Systemic Efficacy of Sirolimus via the ERBB Signaling Pathway in Breast Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Computational Methods for Obtaining Pathway and Gene Interactions
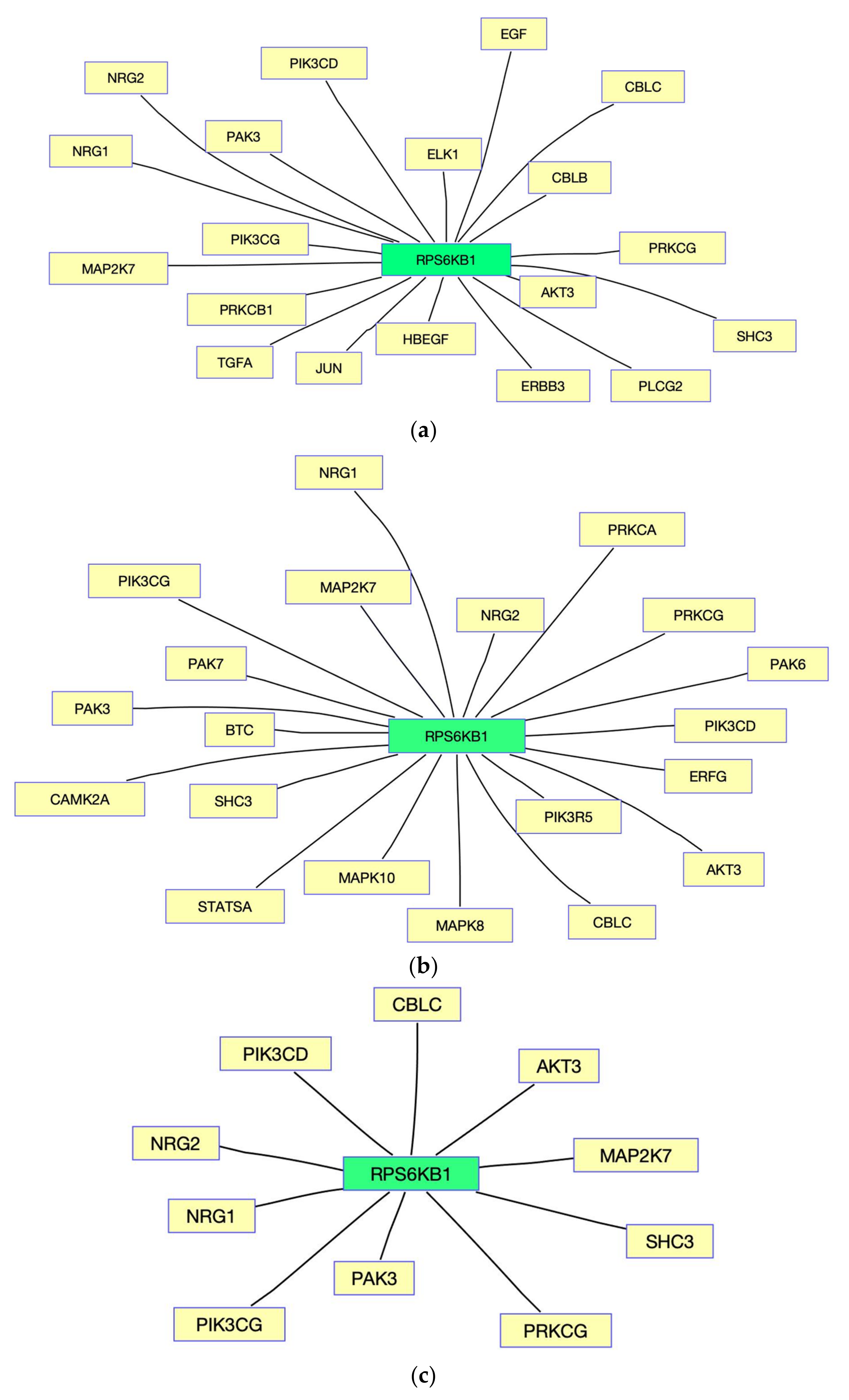
3. Results
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One drug, many effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 169, 361–371. [Google Scholar] [CrossRef]
- Chung, J.; Kuo, C.J.; Crabtree, G.R.; Blenis, J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 1992, 69, 1227–1236. [Google Scholar] [CrossRef]
- Kuo, C.J.; Chung, J.; Fiorentino, D.F.; Flanagan, W.M.; Blenis, J.; Crabtree, G.R. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature 1992, 358, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Price, D.J.; Grove, J.R.; Calvo, V.; Avruch, J.; Bierer, B.E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science 1992, 257, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [Green Version]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [Green Version]
- Malaguti, P.; Vari, S.; Cognetti, F.; Fabi, A. The Mammalian target of rapamycin inhibitors in breast cancer: Current evidence and future directions. Anticancer Res. 2013, 33, 21–28. [Google Scholar]
- Wander, S.A.; Hennessy, B.T.; Slingerland, J.M. Next-generation mTOR inhibitors in clinical oncology: How pathway complexity informs therapeutic strategy. J. Clin. Investig. 2011, 121, 1231–1241. [Google Scholar] [CrossRef] [Green Version]
- Heinonen, H.; Nieminen, A.; Saarela, M.; Kallioniemi, A.; Klefstrom, J.; Hautaniemi, S.; Monni, O. Deciphering downstream gene targets of PI3K/mTOR/p70S6K pathway in breast cancer. BMC Genom. 2008, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Hare, S.H.; Harvey, A.J. mTOR function and therapeutic targeting in breast cancer. Am. J. Cancer Res. 2017, 7, 383–404. [Google Scholar] [PubMed]
- Frogne, T.; Benjaminsen, R.V.; Sonne-Hansen, K.; Sorensen, B.S.; Nexo, E.; Laenkholm, A.V.; Rasmussen, L.M.; Riese, D.J., 2nd; de Cremoux, P.; Stenvang, J.; et al. Activation of ErbB3, EGFR and Erk is essential for growth of human breast cancer cell lines with acquired resistance to fulvestrant. Breast Cancer Res. Treat. 2009, 114, 263–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Block, M.; Grundker, C.; Fister, S.; Kubin, J.; Wilkens, L.; Mueller, M.D.; Hemmerlein, B.; Emons, G.; Gunthert, A.R. Inhibition of the AKT/mTOR and erbB pathways by gefitinib, perifosine and analogs of gonadotropin-releasing hormone I and II to overcome tamoxifen resistance in breast cancer cells. Int. J. Oncol. 