3D-QSAR, ADME-Tox In Silico Prediction and Molecular Docking Studies for Modeling the Analgesic Activity against Neuropathic Pain of Novel NR2B-Selective NMDA Receptor Antagonists
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Database
2.2. Optimisation and Alignment
2.3. Development of 3D-QSAR Models
2.4. Partial Least Squares (PLS) Analysis
2.5. ADMET in Silico Pharmacocinetics
2.6. Molecular Docking Modeling
3. Results and Discussion
3.1. Molecular Alignement
3.2. 3D-QSAR Analysis
3.3. A Graphical Explanation of CoMSIA/EDA and CoMFA Models
3.4. ADMET in Silico Pharmacocinetics
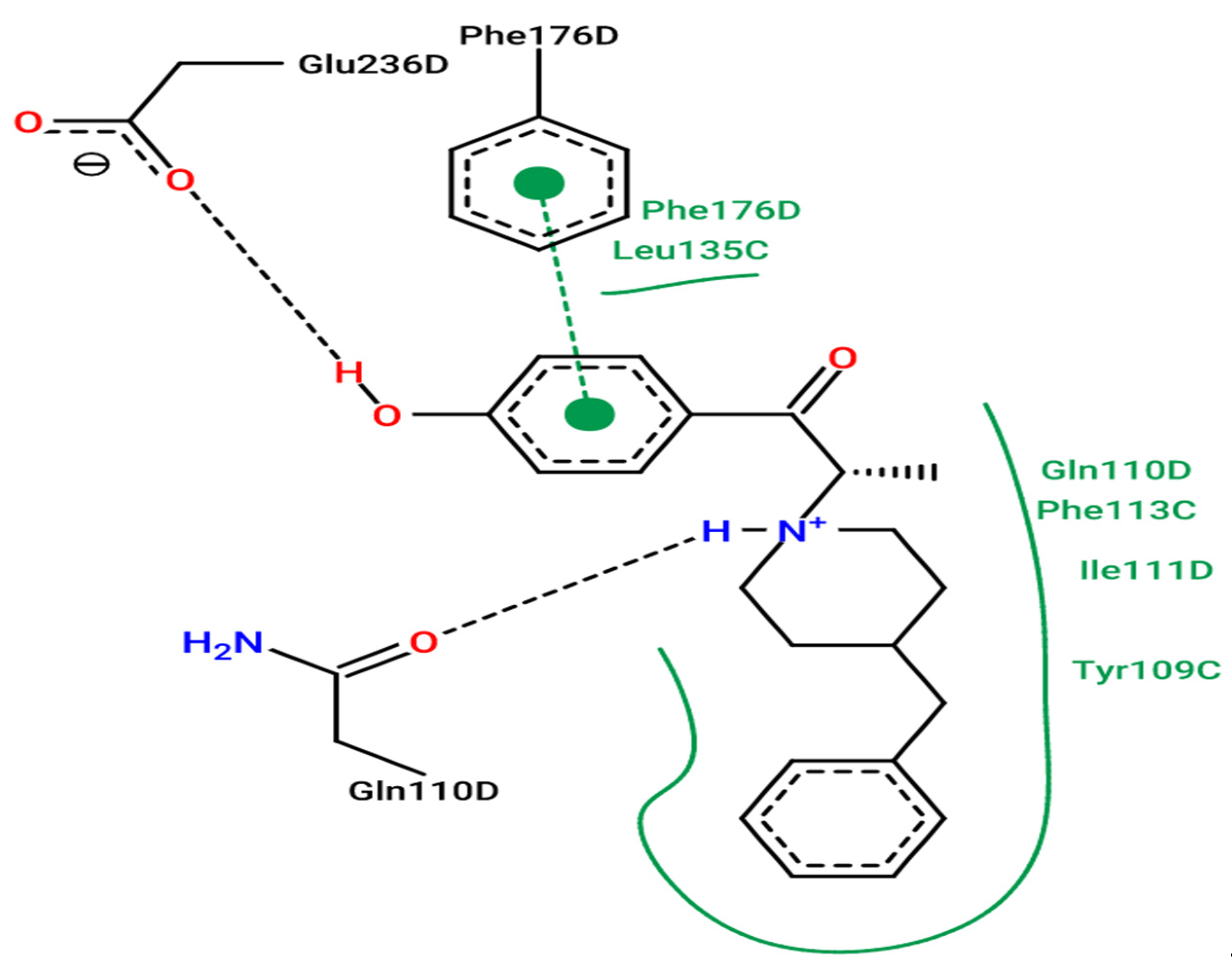
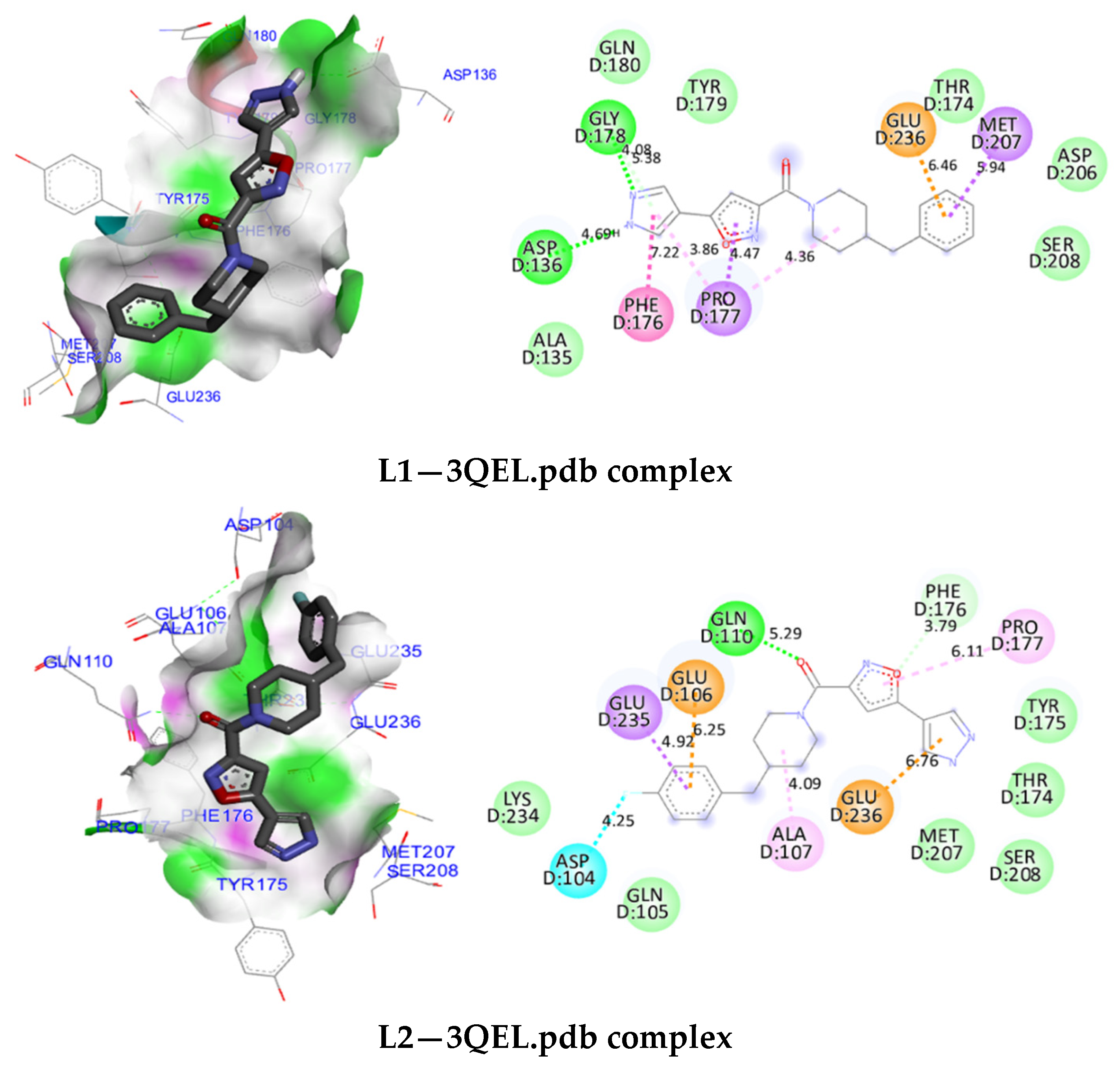
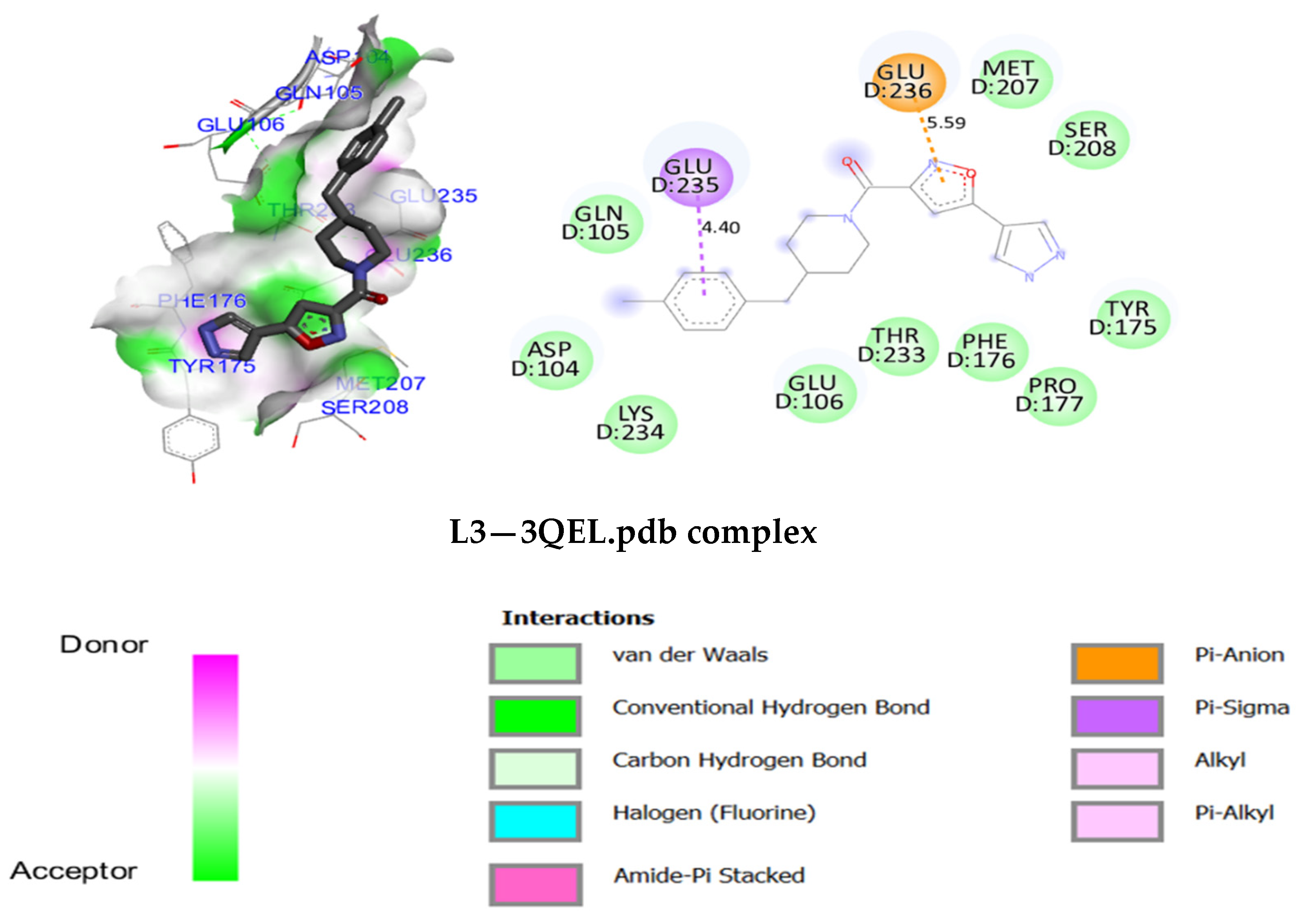
3.5. Molecular Docking
3.6. Docking Validation Protocol
3.7. Molecular Docking Modeling for L1, L2 and L3 Ligands
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bertin, C.; Delage, N.; Rolland, B.; Pennel, L.; Fatseas, M.; Trouvin, A.-P.; Delorme, J.; Chenaf, C.; Authier, N. Analgesic Opioid Use Disorders in Patients with Chronic Non-Cancer Pain: A Holistic Approach for Tailored Management. Neurosci. Biobehav. Rev. 2021, 121, 160–174. [Google Scholar] [CrossRef]
