Abstract
Akebia trifoliata (A. trifoliata) is a significant medicinal and edible fruit crop and has some important bioactivities. However, there are few studies on the bacteriostatic activity of A. trifoliata, and the underlying mechanism of A. trifoliata for antibacterial activity is still unknown. Therefore, the bacteriostatic activity and antibacterial mechanism of A. trifoliata were investigated by a combination of chemical assays, using the UHPLC-TOF-MS/MS technique. The results indicated that alkaloids, triterpenoids, and flavonoids are the major secondary bioactive compounds in A. trifoliata that play a crucial role in antibacterial activity. We found that EEPA exhibited both bacteriostatic and bactericidal effects against all Gram-positive and Gram-negative bacteria tested, with IZDs ranging from 13.80 ± 0.79 to 17.00 ± 0.58 mm. Significant differences in terms of sensitivity between Gram-positive and Gram-negative bacteria were not observed. In contrast, both antibiotics (kanamycin sulfate and ampicillin sodium salt) exhibited much better antimicrobial activity against Gram-positive bacteria than Gram-negative bacteria. In addition, the primary antimicrobial mechanism was that EEPA increased cellular content leakage, altered the cell morphology, and destroyed the internal cell structure. Meanwhile, MA, UA, and OA, as the common triterpenoid components existing in plants, were used to analyze the relationships between the structures and the antimicrobial activities among homologous compounds, to determine the key functional group that plays an antibacterial role in MA, UA, and OA. As result, it was found that both the hydroxide and methyl groups present are important for their antibacterial activity. These findings suggested that EEPA exerted significant antimicrobial activity against S. aureus, E. coli, B. subtilis, and P. aeruginosa and might be a potential natural antibacterial.
1. Introduction
Akebia trifoliata (Thunb.) Koidz is a significant medicinal and edible fruit crop that belongs to the family Lardizabalaceae. It is widely cultivated in China and Japan, especially in the southern areas of China [1]. Traditionally, the raw materials (stem and fruit) of A. trifoliata have been used in traditional Chinese medicine [2]. It is used for diuresis and analgesia, as well as to treat heart disease as a cardiostimulator. It is also valued in the treatment of inflammation and for its anti-aging properties [3]. This herb has also been used traditionally as an antibacterial medicine [4]. In traditional Chinese medicine, A. trifoliata extract is used to treat neuralgia, gastric ulcers, viral hepatitis, mouth wounds and ulcers, burning and pain in the tongue, acute pharyngitis, circulatory failure, oedema, and infections and inflammation of the kidneys and urinary tract. There are also data in the literature supporting the antibacterial activity of this herb. For example, citric-acid-extracted pectin from A. trifoliata var. australis peel effectively facilitated a moist environment with bacterial disinfection capability that accelerated the healing of infected wounds [5]. Jiang et al. indicated that A. trifoliata peel extract had the potential to improve the antibacterial activity of chitosan films [6].
Traditional use and previous literature data show that A. trifoliata has antibacterial activity. However, there is little scientific evidence directly testing A. trifoliata’s antibacterial activity and systematically elucidating the antibacterial mechanisms. Therefore, the potential value of A. trifoliata as a natural antibacterial herb will be expounded with regard to metabolite profiles, antibacterial activity, antibacterial spectrum, and bactericidal mechanism in this study, and the feasibility of A. trifoliata as a natural bactericidal herb will be more fully clarified. Two Gram-positive bacteria (Staphylococcus aureus, Bacillus subtilis) and two Gram-negative bacteria (Escherichia coli, Pseudomonas aeruginosa) were selected to investigate the antimicrobial bioactivity and antibacterial spectrum of an ethanol extract from the pericarp of A. trifoliata (EEPA) with IZDs and MIC (the minimum concentration), and the antibacterial action of EEPA was also explored using morphological indices, to research the potential of A. trifoliata as an antimicrobial herb.
Triterpenoids and triterpenoid saponins are very important types of antibacterial bioactive compounds among which MA, UA, and OA are the common triterpenoid components existing in plants [7,8]. Based on the metabolite profiles of A. trifoliata, we conclude that MA, UA, and OA are the secondary metabolites of A. trifoliata, and they exist differently in different tissues. OA and UA are isomers, and MA differs from them by one oxygen atom [9]. The structural characteristics of MA, UA, and OA are more conducive to the analysis of the relationship between structures and their bioactivities. The structural characteristics of bioactive substances determine their bioactivity, and knowledge of the key functional group of bioactive substances is of great significance for the development of new antimicrobial products. Therefore, we studied the antimicrobial activities of MA, UA, and OA to determine the key functional group that plays an antibacterial role in these components. This will lay a good foundation for the study of the molecular mechanism of new antibiotic molecules and help research into new triterpenoid derivatives of A. trifoliata.
2. Materials and Methods
2.1. Materials
A. trifoliata fruits were obtained from planting bases (Changsha, Hunan, China). The samples were identified by Professor Liu Keming of Hunan Normal University as A. trifoliata, and voucher specimens were kept at the Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences. The samples were stored at −80 °C. MA, OA, and UA were purchased from Beijing Solarbio Science & Technology Co., Ltd. (Solarbio, Beijing, China). Kanamycin sulfate and ampicillin sodium salt were also purchased from Beijing Solarbio Science & Technology.
2.2. Extraction
A. trifoliata pericarp (5.0 g) was extracted in 100 mL 60% (v/v) of ethanol at room temperature for 120 min each time, using ultrasound extraction. The process was repeated three times. The obtained extract solution was combined and concentrated by vacuum to yield 1.08 g of the dried solid matter, which was dissolved in an aqueous solution at 10.0 mg·mL−1. The extract solution was stored at 4 °C.
2.3. Quantitative Analysis by UHPLC-TOF -MS/MS
2.3.1. Liquid Chromatography Conditions
The quantitative analysis of A. trifoliata extracts was performed using an Agilent 1290 Infinity UHPLC system equipped with a ZORBAX Eclipse Plus C18 HD (2.1 × 50 mm, 1.8 μm) column and an autosampler. The mobile phase consisted of A (water containing 0.5% formic acid and 25 mM ammonium acetate) and B (methanol). The flow rate was 0.4 mL min−1, and the injection volume was 2 µL. The elution condition was applied with a linear gradient as follows: 0–0.5 min, 5% B; 0.5–10 min, 5–100% B; 10–12 min, 100% B; 12–12.1 min, 100–5% B; 12.1–16 min, 5% B. During the whole analysis process, the samples were placed in 4 automatic samplers. To avoid the influence of fluctuation of the instrument detection signal, samples were continuously analyzed at random. QC samples were inserted into the sample queue to monitor and evaluate the stability of the system and the reliability of the experimental data.
2.3.2. Mass Spectrometry Conditions
An AB triple TOF 6600 mass spectrometer was used to collect the first-order and second-order spectrograms of the samples. The ESI source conditions for HILIC chromatography were as follows: ion source gas 1: 60, ion source gas 2: 60, curtain gas: 30, source temperature: 600 °C; ion sapary voltage floating (ISVF ± 5500 V); TOF MS scan m/z range: 60–1000 Da, product ion scan m/z range: 25–1000 Da, TOF MS scan accumulation time 0.20 s/spectra, product ion scan accumulation time 0.05 s/spectra. Secondary mass spectrograms were obtained by information-dependent acquisition (IDA) in high-sensitivity mode, with decluttering potential (DP): ±60 V, collision energy: 35 ± 15 eV, IDA: excluding isotopes within 4 Da, candidate ions to monitor per cycle: 10.
2.4. Bacterial Strains and Culture
Staphylococcus aureus ATCC25923 was purchased from Yuanye Biotechnology, Shanghai, China. Bacillus subtilis, Escherichia coli, and Pseudomonas aeruginosa were obtained from the College of Biology, Hunan University. The strains were inoculated on nutrient broth (NB) or nutrient agar (NA) media at 37 °C for 16–24 h. The cultures were diluted to 105–108 colony-forming units (CFU)/mL using sterile 0.9% saline solution for further study.
2.5. Antimicrobial Assay
The bacteriostatic activities of the samples (EEPA, MA, OA, UA, kanamycin sulfate, and ampicillin sodium salt) were evaluated by measuring the diameters of the bacteriostatic circles. The diluted inoculum (100 µL, 105–108 CFU/mL) was uniformly smeared onto individual NA plates using a triangle coater. The filter paper disks (6.0 mm in diameter) were placed on the NA plates and filled with the samples. Sterile water was used as a negative control. The plates were cultured for 16–24 h at 37 °C in an incubator [10].
