Imaging Method by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS) for Tissue or Tumor: A Mini Review
Abstract
:1. Introduction
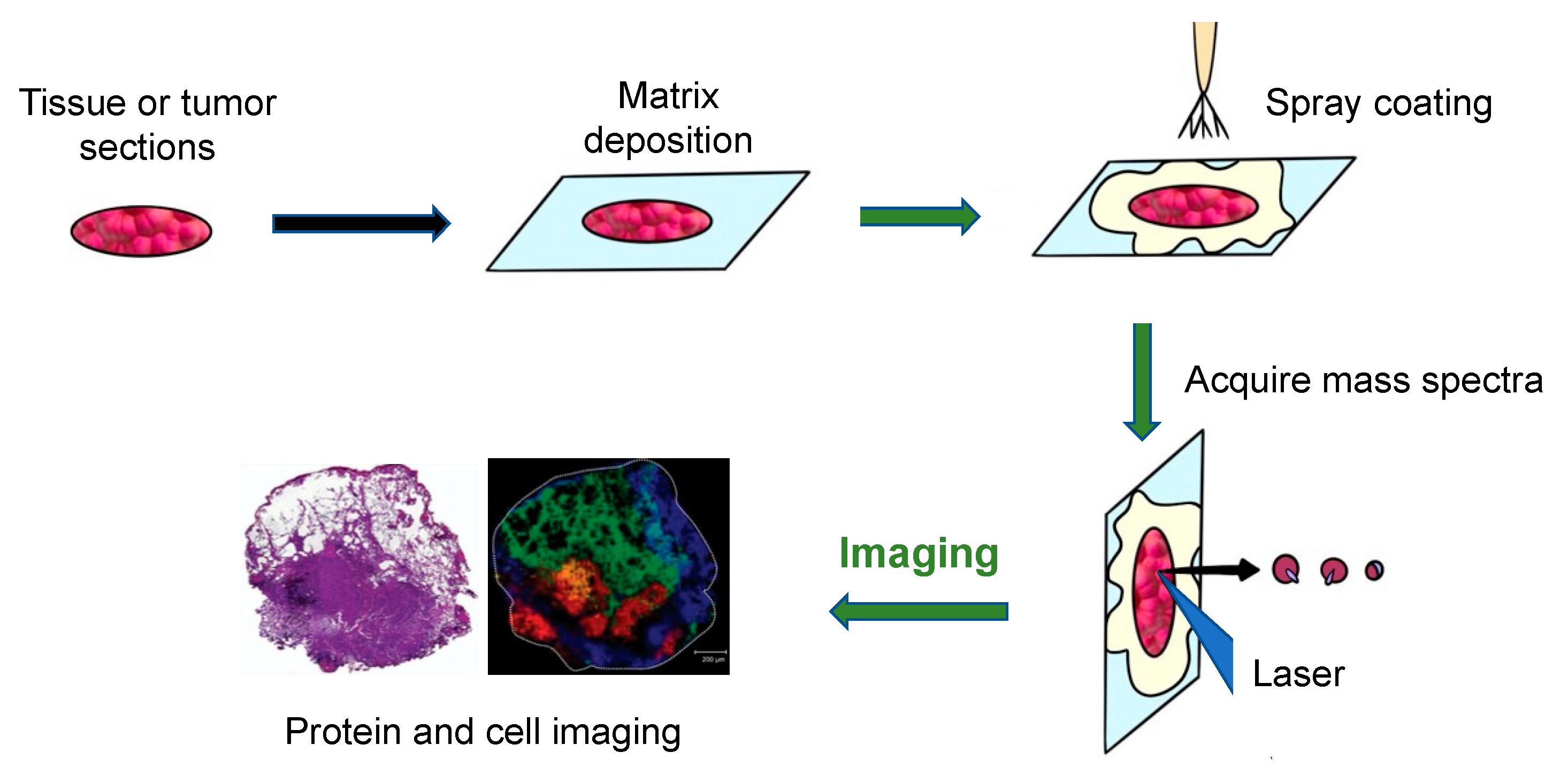
2. Sample Handling and Processing
2.1. Tissue Storage
2.2. Sectioning
2.3. Mounting
2.4. Pretreatment of Samples
2.5. Matrix Deposition
2.6. Staining
3. Instrumentation
3.1. Ion Source (MALDI)
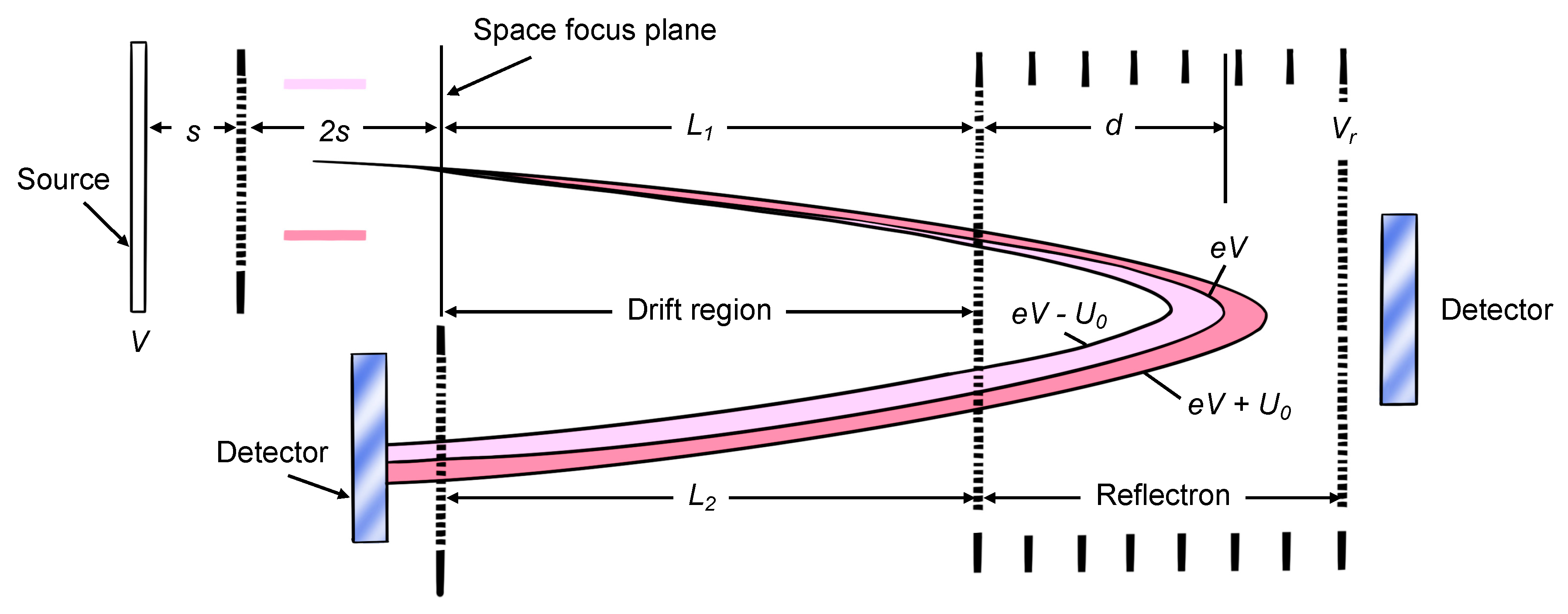
3.2. Mass Analyzer
3.3. Detector
4. Principle of MALDI-MS for Tissue or Tumor Imaging
5. Limitations and Future Applications
5.1. Diagnostic and Prognostic Assessment in Clinical Pathology
5.2. Tumor Removal
5.3. Drug Development
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gessel, M.; Norris, J.; Caprioli, R.M. MALDI imaging mass spectrometry: Spatial molecular analysis to enable a new age of discovery. J. Proteom. 2014, 107, 71–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillenkamp, F.; Karas, M.; Beavis, R.C.; Chait, B.T. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991, 63, 1193–1203. [Google Scholar] [CrossRef]
- Oppenheimer, S.R.; Mi, D.; Sanders, M.E.; Caprioli, R.M. Molecular Analysis of Tumor Margins by MALDI Mass Spectrometry in Renal Carcinoma. J. Proteome Res. 2010, 9, 2182–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Y.; Du, Y.; Cui, M.; Liu, Z. Characterization of the Noncovalent Interactions between Lysozyme and Panaxadiol Glycosides by Intensity-Fading—Matrix-Assisted Laser Desorption Ionization—Mass Spectrometry (IF-MALDI-MS). Anal. Lett. 2021, 54, 2387–2394. [Google Scholar] [CrossRef]
- Chaurand, P.; Latham, J.C.; Lane, K.B.; Mobley, J.; Polosukhin, V.V.; Wirth, P.S.; Nanney, L.B.; Caprioli, R.M. Imaging Mass Spectrometry of Intact Proteins from Alcohol-Preserved Tissue Specimens: Bypassing Formalin Fixation. J. Proteome Res. 2008, 7, 3543–3555. [Google Scholar] [CrossRef] [Green Version]
