Cell-Free Escherichia coli Synthesis System Based on Crude Cell Extracts: Acquisition of Crude Extracts and Energy Regeneration
Abstract
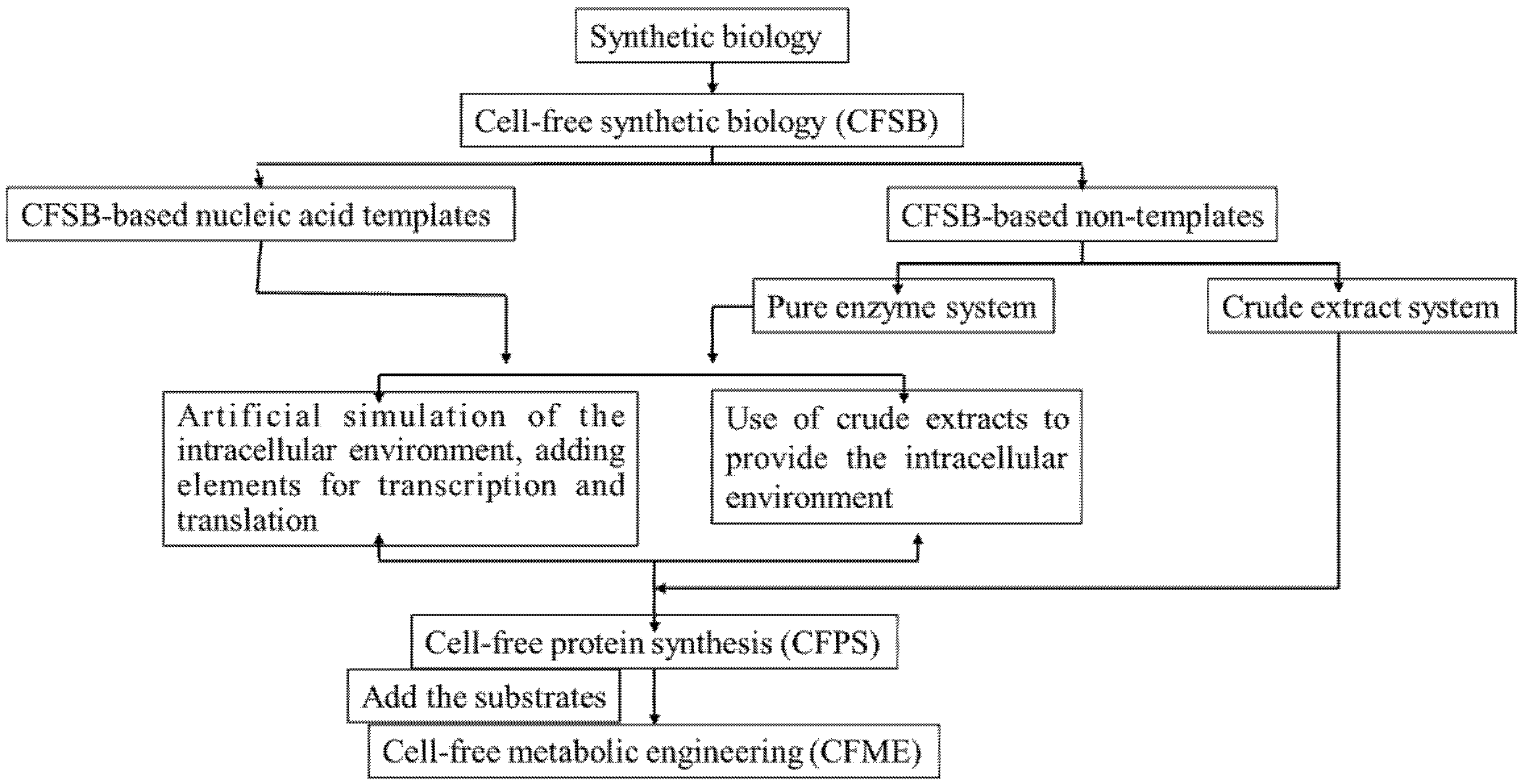
:1. Introduction
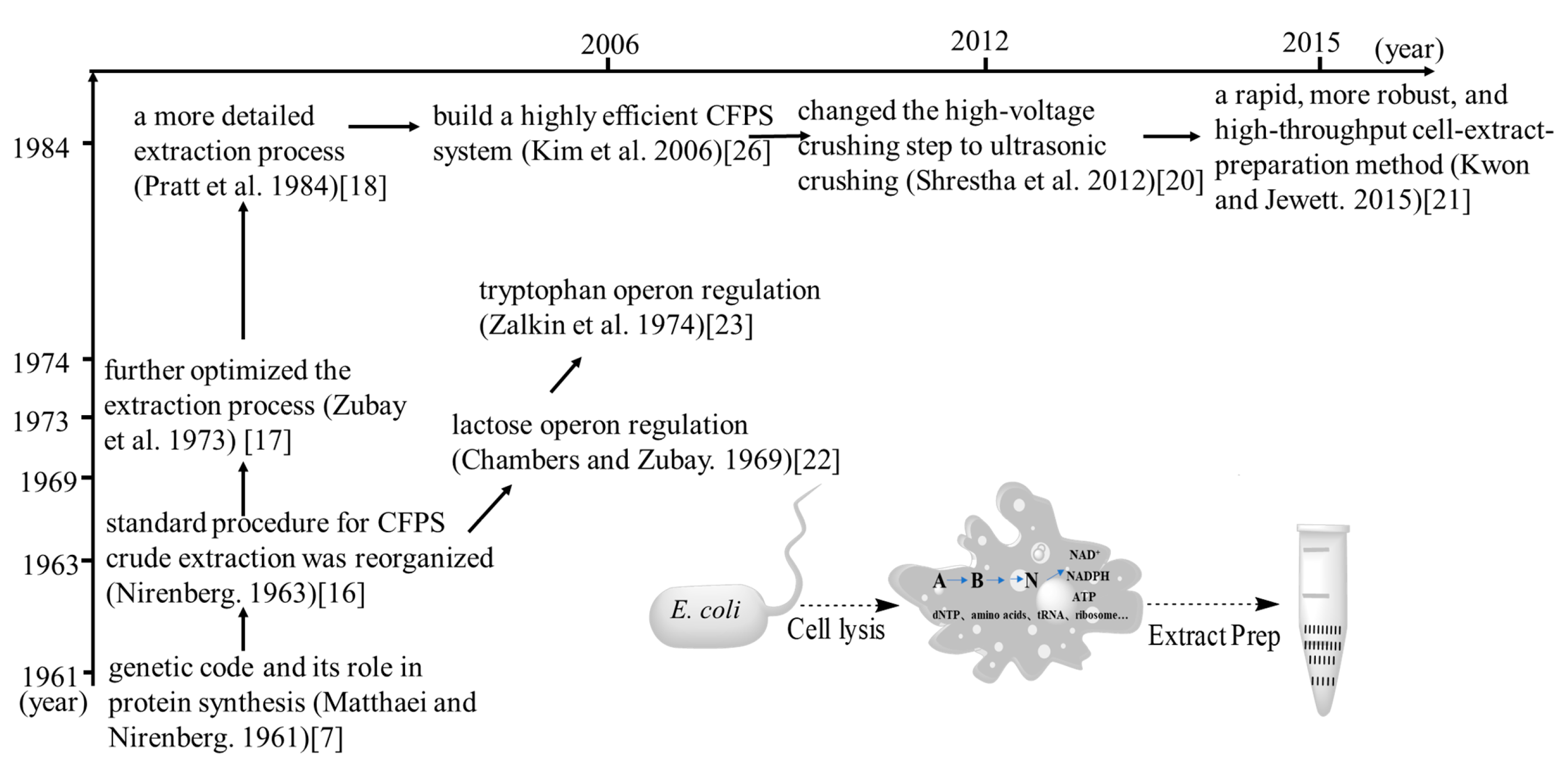
2. The Development of CFSB Based on Crude Cell Extracts
3. Preparation of Crude Extracts from E. coli Cells
4. Energy Regeneration
4.1. Phosphate Bond Breakage to Provide Energy
4.2. Non-Phosphate-Bond Breaks to Provide Energy
4.3. Others
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shi, T.; Han, P.; You, C.; Zhang, Y.H. An in vitro synthetic biology platform for emerging industrial biomanufacturing: Bottom-up pathway design. Synth. Syst. Biotechnol. 2018, 3, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.J.; Kim, D.M. Cell-free metabolic engineering: Recent developments and future prospects. Meth. Protoc. 2019, 2, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y. Cell-free synthetic biology: Engineering in an open world. Synth. Syst. Biotechnol. 2017, 2, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.Z.; Gregorio, N.E.; Jewett, M.C.; Watts, K.R.; Oza, J.P. Escherichia coil-based cell-free protein synthesis: Protocols for a robust, flexible, and accessible platform technology. J. Vis. Exp. 2019, 25, 11. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Karim, A.S.; Jewett, M.C. Cell-free metabolic engineering: Biomanufacturing beyond the cell. Biotechnol. J. 2015, 10, 69–82. [Google Scholar] [CrossRef]
- Hoagland, M.B.; Stephenson, M.L.; Scott, J.F.; Hecht, L.I.; Zamecnik, P.C. Soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 1958, 231, 241–257. [Google Scholar] [CrossRef]
- Matthaei, J.H.; Nirenberg, M.W. Characteristics and stabilization of dnaase-sensitive protein synthesis in E. coli extracts. Proc. Natl. Acad. Sci. USA 1961, 47, 1580–1588. [Google Scholar] [CrossRef] [Green Version]
- So, A.G.; Davie, E.W. The incorporation of amino acids into protein in a cell-free system from yeast. Biochemistry 1963, 2, 132–136. [Google Scholar] [CrossRef]
- Grünberger, D. The incorporation of amino acids-14C into proteins by ribosomes of Bacillus cereus. Collect. Czech. Chem. Commun. 1964, 29, 2400–2405. [Google Scholar] [CrossRef]