- Sheikholeslami, M.A.; Ghafghazi, S.; Parvardeh, S.; Koohsari, S.; Aghajani, S.H.; Pouriran, R.; Vaezi, L.A. Analgesic Effects of Cuminic Alcohol (4-Isopropylbenzyl Alcohol), a Monocyclic Terpenoid, in Animal Models of Nociceptive and Neuropathic Pain: Role of Opioid Receptors, L-Arginine/NO/CGMP Pathway, and Inflammatory Cytokines. Eur. J. Pharmacol. 2021, 900, 174075. [Google Scholar] [CrossRef]
- Noguchi, T.; Umezaki, Y.; Takano, R.; Fujimoto, T.; Domon, Y.; Kubota, K.; Ueda, K.; Kawase, Y.; Yabe, K.; Yokoyama, M.; et al. A Novel [5.2.1]Bicyclic Amine Is a Potent Analgesic without µ Opioid Activity. Bioorganic Med. Chem. Lett. 2021, 36, 127790. [Google Scholar] [CrossRef]
- Villarreal, Y.R.; Stotts, A.L.; Paniagua, S.M.; Rosen, K.; Eckmann, M.; Suchting, R.; Potter, J.S. Mindfulness Predicts Current Risk of Opioid Analgesic Misuse in Chronic Low Back Pain Patients Receiving Opioid Therapy. J. Contextual Behav. Sci. 2020, 18, 111–116. [Google Scholar] [CrossRef]
- Wtorek, K.; Piekielna-Ciesielska, J.; Janecki, T.; Janecka, A. The Search for Opioid Analgesics with Limited Tolerance Liability. Peptides 2020, 130, 170331. [Google Scholar] [CrossRef]
- Arita, T.; Asano, M.; Kubota, K.; Domon, Y.; Machinaga, N.; Shimada, K. Discovery of DS34942424: An Orally Potent Analgesic without Mu Opioid Receptor Agonist Activity. Bioorganic Med. Chem. 2020, 28, 115714. [Google Scholar] [CrossRef]