- Treu, A.; Römpp, A. Matrix ions as internal standard for high mass accuracy matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom. 2021, 35, e9110. [Google Scholar] [CrossRef]
- Wang, C.; Bi, H.; Xie, J. Visualization of the Distance among Fishes by MALDI MS for Rapid Determination of the Taxonomic Status of Fish Fillets. J. Agric. Food Chem. 2020, 68, 8438–8446. [Google Scholar] [CrossRef]
- Norris, J.; Caprioli, R.M. Analysis of Tissue Specimens by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry in Biological and Clinical Research. Chem. Rev. 2013, 113, 2309–2342. [Google Scholar] [CrossRef] [Green Version]
- Lemaire, R.; Desmons, A.; Tabet, J.C.; Day, R.; Salzet, M.; Fournier, I. Solid Ionic Matrixes for Direct Tissue Analysis and MALDI Imaging. Anal. Chem. 2006, 78, 809–819. [Google Scholar] [CrossRef] [Green Version]
- Karas, M.; Krüger, R. Ion Formation in MALDI: The Cluster Ionization Mechanism. Chem. Rev. 2003, 103, 427–440. [Google Scholar] [CrossRef]
- Tsuchida, S.; Umemura, H.; Nakayama, T. Current Status of Matrix-Assisted Laser Desorption/Ionization–Time-of-Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Diagnostic Microbiology. Molecules 2020, 25, 4775. [Google Scholar] [CrossRef] [PubMed]
- Beavis, R.C.; Chaudhary, T.; Chait, B.T. α-Cyano-4-hydroxycinnamic acid as a matrix for matrixassisted laser desorption mass spectrometry. Org. Mass Spectrom. 1992, 27, 156–158. [Google Scholar] [CrossRef]
- Beavis, R.C.; Chait, B.T.; Standing, K.G. Matrix-assisted laser-desorption mass spectrometry using 355 nm radiation. Rapid Commun. Mass Spectrom. 1989, 3, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, C.; Tan, W. Brain Lipid Dynamics in Amyloid Precursor Protein/Presenilin 1 Mouse Model of Early Alzheimer’s Disease by Desorption Electrospray Ionization and Matrix Assisted Laser Desorption Ionization–Mass Spectrometry Imaging Techniques. J. Proteome Res. 2021, 20, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Chaurand, P.; Norris, J.L.; Cornett, D.S.; Mobley, J.A.; Caprioli, R.M. New Developments in Profiling and Imaging of Proteins from Tissue Sections by MALDI Mass Spectrometry. J. Proteome Res. 2006, 5, 2889–2900. [Google Scholar] [CrossRef]