2.6. Broth Dilution MIC Assay
EEPA was diluted in nutrient broth (NB) using the half dilution method, to yield concentrations ranging from 5 to 0.156 mg/mL. The S. aureus inoculums (5.0 µL, 105–108 CFU/mL) were added to the above NB and incubated at 37 °C for 16–24 h. The culture strains (100 µL), prepared in triplicate, were smeared onto individual nutrient agar (NA) plates with a triangle coater and incubated at 37 °C for 16–24 h. The concentration of EEPA at which no growth occurred on two of the three plates was defined as the MIC. Sterilized distilled water was used as a negative control [11].
2.7. Resazurin Microplate Assay
Different concentrations of OA solution, etc. (0.5 mg/mL, 0.25 mg/mL, … 0.00187 mg/mL, 0.0009 mg/mL) were obtained using the double dilution method. Then, the inoculums (50 µL, 108 CFU/mL) and the resazurin (50 µL, 100 ug/mL) were added into 1–10 columns one by one. The 11th column, in which the same dosage inoculums and indicators were added as above but without test samples, was defined as the growth column. In addition, the 12th column, with the indicator (50 µL, 100 ug/mL), had the same volume of solution as columns 1–11. Then, the plate was incubated at 37 °C for 5–6 h until the growth wells became pink in color. The concentration of OA solution, etc., at which no pink color occurred on two of the three wells was defined as the minimum inhibitory concentration [12].
2.8. Determination of Bacterial Mortality
The bacteria were mixed with different concentrations of EEPA solution and incubated at 37 °C for 18 h. After discarding the supernatant, the sediment was washed and dissolved in a sterile phosphate-buffered solution (PBS). MTT solution (5 µL of 5.0 mg·mL−1, dissolved in PBS medium) was added to each well, and the plate was further incubated at 37 °C for 4 h. Next, 100 µL of dimethyl sulfoxide was added to solubilize the crystalline formazan. The absorbance values were determined at 515 nm [13,14]. All experiments were performed in triplicate in three independent experiments. Cell mortality was calculated using the following equation:
where the positive control (pos control) was the bacterium grown in NB medium (without EEPA solution).
2.9. Bacterial Cell Membrane Permeability
After incubation at 37 °C overnight, 1 mL of fresh bacterial culture was centrifuged at 16,000× g for 1 min, and the supernatant was discarded. Buffer solution (1.0 mL) was added to the tube, mixed uniformly, and centrifuged at 16,000× g for 1 min. These steps were repeated 4–6 times. The obtained bacterial solution was re-suspended in 1 mL of buffer solution. Next, 150 µL of the bacterial solution was transferred to a plate, followed by the addition of EEPA (45 µL) and dye liquor (5 µL). The bacterial solutions were incubated at 25 °C for 5 min and then promptly evaluated using a microplate reader (Infinite M200PRO, Grodig, Austria) at excitation and emission wavelengths of 355 and 460 nm. A bacterial solution without EEPA was used as a negative control [5,15].
2.10. TEM