2012, 41, 1845–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaturvedi, D.; Gao, X.; Cohen, M.S.; Taunton, J.; Patel, T.B. Rapamycin induces transactivation of the EGFR and increases cell survival. Oncogene 2009, 28, 1187–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, 1–16. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [Green Version]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef] [Green Version]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Pammolli, F.; Magazzini, L.; Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 2011, 10, 428–438. [Google Scholar]
- Yeu, Y.; Yoon, Y.; Park, S. Protein localization vector propagation: A method for improving the accuracy of drug repositioning. Mol. Biosyst. 2015, 11, 2096–2102. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, S.; Zhang, X.; Li, J. Drug repositioning by integrating target information through a heterogeneous network model. Bioinformatics 2014, 30, 2923–2930. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, F.; Carrella, D.; Mandriani, B.; Pisonero-Vaquero, S.; Sirci, F.; Medina, D.L.; Brunetti-Pierri, N.; di Bernardo, D. gene2drug: A computational tool for pathway-based rational drug repositioning. Bioinformatics 2018, 34, 1498–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laenen, G.; Thorrez, L.; Bornigen, D.; Moreau, Y. Finding the targets of a drug by integration of gene expression data with a protein interaction network. Mol. Biosyst. 2013, 9, 1676–1685. [Google Scholar] [CrossRef]
- Isik, Z.; Baldow, C.; Cannistraci, C.V.; Schroeder, M. Drug target prioritization by perturbed gene expression and network information. Sci. Rep. 2015, 5, 17417. [Google Scholar] [CrossRef] [PubMed]
- Kim, S. A New Computational Approach to Evaluating Systemic Gene–Gene Interactions in a Pathway Affected by Drug LY294002. Processes 2020, 8, 1230. [Google Scholar] [CrossRef]
- Badkas, A.; De Landtsheer, S.; Sauter, T. Topological network measures for drug repositioning. Brief. Bioinform. 2021, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Monks, A.; Zhao, Y.; Hose, C.; Hamed, H.; Krushkal, J.; Fang, J.; Sonkin, D.; Palmisano, A.; Polley, E.C.; Fogli, L.K.; et al. The NCI Transcriptional Pharmacodynamics Workbench: A Tool to Examine Dynamic Expression Profiling of Therapeutic Response in the NCI-60 Cell Line Panel. Cancer Res. 2018, 78, 6807–6817. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Press, W.H. Numerical Recipes in C: The Art of Scientific Computing, 2nd ed.; Cambridge University Press: Cambridge, UK, 1992; pp. 425–430. [Google Scholar]