- Rossato, M.F.; Oliveira, S.M.; Trevisan, G.; Rotta, M.; Machado, P.; Martins, M.A.P.; Ferreira, J. Structural Improvement of Compounds with Analgesic Activity: AC-MPF4, a Compound with Mixed Anti-Inflammatory and Antinociceptive Activity via Opioid Receptor. Pharmacol. Biochem. Behav. 2015, 129, 72–78. [Google Scholar] [CrossRef]
- Anan, K.; Masui, M.; Tazawa, A.; Tomida, M.; Haga, Y.; Kume, M.; Yamamoto, S.; Shinohara, S.; Tsuji, H.; Shimada, S.; et al. Discovery of NR2B-Selective Antagonists via Scaffold Hopping and Pharmacokinetic Profile Optimization. Bioorganic Med. Chem. Lett. 2019, 29, 1143–1147. [Google Scholar] [CrossRef]
- Temnyakova, N.S.; Vasilenko, D.A.; Barygin, O.I.; Dron, M.Y.; Averina, E.B.; Grishin, Y.K.; Grigoriev, V.V.; Palyulin, V.A.; Fedorov, M.V.; Karlov, D.S. Ifenprodil-like NMDA Receptor Modulator Based on the Biphenyl Scaffold. Mendeleev Commun. 2020, 30, 342–343. [Google Scholar] [CrossRef]
- Wollmuth, L.P.; Chan, K.; Groc, L. The Diverse and Complex Modes of Action of Anti-NMDA Receptor Autoantibodies. Neuropharmacology 2021, 194, 108624. [Google Scholar] [CrossRef]
- Zambre, V.P.; Patil, R.B.; Sangshetti, J.N.; Sawant, S.D. Comprehensive QSAR Studies Reveal Structural Insights into the NR2B Subtype Selective Benzazepine Derivatives as N-Methyl-d-Aspartate Receptor Antagonists. J. Mol. Struct. 2019, 1197, 617–627. [Google Scholar] [CrossRef]