Bacteria mixed with EEPA were cultured at 3 °C for 16 h. The bacterial cells were collected by centrifugation at 8000× g for 10 min at 4 °C, and the pellet was fixed with 2.5% glutaraldehyde at 4 °C for 24 h. The cells were washed with phosphate-buffered solution (PBS) (PH 7.0) and post-fixed with 1% osmic acid at 4 °C for 2 h. After embedding, solidifying, sectioning, and dyeing, the cell morphology was observed using transmission electron microscopy (JEOL, Tokyo, Japan). Bacterial cells that had not been treated with EEPA were used as a control [16].
2.11. Data Analysis
IBM SPSS Statistics 25 was used to analyze all data with a one-way analysis of variance (ANOVA). Duncan’s multiple range tests were applied to determine a significant difference (p < 0.05). Data were obtained from three independent experiments and were expressed using the mean ± standard deviation.
3. Results
3.1. Metabolomic Profile Analysis of A. trifoliata Extract
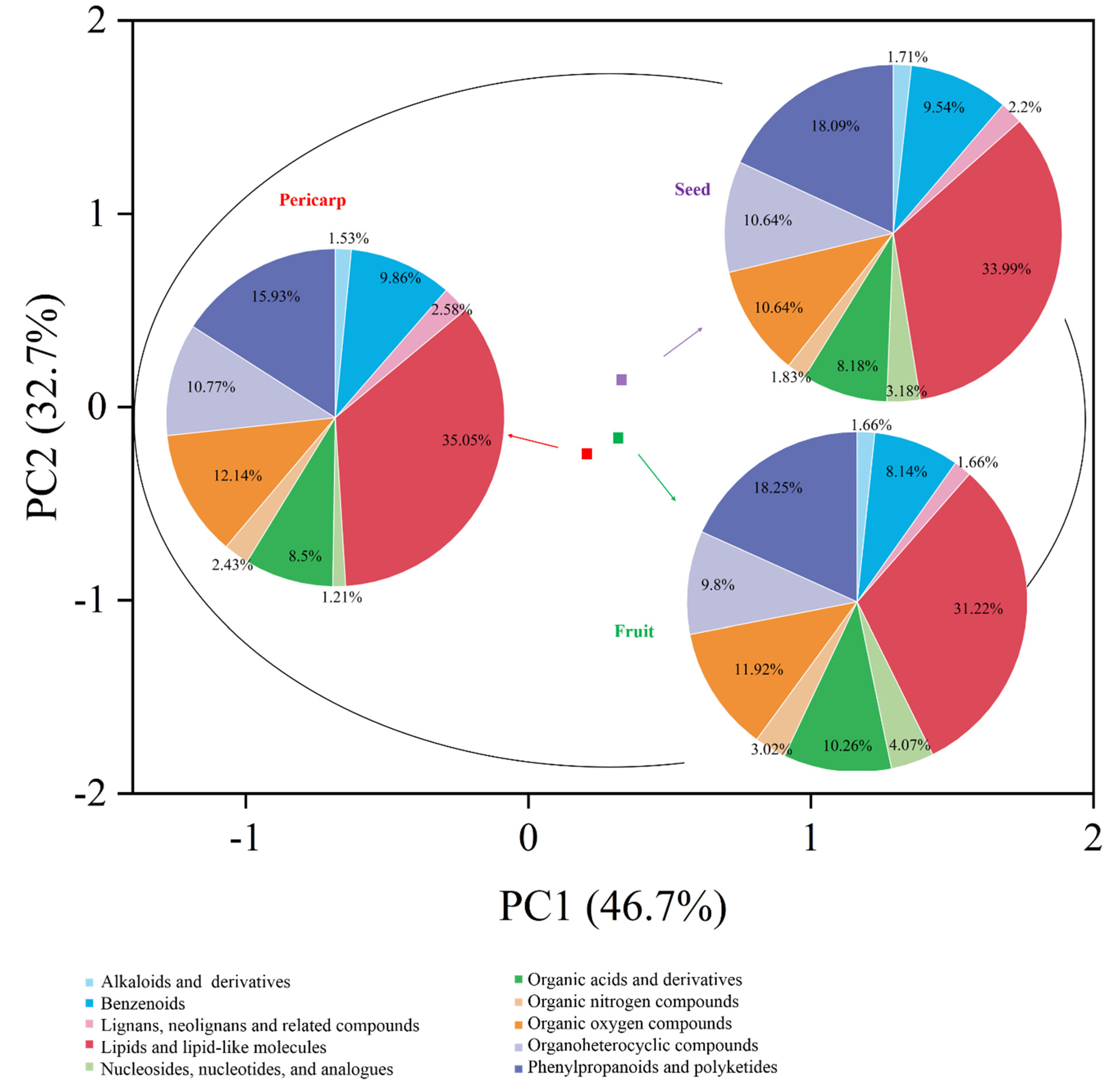
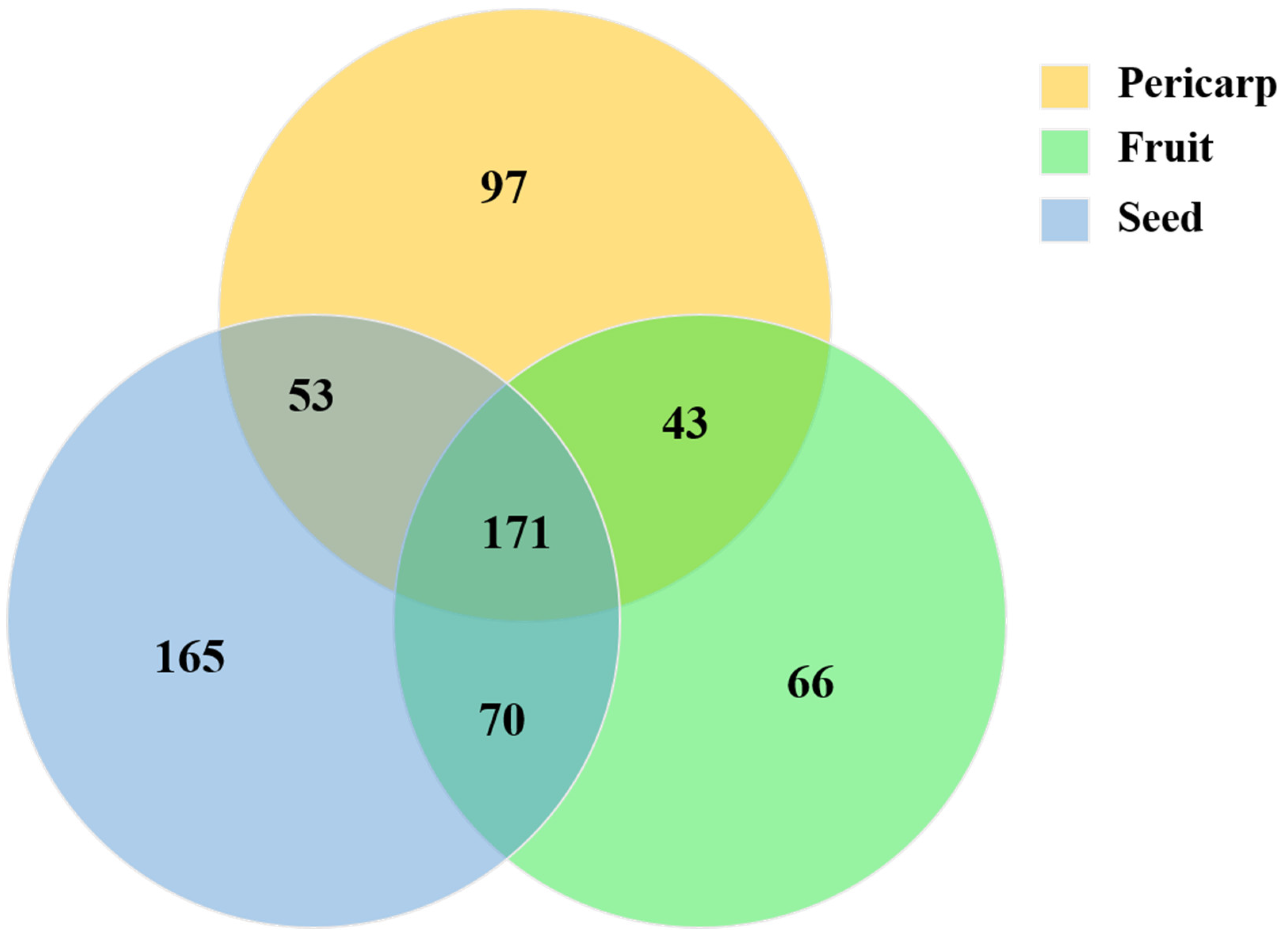
Metabolomic profiling of the extract from A. trifoliata was constructed using UHPLC-QTOF-MS/MS, and 352, 342, and 447 secondary metabolites were identified in the pericarp, fruit, and seed extracts, respectively (Tables S1–S3 in the Supplementary Material). Alkaloids, triterpenoids, and flavonoids, together with their derivatives, were the main secondary metabolites that might provide the crucial bioactivity (Figure 1). Different secondary metabolites were observed in the fruit, pericarp, and seed, although the presence of alkaloids, triterpenoids, and flavonoids was identified in the main compounds. The seeds were richest in secondary metabolites, followed by the pericarp and fruit (Figure 2). The phytochemical profiles of A. trifoliata showed that triterpenoids in particular were the most important bioactive compounds. In this study, 45 triterpenoids were identified in the fruit, pericarp, and seed (Tables S1–S3 and Table 1). OA, UA, and MA, as the common triterpenoids existing in plants, were also identified in the seed and pericarp, but only OA was identified in the fruit. Contrary to reports in the literature, this is the first time that UA has been identified in A. trifoliata. The content of OA was highest in pericarp and seed, followed by UA and MA. The content of OA in pericarp was higher than that in seed, but the contents of UA and MA showed the opposite trend.
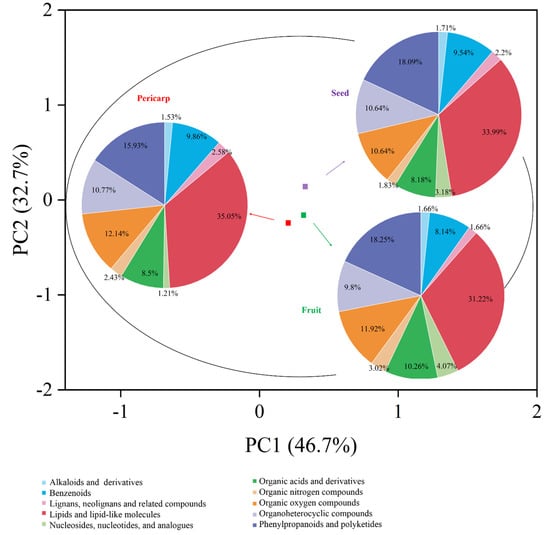
Figure 1.
PCA plot of A. trifoliata fruit, pericarp, and seed extracts. PC1 (46.7%), PC2 (32.7%). Within the PCA plot, fruit extract is represented in green, pericarp extract in red, and seed extract in purple. Different classes of compounds identified in each sample are represented using a pie chart.
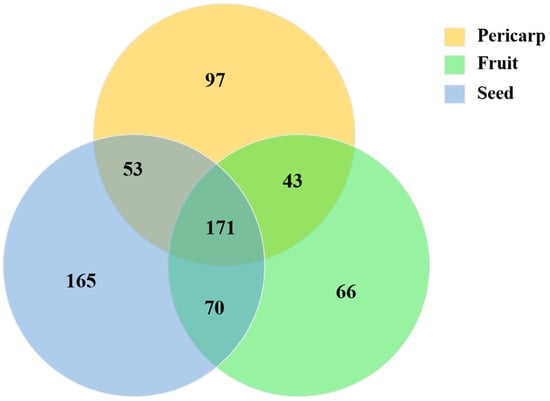
Figure 2.
Venn diagram showing the main secondary metabolites (alkaloids, lignans and neolignans, lipids and lipid-like molecules, phenylpropanoids, and polyketides) among the fruit, pericarp, and seed.

Table 1.
Triterpenoid compounds identified in the fruit, pericarp, and seed of A. trifoliata (UHPLC-TOF-MS/MS) and metabolomic pathways.