- Han, X. Lipidomics: Comprehensive Mass Spectrometry of Lipids; Wiley: Hoboken, NJ, USA, 2016. [Google Scholar]
- Harvey, J. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2013–2014. Mass Spectrom. Rev. 2018, 37, 353–491. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Zenobi, R. Characterizing the iron loading pattern of ferritin using high-mass matrix-assisted laser desorption ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2019, 33, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- Mamyrin, B.A.; Karataev, V.I.; Shmikk, D.V.; Zagulin, V.A. Mass reflection: A new nonmagnetic time-of-flight high resolution mass- spectrometer. Sov. Phys. Tech. JETP 1973, 37, 5. [Google Scholar]
- Khatri, N.; Gupta, A.; Taneja, R.; Bilandi, A.; Beniwal, P. A Review on Mass Spectrometry Detectors. Int. Res. J. Pharm. 2012, 3, 33–42. [Google Scholar]
- Thurner, G.C.; Debbage, P. Molecular imaging with nanoparticles: The dwarf actors revisited 10 years later. Histochem. Cell Biol. 2018, 150, 733–794. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Caprioli, R.M. Matrix pre-coated targets for high throughput MALDI imaging of proteins. Biol. Mass Spectrom. 2014, 49, 417–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaepe, K.; Bhandari, D.R.; Werner, J.; Henss, A.; Pirkl, A.; Kleine-Boymann, M.; Rohnke, M.; Wenisch, S.; Neumann, E.; Janek, J.; et al. Imaging of Lipids in Native Human Bone Sections Using TOF–Secondary Ion Mass Spectrometry, Atmospheric Pressure Scanning Microprobe Matrix-Assisted Laser Desorption/Ionization Orbitrap Mass Spectrometry, and Orbitrap–Secondary Ion Mass Spectrometry. Anal. Chem. 2018, 90, 8856–8864. [Google Scholar] [CrossRef] [PubMed]