- Shiio, T.; McFadden, B.A. Cell-free amino acid-incorporating system from Pseudomonas indigofera. J. Bacterial. 1965, 90, 978–983. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, J.; Abraham, E.P. The conversion of cephalosporins to 7′-alpha-methoxycephalosporins by cell-free extracts of Streptomyces clavuligerus. Biochem. J. 1980, 186, 613. [Google Scholar] [CrossRef] [PubMed]
- Failmezger, J.; Scholz, S.; Blombach, B.; Siemann-Herzberg, M. Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 2018, 9, 1146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchner, E. Alkoholische gährung ohne hefezellen. J. Inorg. Chem. 1897, 30, 117–124. [Google Scholar] [CrossRef] [Green Version]
- Bujara, M.; Schuemperli, M.; Billerbeek, S.; Heinemann, M.; Panke, S. Exploiting cell-free systems: Implementation and debugging of a aystem of biotransformations. Biotechnol. Bioeng. 2010, 106, 376–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swartz, J.R. Transforming biochemical engineering with cell-free biology. AIChE J. 2012, 58, 5–13. [Google Scholar] [CrossRef]
- Nirenberg, M. Cell-free synthesis directed by messenger RNA. Meth. Enzymol. 1963, 6, 17–23. [Google Scholar] [CrossRef]
- Zubay, G. In vitro synthesis of protein in microbial systems. Annu. Rev. Genet. 1973, 7, 267–287. [Google Scholar] [CrossRef]
- Pratt, J.M. Coupled transcription-translation in prokaryotic cell-free sytems. Transcrip. Transla. A Prac. App. 1984, 179–209. [Google Scholar] [CrossRef]
- Liu, D.V.; Zawada, J.F.; Swartz, J.R. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol. Progress. 2005, 21, 460–465. [Google Scholar] [CrossRef]
- Shrestha, P.; Holland, T.M.; Bundy, B.C. Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. Biotechniques 2012, 53, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Kwon, Y.C.; Jewett, M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 2015, 5, 8663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, D.A.; Zubay, G. The stimulatory effect of cyclic adenosine 3’5’-monophosphate on DNA-directed synthesis of beta-galactosidase in a cell-free system. Proc. Natl. Acad. Sci. USA 1969, 63, 118–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalkin, H.; Yanofsky, C.; Squires, C.L. Regulated in vitro synthesis of Escherichia coli tryptophan operon messenger ribonucleic acid and enzymes. J. Biol. Chem. 1974, 249, 465–475. [Google Scholar] [CrossRef]