3.2. Antimicrobial Activities of Pericarp Extract of A. trifoliata
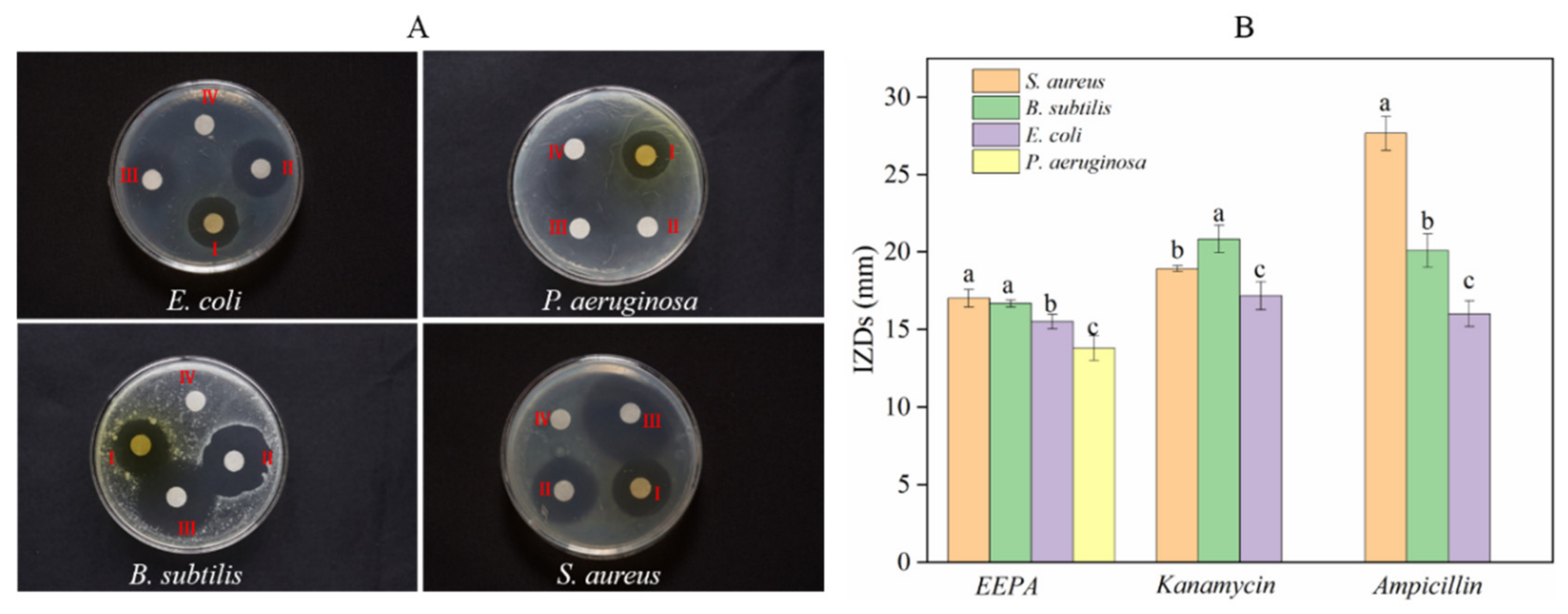
EEPA exhibited both bacteriostatic and bactericidal effects against all Gram-positive and Gram-negative bacteria tested, with IZDs ranging from 13.80 ± 0.79 to 17.00 ± 0.58 mm (Figure 3B). Kanamycin sulfate (aminoglycoside broad-spectrum antibiotic) and ampicillin sodium salt (B-lactam antibiotic) were effective against S. aureus, E. coli, and B. subtilis but not P. aeruginosa, with IZDs ranging from 17.17 ± 0.89 to 20.83 ± 0.89 mm and 16 ± 0.82 to 27.66 ± 1.11 mm, respectively (Figure 3B). Among the Gram-negative bacteria, E. coli showed higher sensitivity towards EEPA than P. aeruginosa. Significant differences in terms of sensitivity between Gram-positive and Gram-negative bacteria were not observed in EEPA (Figure 3A), and this could be attributed to the combined effect of the triterpenoids and triterpenoids saponins or some of their components. Duncan’s test was used to compare the significant differences in IZDs between different bacteria against EEPA, and the different letters appearing on the bars represent these organisms (Figure 3B), showing inconsistency with the results (Figure 3A). Duncan’s multiple ranges were tested at the p < 0.05 level; this may be the cause of the lack of conformity between the IZDs of these organisms in cultures treated with EEPA. Different letters appear on the bars representing these organisms. However, both kanamycin sulfate and ampicillin sodium salt exhibited much better antimicrobial activity against Gram-positive bacteria than Gram-negative bacteria, indicating the limitations of antibiotics with respect to antimicrobial activity compared with EEPA. The antimicrobial activity of EEPA was close to that of kanamycin but weaker than that of ampicillin.
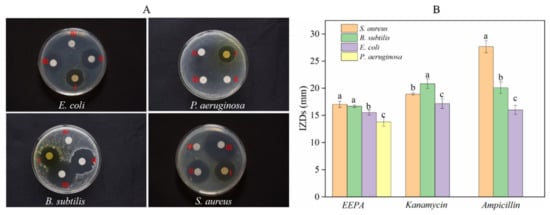
Figure 3.
Antibacterial activity assay of the ethanol extract from pericarp of A. trifoliata. (A) Bacteriostatic circle: (Ⅰ) 30 ug of EEPA; (Ⅱ) 5.0 ug of kanamycin; (Ⅲ) 5.0 ug of ampicillin; (Ⅳ) sterile water. These experiments were performed in triplicate. (B) The inhibition zone diameter. Different lowercase letters a, b, and c represent a significant difference between bacteria against each sample, according to Duncan’s multiple range test (p < 0.05).
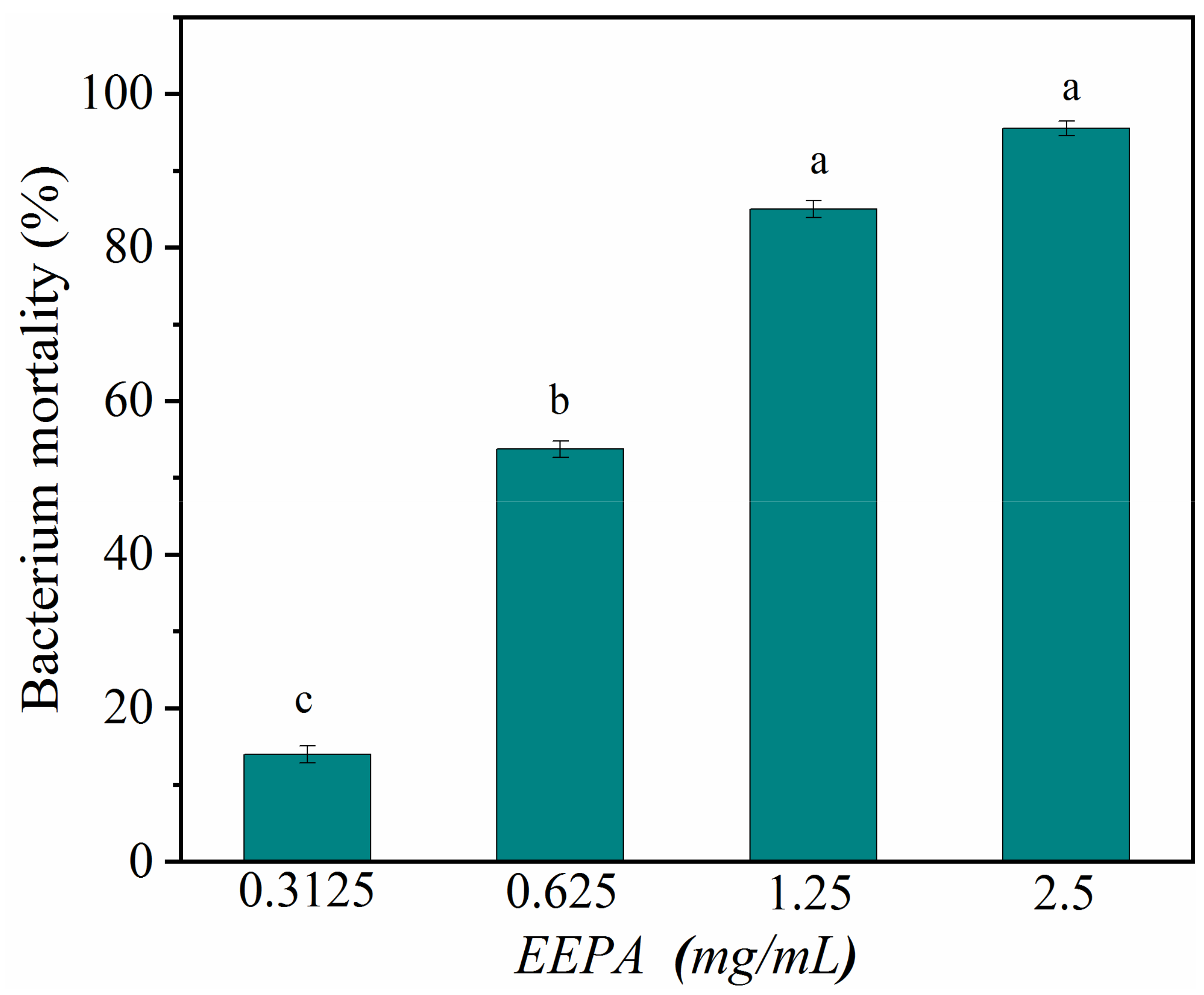
To better understand the antibacterial activity and antibacterial mechanism of EEPA, we further investigated the antibacterial activity and bactericidal mechanism of EEPA against S. aureus. The average IZD of EEPA for Gram-positive S. aureus was 17.00 ± 0.57 mm. The average IZDs of ampicillin sodium salt and kanamycin sulfate were 27.66 ± 1.11 mm and 18.92 ± 0.19 mm, respectively. The results indicated that EEPA had an antimicrobial activity comparable to those of the antibiotics tested. The MIC of EEPA for inhibiting the growth of S. aureus in vitro was 1.25 mg/mL, which is consistent with the results giving an IC50 value of less than 0.625 mg·mL−1 (Figure 4). These results confirmed that EEPA possesses good antibacterial activity against S. aureus.