- Kim, S.; Kim, J.; Cho, K.H. Inferring gene regulatory networks from temporal expression profiles under time-delay and noise. Comput. Biol. Chem. 2007, 31, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kreider, K. Parameter identification for nonlinear elastic and viscoelastic plates. Appl. Numer. Math. 2006, 56, 1538–1554. [Google Scholar] [CrossRef]
- Kim, S. Identifying dynamic pathway interactions based on clinical information. Comput. Biol. Chem. 2017, 68, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Kim, S. A Computational Approach for Pathway-Based Systemic Drug Influence. Processes 2021, 9, 1063. [Google Scholar] [CrossRef]
- Vignot, S.; Faivre, S.; Aguirre, D.; Raymond, E. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann. Oncol. 2005, 16, 525–537. [Google Scholar] [CrossRef]
- Chua, Y.L.; Ito, Y.; Pole, J.C.; Newman, S.; Chin, S.F.; Stein, R.C.; Ellis, I.O.; Caldas, C.; O’Hare, M.J.; Murrell, A.; et al. The NRG1 gene is frequently silenced by methylation in breast cancers and is a strong candidate for the 8p tumour suppressor gene. Oncogene 2009, 28, 4041–4052. [Google Scholar] [CrossRef] [Green Version]
- Ye, D.Z.; Field, J. PAK signaling in cancer. Cell Logist. 2012, 2, 105–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.-X.; Wang, W.-Q.; Ye, L.; Bi, Y.-F.; Fang, H.; Cui, B.; Zhou, W.-W.; Dai, M.; Zhang, J.; Li, X.-Y. p21-activated kinase 3 is overexpressed in thymic neuroendocrine tumors (carcinoids) with ectopic ACTH syndrome and participates in cell migration. Endocrine 2010, 38, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.J.; Yu, T.Y.; Li, Y.R.; Liu, Y.J.; Deng, B.B. Circ_0000190 suppresses gastric cancer progression potentially via inhibiting miR-1252/PAK3 pathway. Cancer Cell Int. 2020, 20, 351. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S. Systemic Efficacy of Sirolimus via the ERBB Signaling Pathway in Breast Cancer. Processes 2022, 10, 552. https://doi.org/10.3390/pr10030552
Kim S. Systemic Efficacy of Sirolimus via the ERBB Signaling Pathway in Breast Cancer. Processes. 2022; 10(3):552. https://doi.org/10.3390/pr10030552
Chicago/Turabian StyleKim, Shinuk. 2022. "Systemic Efficacy of Sirolimus via the ERBB Signaling Pathway in Breast Cancer" Processes 10, no. 3: 552. https://doi.org/10.3390/pr10030552