- Zawada, J.F.; Yin, G.; Steiner, A.R.; Yang, J.; Naresh, A.; Roy, S.M. Microscale to manufacturing scale-up of cell-free cytokine production—A new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011, 108, 1570–1578. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.M.; Swartz, J.R. Regeneration of adenosine triphosphate from glycolytic intermediates for cell-free protein synthesis. Biotechnol. Bioeng. 2001, 74, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Keum, J.W.; Oh, I.S.; Choi, C.Y.; Park, C.G.; Kim, D.M. Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J. Biotechnol. 2006, 126, 554–561. [Google Scholar] [CrossRef]
- Kay, J.E.; Jewett, M.C. Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng. 2015, 32, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Dudley, Q.M.; Nash, C.J.; Jewett, M.C. Cell-free biosynthesis of limonene using enzyme-enriched Escherichia coli lysates. Synth Biol. 2019, 4, ysz003. [Google Scholar] [CrossRef] [Green Version]
- Niu, F.X.; Huang, Y.B.; Shen, Y.P.; Ji, L.N.; Liu, J.Z. Enhanced production of pinene by using a cell-free system with modular cocatalysis. J. Agric. Food Chem. 2020, 68, 2139–2145. [Google Scholar] [CrossRef]
- Kang, S.H.; Oh, T.J.; Kim, R.G.; Kang, T.J.; Hwang, S.H.; Lee, E.Y. An efficient cell-free protein synthesis system using periplasmic phosphatase-removed S30 extract. J. Microbiol. Methods 2000, 43, 91–96. [Google Scholar] [CrossRef]
- Kim, R.G.; Choi, C.Y. Expression-independent consumption of substrates in cell-free expression system from Escherichia coli. J. Biotechnol. 2000, 84, 27–32. [Google Scholar] [CrossRef]