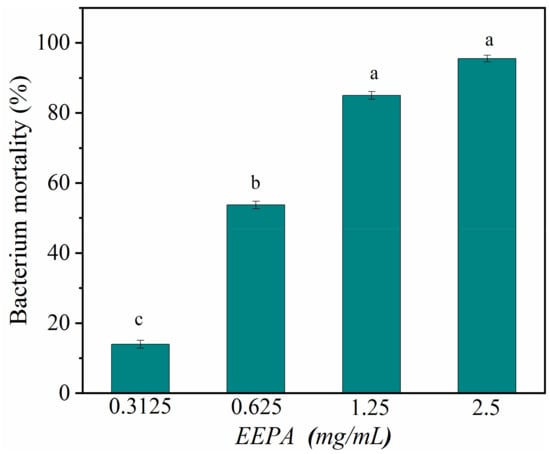
Figure 4.
Bacterium mortality of S. aureus after treatment with EEPA at concentrations of 0.3125–2.5 mg·mL−1. Different lowercase letters indicate a significant difference in bacterium mortality at different concentrations, according to Duncan’s multiple range test (p < 0.05).
3.3. Antimicrobial Mechanism
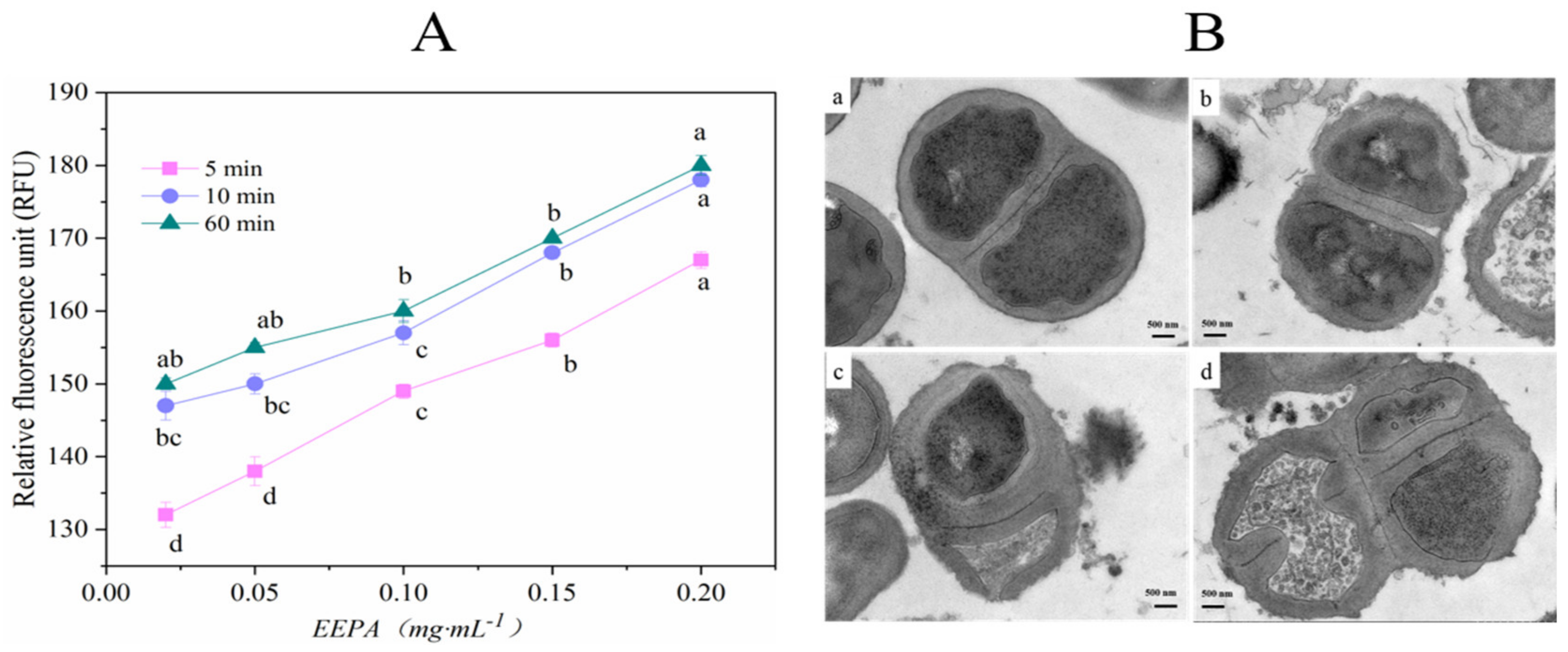
N-Phenyl-naphtha amine, a hydrophobic fluorescence probe, interacts with the phospholipids layer of the damaged bacterial membrane, resulting in increased fluorescence intensity. As shown in Figure 5A, the degree of bacterial membrane injury was dependent on the concentration of EEPA. The fluorescence intensities of bacterial solutions treated with EEPA were determined at different times. The fluorescence intensity significantly increased between incubation times of 5 and 10 min. After 60 min, the fluorescence intensity remained stable.
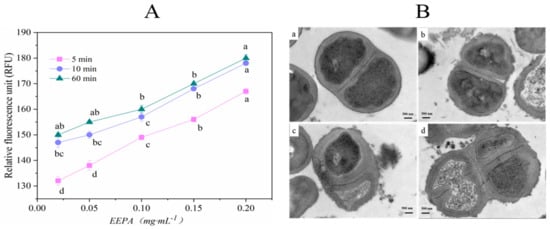
Figure 5.
The bactericidal mechanism analysis of EEPA against S. aureus. (A) Membrane permeability analysis. These experiments were performed in triplicate. Values with different letters as the label are significantly different for different concentrations (p < 0.05). (B) TEM images of S. aureus cells: (a) control; (b–d) treated with MIC of EEPA and S. aureus cells.
TEM (transmission electron microscopy) was performed to analyze the morphological features of S. aureus treated with EEPA. Untreated S. aureus cells retained their regular morphology with a smooth surface and spherical shape (Figure 5(Ba)). However, S. aureus cells treated with EEPA displayed an irregular rope-like shape with a wrinkled surface (Figure 5(Bd)). As shown in Figure 5(Bb), partial disappearance of the cell wall and membrane of S. aureus cells was observed following treatment with EEPA. In contrast, untreated S. aureus cells showed an intact cell wall and membrane (Figure 5(Ba)). Staphylococcus aureus cells treated with EEPA appeared to undergo lysis, resulting in the release of their cellular contents into the surrounding environment and eventual disruption (Figure 5(Bc)). As shown in Figure 5(Bd), S. aureus treated with sample displayed cell distortion and nonuniform distribution of the endochylema. Cells incubated with EEPA suffered cell wall degradation, cytoplasmic membrane deterioration, and damage to the interior structure, inducing bacterial metabolism dysfunction.
