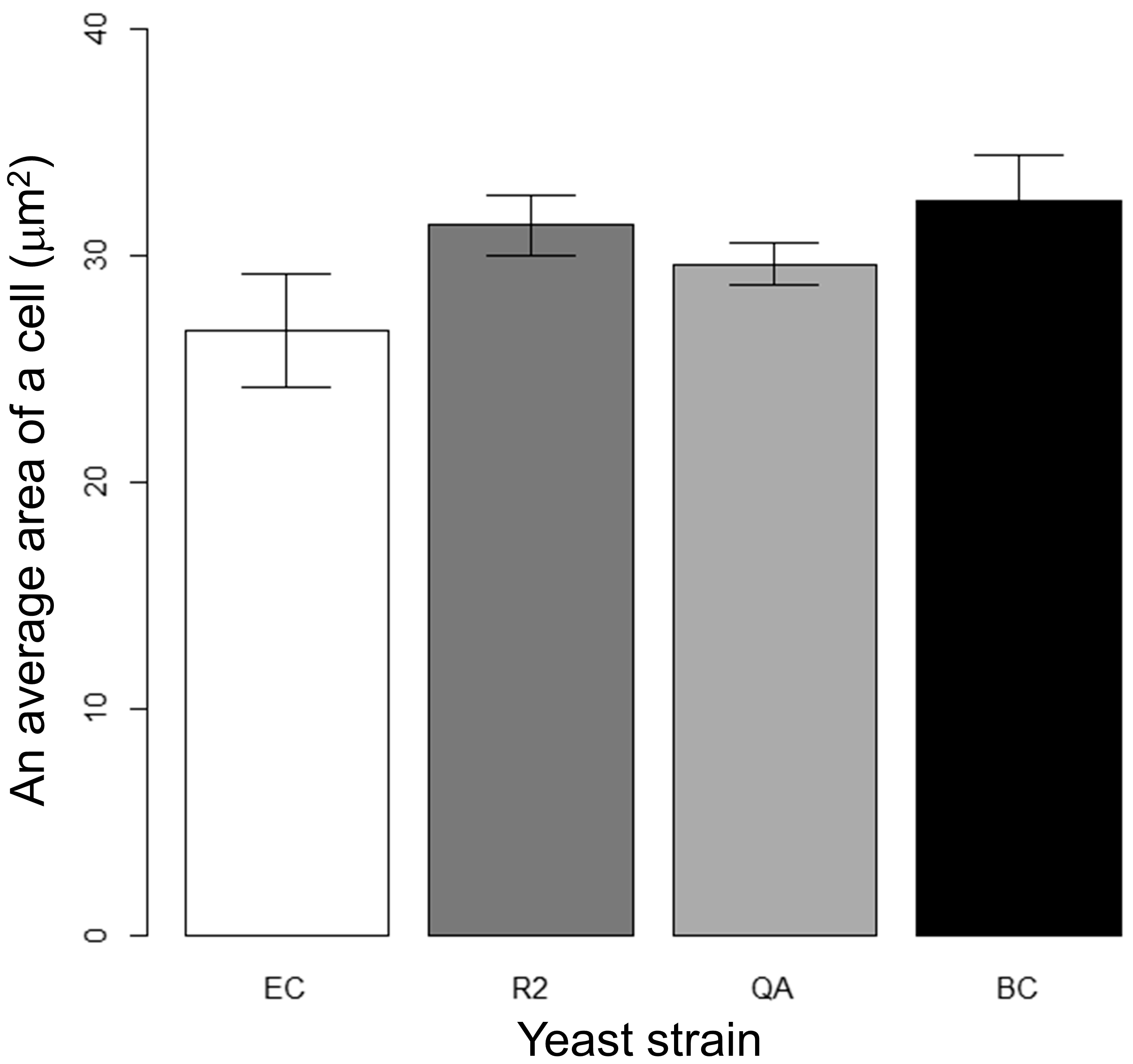


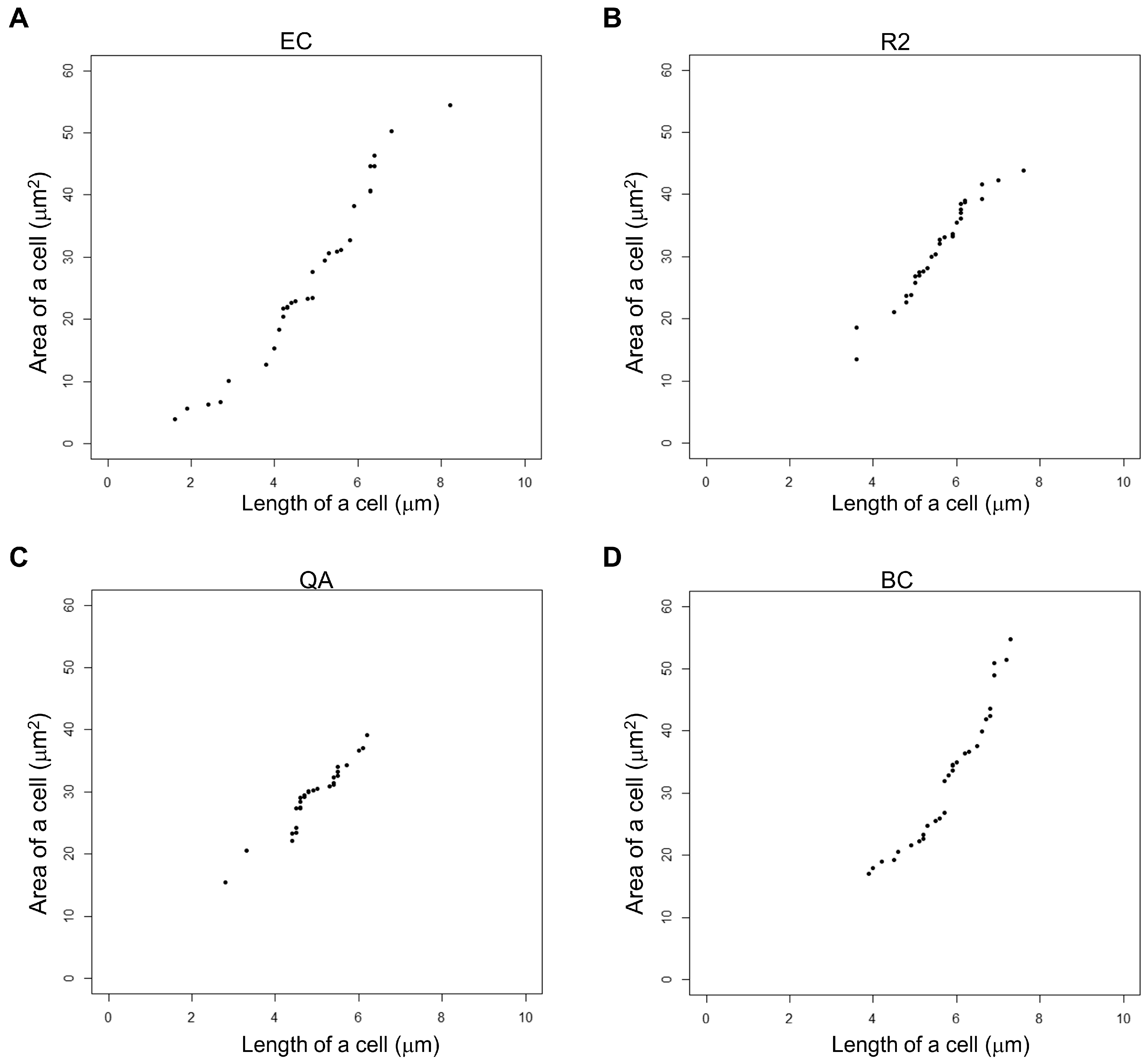
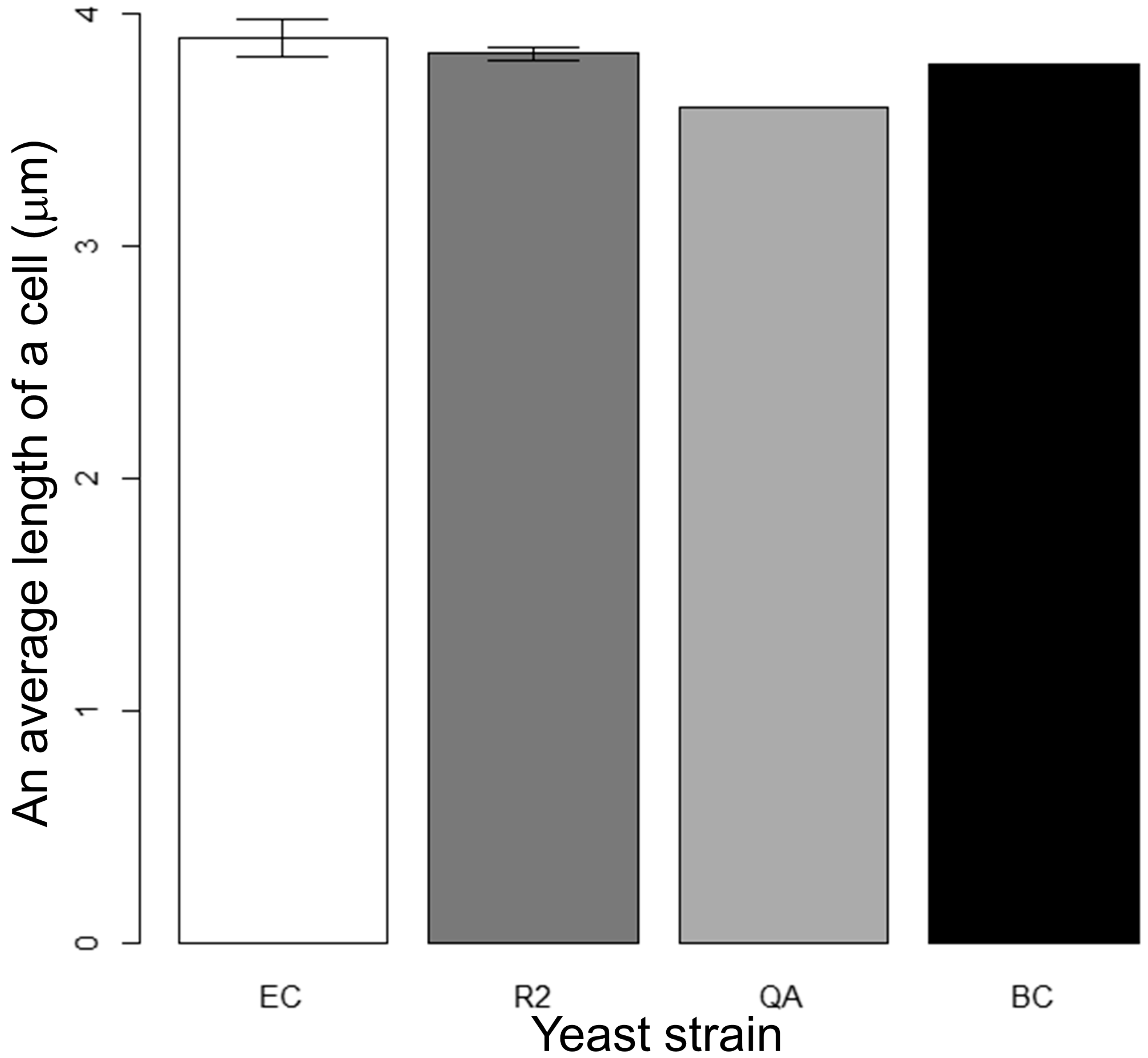
| Pairs for Strains | p-Values (Area) for t-Test | ||
|---|---|---|---|
| Proposed Measurements for Area | Proposed Measurements for Length | ||
| EC | R2 | 0.001 | 9.7 × 10−7 |
| EC | QA | 0.08 | 0.28 |
| EC | BC | 0.21 | 0.04 |
| R2 | QA | 0.001 | 5.9 × 10−15 |
| R2 | BC | 0.74 | 0.55 |
| QA | BC | 0.33 | 0.01 |
| Pairs for Strains | p-Values (Length) for t-Test | |
|---|---|---|
| EC | R2 | 0.67 |
| EC | QA | 0.10 |
| EC | BC | 0.42 |
| R2 | QA | 0.02 |
| R2 | BC | Uncalculated |
| QA | BC | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoda, T. Measurement and Characterization of Yeast Cell Size Using a Digital Optical Microscope. Processes 2022, 10, 2396. https://doi.org/10.3390/pr10112396
Yoda T. Measurement and Characterization of Yeast Cell Size Using a Digital Optical Microscope. Processes. 2022; 10(11):2396. https://doi.org/10.3390/pr10112396
Chicago/Turabian StyleYoda, Tsuyoshi. 2022. "Measurement and Characterization of Yeast Cell Size Using a Digital Optical Microscope" Processes 10, no. 11: 2396. https://doi.org/10.3390/pr10112396