- Kigawa, T.; Yabuki, T.; Matsuda, N.; Matsuda, T.; Nakajima, R.; Tanaka, A. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004, 5, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.C.; Patel, K.G.; Wong, H.E.; Swartz, J.R. Simplifying and streamlining Escherichia coli-based cell-free protein synthesis. Biotechnol. Prog. 2012, 28, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, N.E.; Kao, W.Y.; Williams, L.C.; Hight, C.M.; Patel, P.; Watts, K.R. Unlocking applications of cell-free biotechnology through enhanced shelf life and productivity of E. coli extracts. ACS Synth. Biol. 2020, 9, 766–778. [Google Scholar] [CrossRef]
- Mortenson, L.E. Ferredoxin and ATP, requirements for nitrogen fixation in cell-free of Clostridium pasteurianum. Proc. Natl. Acad. Sci. USA 1964, 52, 272–279. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, H.; Kwon, Y.C.; Jewett, M.C. Establishing a high yielding streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng. 2017, 114, 1343–1353. [Google Scholar] [CrossRef]
- Ryabova, L.A.; Vinokurov, L.M.; Shekhovtsova, E.A.; Alakhov, Y.B.; Spirin, A.S. Acetyl phosphatase as an energy source for bacterial cell-free translation systems. Anal. Biochem. 1995, 226, 184–186. [Google Scholar] [CrossRef]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 2004, 86, 19–26. [Google Scholar] [CrossRef]
- Kelwick, R.; Webb, A.J.; MacDonald, J.T.; Freemont, P.S. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng. 2016, 38, 370–381. [Google Scholar] [CrossRef] [Green Version]
- Jewett, M.C.; Calhoun, K.A.; Voloshin, A.; Wuu, J.J.; Swartz, J.R. An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 2008, 4, 220. [Google Scholar] [CrossRef]
- Khattak, W.A.; Ul-Islam, M.; Ullah, M.W.; Yu, B.; Khan, S.; Park, J.K. Yeast cell-free enzyme system for bio-ethanol production at elevated temperatures. Process. Biochem. 2014, 49, 357–364. [Google Scholar] [CrossRef]
- Niu, F.X.; Yan, Z.B.; Huang, Y.B.; Liu, J.Z. Cell-free biosynthesis of chlorogenic acid using a mixture of chassis cell extracts and purified spy-cyclized enzymes. J. Agric. Food Chem. 2021, 69, 7938–7947. [Google Scholar] [CrossRef] [PubMed]

| [16] | [17] | [18] | [20] | [21] | |
|---|---|---|---|---|---|