3.4. Antimicrobial Activities of OA, UA, and MA
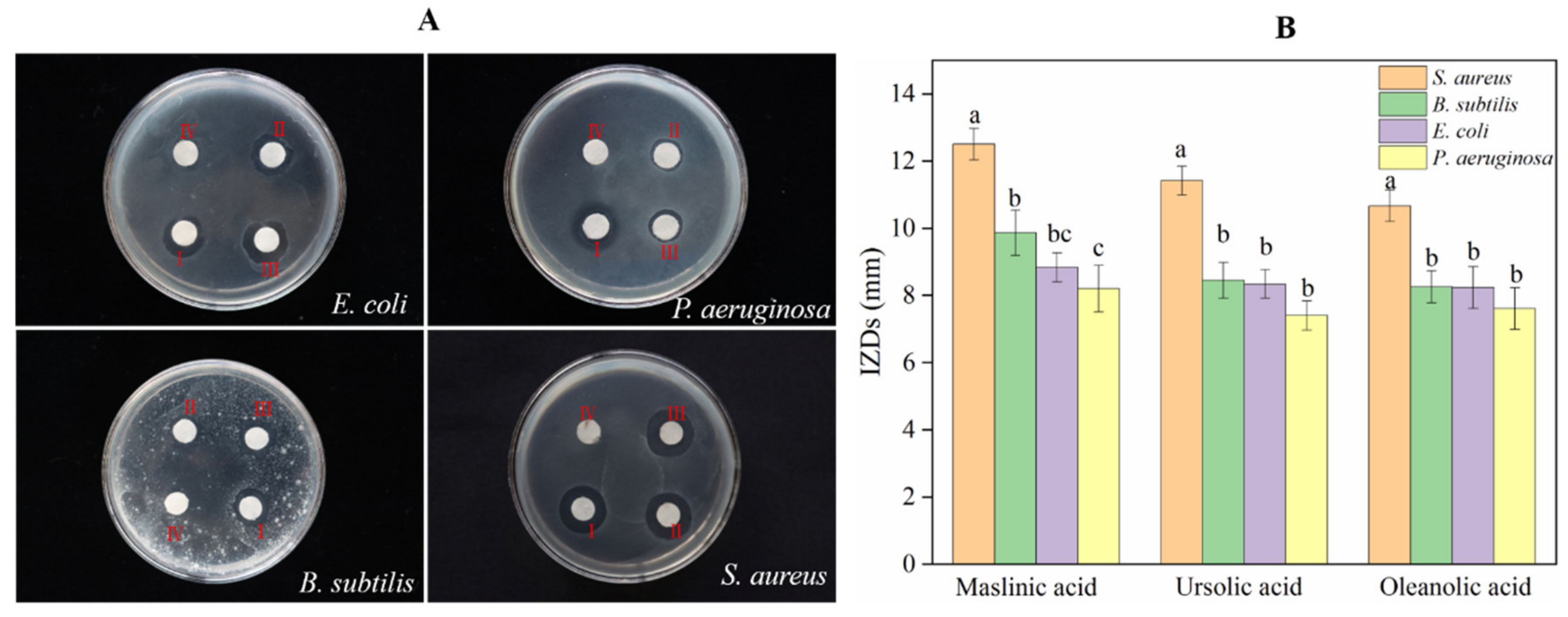
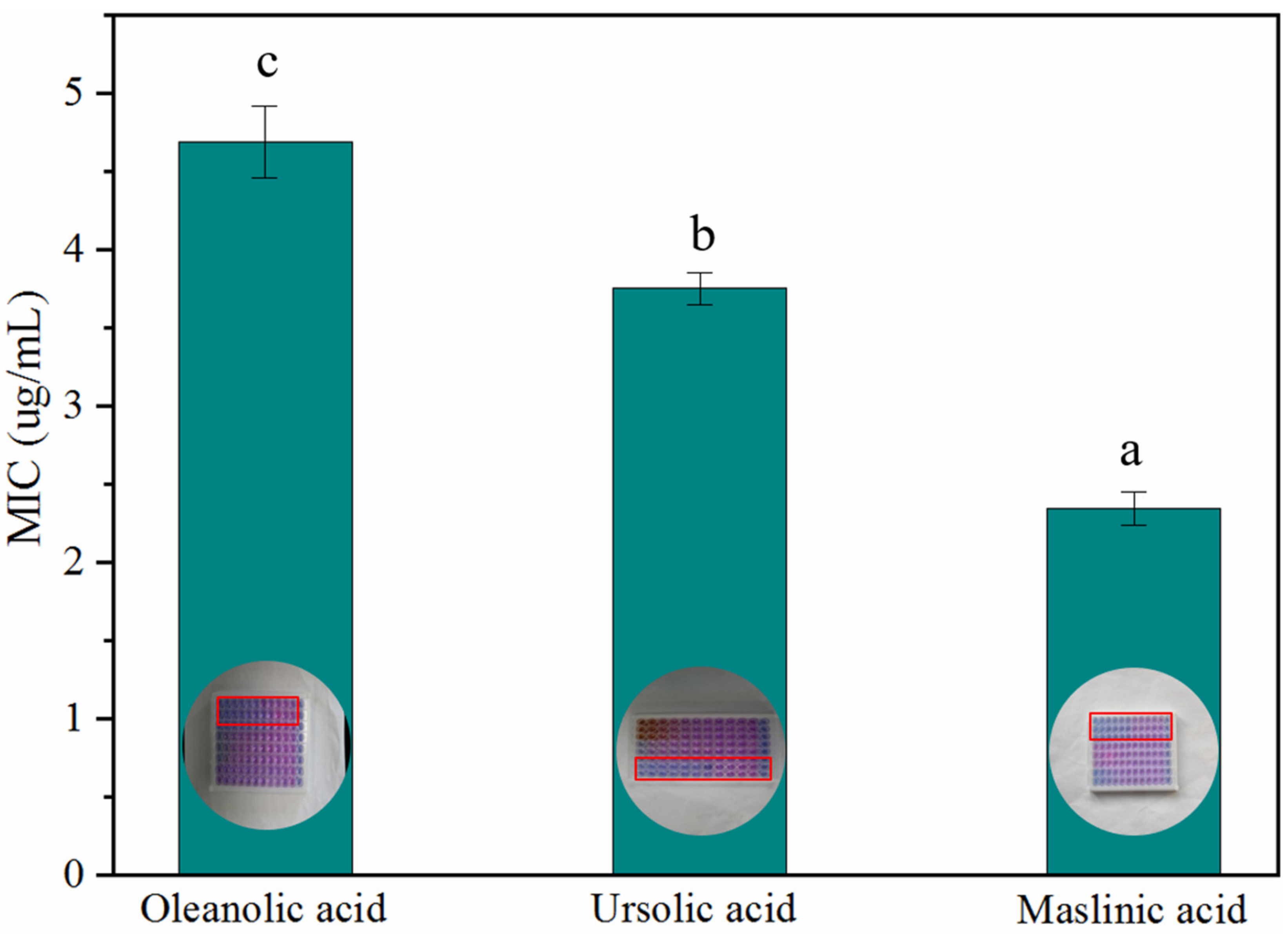
As the most important group of secondary metabolites in A. trifoliata, triterpenoids and triterpenoid saponins exhibited antimicrobial activity against bacteria such as Bacillus thuringiensis, Escherichia coli, Salmonella enterica, and Shigella dysenteriae [8]. However, the antimicrobial activities of OA, MA, and UA against S. aureus, E. coli, B. subtilis, and P. aeruginosa were not identified simultaneously in this research. In the paper, the assay IZDs of the above three triterpenoids against S. aureus, E. coli, B. subtilis, and P. aeruginosa were determined, in order to compare the variation of bactericidal activity among homologous compounds. As result, the three triterpenoids exhibited antimicrobial activities against S. aureus, E. coli, B. subtilis, and P. aeruginosa at different levels; they did not show Gram-positive or Gram-negative specificity in antibacterial activity and MA showed the best bactericidal activity for S. aureus, E. coli, B. subtilis, and P. aeruginosa. S. aureus was more sensitive to MA, UA, and OA than the other three bacteria and showed a significant variation in resistance against MA, UA, and OA (Figure 6). The results of the MIC assay (Figure 7) indicated that these triterpenoids could effectively hinder the normal growth of S. aureus, and this is consistent with the results of the bacteriostatic circle assay. MA showed the highest bactericidal activity for S. aureus, followed by UA and finally OA (Figure 7).
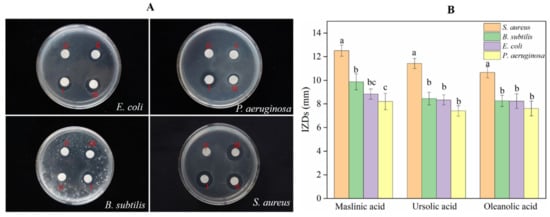
Figure 6.
Antibacterial activity assay for OA, UA, and MA. (A) Bacteriostatic circles for 10 ug of: (Ⅰ) MA; (Ⅱ) OA; (Ⅲ) UA; (Ⅳ) control, where 50% of DMSO was used as the control. These experiments were performed in triplicate. (B) The inhibition zone diameter. Values with different letters on the label are significantly different between bacteria against each compound (p < 0.05).
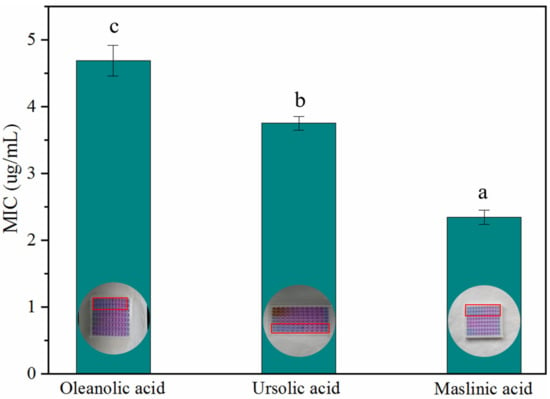
Figure 7.
MIC of OA, UA, and MA against S. aureus. Values with different letters as the label are significantly different between compounds (p < 0.05).