- Lill, M.A. Multi-Dimensional QSAR in Drug Discovery. Drug Discov. Today 2007, 12, 1013–1017. [Google Scholar] [CrossRef]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef]
- Halder, A.K.; Jha, T. Validated Predictive QSAR Modeling of N-Aryl-Oxazolidinone-5-Carboxamides for Anti-HIV Protease Activity. Bioorganic Med. Chem. Lett. 2010, 20, 6082–6087. [Google Scholar] [CrossRef]
- Roy, K.; Kar, S.; Das, R.N. Validation of QSAR Models. In Understanding the Basics of QSAR for Applications in Pharmaceutical Sciences and Risk Assessment; Elsevier: Amsterdam, The Netherlands, 2015; pp. 231–289. ISBN 978-0-12-801505-6. [Google Scholar]
- El Mchichi, L.; El Aissouq, A.; Kasmi, R.; Belhassan, A.; El-Mernissi, R.; Ouammou, A.; Lakhlifi, T.; Bouachrine, M. In Silico Design of Novel Pyrazole Derivatives Containing Thiourea Skeleton as Anti-Cancer Agents Using: 3D QSAR, Drug-Likeness Studies, ADMET Prediction and Molecular Docking. Mater. Today Proc. 2021, 45, 7661–7674. [Google Scholar] [CrossRef]
- Bank, R.P.D. RCSB PDB-QEL: Crystal Structure of Amino Terminal Domains of the NMDA Receptor Subunit GluN1 and GluN2B in Complex with Ifenprodil. Available online: https://www.rcsb.org/structure/3QEL (accessed on 11 May 2022).