| buffer | standard buffer (0.01 M tris-Ac, pH 7.8, 0.06 M potassium chloride, 0.014 M magnesium acetate, 0.006 M 2-mercaptoethanol) | S-30 buffer (0.01 M tris-Ac, pH 8.2, 0.06 M potassium chloride, 0.014 M magnesium acetate, 0.001 M dithiothreitol) | buffer S30 (0.01 M Tris/acetic acid, pH 8.2; 0.014 M magnesium acetate; 0.06 M potassium acetate; 0.002 M dithiothreitol) | buffer A (0.01 M Tris; 0.014 M magnesium acetate; 0.06 M potassium glutamate; 0.001 M dithiothreitol) | buffer A (0.01 M tris-Ac, pH 8.2, 60 mM potassium glutamate, 0.014 M magnesium acetate, 0.002 M dithiothreitol) |
| ways | Alumina grinding method and high-pressure crushing method | High-pressure crushing method | High-pressure crushing method | High-pressure crushing method and ultrasonic crushing method | High-pressure crushing method and ultrasonic crushing method |
| preculture (run-off reaction) | Yes | Yes | Yes | No | Not essential |
| Energy regeneration | standard buffer (0.01 M tris-Ac, pH 7.8, 0.06 M potassium chloride, 0.014 M magnesium acetate, 0.006 M 2-mercaptoethanol) | S-30 buffer (0.01 M tris-Ac, pH 8.2, 0.06 M potassium chloride, 0.014 M magnesium acetate, 0.001 M dithiothreitol) | buffer S30 (0.01 M Tris/acetic acid, pH 8.2; 0.014 M magnesium acetate; 0.06 M potassium acetate; 0.002 M dithiothreitol) | - | - |
| dialysis | Yes | Yes | Yes | No | Not essential |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.; Wang, W.; Guo, T.; Long, X.; Niu, F. Cell-Free Escherichia coli Synthesis System Based on Crude Cell Extracts: Acquisition of Crude Extracts and Energy Regeneration. Processes 2022, 10, 1122. https://doi.org/10.3390/pr10061122
Huang M, Wang W, Guo T, Long X, Niu F. Cell-Free Escherichia coli Synthesis System Based on Crude Cell Extracts: Acquisition of Crude Extracts and Energy Regeneration. Processes. 2022; 10(6):1122. https://doi.org/10.3390/pr10061122
Chicago/Turabian StyleHuang, Mingyue, Weiyang Wang, Tingting Guo, Xiufeng Long, and Fuxing Niu. 2022. "Cell-Free Escherichia coli Synthesis System Based on Crude Cell Extracts: Acquisition of Crude Extracts and Energy Regeneration" Processes 10, no. 6: 1122. https://doi.org/10.3390/pr10061122
APA StyleHuang, M., Wang, W., Guo, T., Long, X., & Niu, F. (2022). Cell-Free Escherichia coli Synthesis System Based on Crude Cell Extracts: Acquisition of Crude Extracts and Energy Regeneration. Processes, 10(6), 1122. https://doi.org/10.3390/pr10061122