4. Discussion
Bacterial resistance to antibiotics is becoming increasingly serious, and numerous commonly used antibiotics have a poor effect on most bacteria [17]. It is urgently necessary to discover new and efficient antimicrobial alternatives, in order to resolve this dangerous situation. Phytochemical studies have revealed that bioactive compounds of plant origin have antibacterial activity, including phenolics and polyphenols (including simple phenols and phenolic acids, quinines, flavones, flavonoids and flavonols, tannins, and coumarins), terpenoids and essential oils, alkaloids, lectins, and polypeptides [18,19]. In this study, 335 secondary metabolites of A. trifoliata were identified, including alkaloids, phenols, phenolic acids, flavonoids, and triterpenoids. The metabolomic analysis indicated that A. trifoliata was rich in bioactive compounds and had important bioactivity potential. EEPA could efficiently inhibit the growth of S. aureus, E. coli, B. subtilis, and P. aeruginosa (Figure 3). The MIC of EEPA for inhibiting the growth of S. aureus in vitro was 1.25 mg/mL, and the inhibitory efficiency was positively correlated with EEPA concentrations (Figure 4). Similar results showed that the growth of E. coli, S. aureus, B. subtilis, and P. aeruginosa was significantly inhibited by Vernonia amygdalina leaf extract [20]. A. trifoliata exhibited bacteriostatic and bactericidal performance on all bacteria tested with a good antimicrobial spectrum, and it may be a candidate plant for use in developing new antimicrobial alternatives.
Notably, EEPA caused cell membrane disruption, abnormal cell morphology, and cellular content leakage in S. aureus. The cytoplasm membrane may be involved in bacterial defense. Intact cell membrane structure is an important indicator of cell vitality, as this is where some crucial physiological processes are completed, including substance transportation, energy transition, and information communication [21,22]. The fluorescence intensity of S. aureus cells treated with EEPA increased distinctly in 10 min, which indicated that exposure of the cells to EEPA led to the rapid destruction of the cell membrane. Therefore, the cellular content and small ions leaking out from S. aureus cells resulted in abnormal cell morphology (Figure 5B). This was also reported in cells of E. coli, S. aureus, B. subtilis, and P. aeruginosa after exposure to water-soluble carboxymethyl chitosan/copper [23]. Thus, it can be speculated that the primary antimicrobial mechanism of EEPA against S. aureus may involve cell membrane penetration and destruction, and loss of osmotic balance. However, the specific antimicrobial mechanism still requires further investigation.
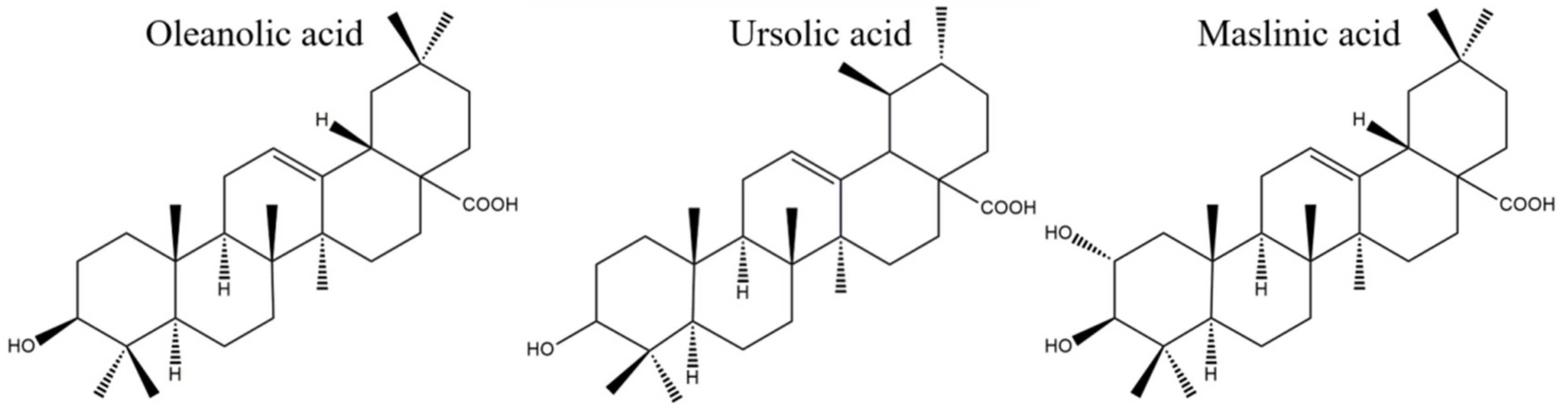
OA, UA, and MA had different antibacterial activity against the four strains tested in the study. The diversities of their chemical structures are strongly associated with their different antibacterial performances. OA (oleanane-type), UA (ursane-type), and MA (oleanane-type), representatives of the pentacyclic triterpenoids are based on the structure of isoprene and contain 30 carbon atoms and oxygen. Their chemical structures are shown in Figure 8. MA derives from OA after substitution of the hydrogen atom in the A ring with a hydroxide radical, which indicates that the hydroxide radical might play an essential role in improving the antimicrobial activity. It was found that the presence of a hydroxyl group in the A ring combined with a carboxylic group in the E ring was associated with the observed anti-tubercular activity of OA [24]. These results were consistent with our conclusion that the hydroxide radical in the A ring might have close relationship with the antimicrobial activity. OA and its isomer UA both have a familiar carbon skeleton but have different structures in the E ring. The two methyl groups in the E ring of UA are in the C-19 and C-20 positions, while they are in the C-20 position in OA. The favorable structure of UA over OA probably contributes to the better antimicrobial activity of UA. A negative effect on the antimicrobial activity was confirmed when the methyl group was replaced by a -CH2OH, a -CHO, or a -COOH group [25], indicating that the methyl group appears to be necessary and sufficient for antibacterial activity. This further proves our deduction that a methyl group may be an important group that plays an antibacterial role. The structure–activity relationship analysis of OA and UA indicated that methyl groups at C-19 and C-20 and the orientation of the hydroxyl group at C-3 were essential for this type of UA to show strong antibacterial activity. A positive effect on the antimicrobial activity was evident when the hydrogen atom of C-2 was replaced by an -OH group, which was verified by comparison of the structures and activities of OA and MA. Regarding the structure–activity relationships of OA, MA, and UA, it was suggested that both the hydroxide and methyl groups present in triterpenes are important for their antibacterial activity. The structure–activity relationship analysis is important for identifying functional groups with antibacterial activity and improving biological activity.
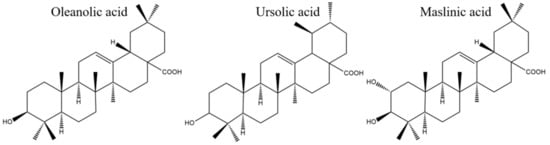
Figure 8.
Structures of OA, UA, and MA.
In recent years, research on A. trifoliata has been undertaken in many fields, including botany, Chinese medicine, analytical chemistry, etc. [26,27], since it is a plant with high commercial value. This study lays a theoretical foundation for the development of A. trifoliata in the food industry and in Chinese-herbal-medicine-related products such as natural preservatives and cavity-cleaning bacteriostatic fluids. At present, the theoretical research work on A. trifoliata is still insufficient; there are only a limited number of terpenoids in A. trifoliata, and a large number of unknown bioactive substances and unknown biological activities are still under exploration.
5. Conclusions
In this study, EEPA exhibited significant antibacterial activities against all Gram-positive and Gram-negative bacteria tested, with IZDs ranging from 13.80 ± 0.79 to 17.00 ± 0.58 mm, and it had a much broader bactericidal spectrum compared with antibiotics. In addition, the primary antimicrobial mechanism was that EEPA increased cellular content leakage, altered the cell morphology, and destroyed the internal cell structure. Finally, OA, MA, and UA could effectively hinder the growth of S. aureus. The bactericidal activities against S. aureus were in the order MA > UA > OA. In addition, both the hydroxide and methyl groups present in OA, MA, and UA were important for their antibacterial activities against S. aureus. The results of this study show that A. trifoliata has excellent prospects for use as natural antibacterial herb, based on good antimicrobial activity and the antimicrobial mechanism.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pr10071394/s1.
Author Contributions
J.C. (Jing Chen) conceived the experiments and wrote the manuscript. Z.S. was in charge of sample collection. J.C. (Jing Chen) and M.L. analyzed the data and prepared the figures. J.C. (Jianhua Chen) and all authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported financially by the Central Public-interest Scientific Institution Basal Research Fund (1610242020010), Agricultural Science and Technology Innovation Program (ASTIP) of CAAS (Grant No. 2017IBFC), Yichun Key Research and Development Projects (20211YFH4179), Natural Science Fund of Changsha (kq202322), and Provincial Key Laboratory of Woody Oil Resource Utilization (GZKF202104).