- Balluff, B.; Schöne, C.; Höfler, H.; Walch, A. MALDI imaging mass spectrometry for direct tissue analysis: Technological advancements and recent applications. Histochem. Cell Biol. 2011, 136, 227–244. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Jiao, L.; Bütikofer, M.; Zeng, Z.; Zenobi, R. High-Mass Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry for Absolute Quantitation of Noncovalent Protein–Protein Binding Interactions. Anal. Chem. 2021, 93, 10982–10989. [Google Scholar] [CrossRef]
- Römpp, A.; Spengler, B. Mass spectrometry imaging with high resolution in mass and space. Histochem. Cell Biol. 2013, 139, 759–783. [Google Scholar] [CrossRef] [Green Version]
- Yasunaga, M.; Furuta, M.; Ogata, K.; Koga, Y.; Yamamoto, Y.; Takigahira, M.; Matsumura, Y. The significance of microscopic mass spectrometry with high resolution in the visualisation of drug distribution. Sci. Rep. 2013, 3, 3050. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Rong, Z.; Xiao, P.; Li, Y. Imaging Method by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS) for Tissue or Tumor: A Mini Review. Processes 2022, 10, 388. https://doi.org/10.3390/pr10020388
Wu J, Rong Z, Xiao P, Li Y. Imaging Method by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS) for Tissue or Tumor: A Mini Review. Processes. 2022; 10(2):388. https://doi.org/10.3390/pr10020388
Chicago/Turabian StyleWu, Jiawen, Ze Rong, Peng Xiao, and Yuanzhe Li. 2022. "Imaging Method by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS) for Tissue or Tumor: A Mini Review" Processes 10, no. 2: 388. https://doi.org/10.3390/pr10020388
APA StyleWu, J., Rong, Z., Xiao, P., & Li, Y. (2022). Imaging Method by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-MS) for Tissue or Tumor: A Mini Review. Processes, 10(2), 388. https://doi.org/10.3390/pr10020388