Acknowledgments
We are grateful to Jixiang Tian Biological Co., Ltd. for providing the A. trifoliata samples. The authors thank the College of Biology, Hunan University for providing strains and Scientific Compass Technology Co., Ltd. for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, L.; Chen, X.; Yao, X.; Tian, H.; Huang, H. Geographic distribution and resource status of three important Akebia species. J. Wuhan Bot. Res. 2010, 28, 497–506. [Google Scholar]
- Kitaoka, F.; Kakiuchi, N.; Long, C.; Itoga, M.; Mitsue, A.; Mouri, C.; Mikage, M. Molecular characterization of akebia plants and the derived traditional herbal medicine. Biol. Pharm. Bull. 2009, 32, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, N.; Wang, Z.; Qiu, T.; Jiang, L.; Zhu, X.; Sun, Y.; Xiong, H. Akebia trifoliata pericarp extract ameliorates inflammation through NF-κB/MAPK signaling pathways and modifies gut microbiota. Food Funct. 2020, 11, 4682–4696. [Google Scholar] [CrossRef] [PubMed]
- Maciag, D.; Dobrowolska, E.; Sharafan, M.; Ekiert, H.; Tomczyk, M.; Szopa, A. Akebia quinata and Akebia trifoliata—A review of phytochemical composition, ethnopharmacological approaches and biological studies. J. Ethnopharmacol. 2021, 280, 114486. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Wang, X.; Ning, F.; Jiang, C.; Li, Y.; Peng, H.; Xiong, H. Development of antibacterial pectin from Akebia trifoliata var. australis waste for accelerated wound healing. Carbohydr. Polym. 2019, 217, 58–68. [Google Scholar] [CrossRef]
- Jiang, Y.; Yin, H.; Zhou, X.; Wang, D.; Zhong, Y.; Xia, Q.; Deng, Y.; Zhao, Y. Antimicrobial, antioxidant and physical properties of chitosan film containing Akebia trifoliata (Thunb.) Koidz. peel extract/montmorillonite and its application. Food Chem. 2021, 361, 130111. [Google Scholar] [CrossRef]
- Zhao, W.W.; Zan, K.; Wu, J.Y.; Gao, W.; Yang, J.; Ba, Y.Y.; Wu, X.; Chen, X.Q. Antibacterial triterpenoids from the leaves of Ilex hainanensis Merr. Nat. Prod. Res. 2019, 33, 2435–2439. [Google Scholar] [CrossRef]
- Wang, J.; Ren, H.; Xu, Q.L.; Zhou, Z.Y.; Wu, P.; Wei, X.Y.; Cao, Y.; Chen, X.X.; Tan, J.W. Antibacterial oleanane-type triterpenoids from pericarps of Akebia trifoliata. Food Chem. 2015, 168, 623–629. [Google Scholar] [CrossRef]
- Sommerwerk, S.; Heller, L.; Kuhfs, J.; Csuk, R. Urea derivates of ursolic, oleanolic and maslinic acid induce apoptosis and are selective cytotoxic for several human tumor cell lines. Eur. J. Med. Chem. 2016, 119, 1–16. [Google Scholar] [CrossRef]
- Tong, C.; Zhong, X.; Yang, Y.; Liu, X.; Zhong, G.; Xiao, C.; Liu, B.; Wang, W.; Yang, X. PB@PDA@Ag nanosystem for synergistically eradicating MRSA and accelerating diabetic wound healing assisted with laser irradiation. Biomaterials 2020, 243, 119936. [Google Scholar] [CrossRef]
- Hartman, E.G.; Geryl, J. Comparison between the minimal inhibitory concentration of tilmicosin and oxytetracycline for bovine pneumonic Pasteurella haemolytica isolates. Vet. Q. 1993, 15, 184. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Liang, Z.; Yang, Y.; Liu, H.; Ji, J.; Fan, Y. A resazurin-based, nondestructive assay for monitoring cell proliferation during a scaffold-based 3D culture process. Regen. Biomater. 2020, 7, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, X.; Jin, S.; Zhang, H.; Wang, R.; Pei, L.; Zheng, J. The Anti-Proliferative Effect of Flavonoid Nanoparticles on the Human Ovarian Cancer Cell Line SK0V3. J. Nanosci. Nanotechnol. 2020, 20, 6040–6046. [Google Scholar] [CrossRef] [PubMed]
- Çevik, U.A.; Osmaniye, D.; Levent, S.; Sağlik, B.N.; Çavuşoğlu, B.K.; Karaduman, A.B.; Özkay, Y.; Kaplancikli, Z.A. Synthesis and biological evaluation of novel 1,3,4-thiadiazole derivatives as possible anticancer agents. Acta Pharm. 2020, 70, 499–513. [Google Scholar] [CrossRef]
- Yoo, J.H.; Baek, K.H.; Heo, Y.S.; Yong, H.I.; Jo, C. Synergistic bactericidal effect of clove oil and encapsulated atmospheric pressure plasma against Escherichia coli O157:H7 and Staphylococcus aureus and its mechanism of action. Food Microbiol. 2021, 93, 103611. [Google Scholar] [CrossRef]
- Liao, S.; Zhang, Y.; Pan, X.; Zhu, F.; Jiang, C.; Liu, Q.; Cheng, Z.; Dai, G.; Wu, G.; Wang, L.; et al. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int. J. Nanomed. 2019, 14, 1469–1487. [Google Scholar] [CrossRef]
- Tsakou, F.; Jersie-Christensen, R.; Jenssen, H.; Mojsoska, B. The Role of Proteomics in Bacterial Response to Antibiotics. Pharmaceuticals 2020, 13, 214. [Google Scholar] [CrossRef]
- Nagaraj Sowmya, R.; Osborne Jabez, W. Bioactive compounds from Caulerpa racemosa as a potent larvicidal and antibacterial agent. Front. Biol. 2014, 9, 300–305. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Bama, P.; Deccaraman, M.; Vijayalakshimi, M.; Murugesan, K.; Kalaichelvan, P.T.; Arumugam, P. Isolation and characterization of bioactive and antibacterial compound from Helianthus annuus linn. Indian J. Exp. Biol. 2008, 46, 831–835. [Google Scholar]
- Evbuomwan, L.; Chukwuka, E.P.; Obazenu, E.I.; Ilevbare, L. Antibacterial activity of Vernonia amygdalina leaf extracts against multidrug resistant bacterial isolates. J. Appl. Sci. Environ. Manag. 2018, 22, 17–21. [Google Scholar] [CrossRef][Green Version]
- Wang, C.; Piao, J.; Li, Y.; Tian, X.; Dong, Y.; Liu, D. Construction of Liposomes Mimicking Cell Membrane Structure through Frame-Guided Assembly. Angew. Chem. (Int. Ed. Engl.) 2020, 59, 15176–15180. [Google Scholar] [CrossRef] [PubMed]
- Pei, S.; Liu, R.; Gao, H.; Chen, H.; Wu, W.; Fang, X.; Han, Y. Inhibitory effect and possible mechanism of carvacrol against Colletotrichum fructicola. Postharvest Biol. Technol. 2020, 163, 111126. [Google Scholar] [CrossRef]
- Li, Y.; Li, B.; Wu, Y.; Zhao, Y.; Sun, L. Preparation of carboxymethyl chitosan/copper composites and their antibacterial properties. Mater. Res. Bull. 2013, 48, 3411–3419. [Google Scholar] [CrossRef]
- Wolska, K.; Grudniak, A.; Fiecek, B.; Kraczkiewicz-Dowjat, A.; Kurek, A. Antibacterial activity of oleanolic and ursolic acids and their derivatives. Open Life Sci. 2010, 5, 543–553. [Google Scholar] [CrossRef]
- Chen, Y.; Li, H.; Wu, L.; Zhang, M.; Gao, Y.; Wang, H.; Xu, D.; Chen, W.; Song, G.; Chen, J. Ursolic acid derivatives are potent inhibitors against porcine reproductive and respiratory syndrome virus. RSC Adv. 2020, 10, 22783–22796. [Google Scholar] [CrossRef]
- Huang, H.; Liang, J.; Tan, Q.; Ou, L.; Li, X.; Zhong, C.; Huang, H.; Møller, I.M.; Wu, X.; Song, S. Insights into triterpene synthesis and unsaturated fatty-acid accumulation provided by chromosomal-level genome analysis of Akebia trifoliata subsp. australis. Hortic. Res. 2021, 8, 33. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J.; Yang, Q.; Li, B.; Zhou, W.; Wang, Z. Genome survey sequencing and genetic diversity of cultivated Akebia trifoliata assessed via phenotypes and SSR markers. Mol. Biol. Rep. 2021, 48, 241–250. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).