- Mazigh, M.; El’mbarki, C.; Hadni, H.; Elhallaoui, M. QSAR Studies Combined with DFT-Calculations and Molecular Docking of Polyamine-Sensitive Inhibitors of the NMDA Receptor. Mediterr. J. Chem. 2019, 9, 164–174. [Google Scholar] [CrossRef][Green Version]
- Abdullahi, M.; Shallangwa, G.A.; Uzairu, A. In Silico QSAR and Molecular Docking Simulation of Some Novel Aryl Sulfonamide Derivatives as Inhibitors of H5N1 Influenza A Virus Subtype. Beni-Suef Univ. J. Basic Appl. Sci. 2020, 9, 2. [Google Scholar] [CrossRef]
- Purcell, W.P.; Singer, J.A. A Brief Review and Table of Semiempirical Parameters Used in the Hueckel Molecular Orbital Method. J. Chem. Eng. Data 1967, 12, 235–246. [Google Scholar] [CrossRef]
- Ghaleb, A.; Aouidate, A.; Bouachrine, M.; Lakhlifi, T.; Sbai, A. In Silico Exploration of Aryl Halides Analogues as CheckpointKinase 1 Inhibitors by Using 3D QSAR, Molecular Docking Study, and ADMET Screening. Adv. Pharm. Bull. 2019, 9, 84–92. [Google Scholar] [CrossRef]
- Clark, M.; Cramer, R.D.; Van Opdenbosch, N. Validation of the General Purpose Tripos 5.2 Force Field. J. Comput. Chem. 1989, 10, 982–1012. [Google Scholar] [CrossRef]
- Cramer, R.D.; Patterson, D.E.; Bunce, J.D. Comparative Molecular Field Analysis (CoMFA). 1. Effect of Shape on Binding of Steroids to Carrier Proteins. J. Am. Chem. Soc. 1988, 110, 5959–5967. [Google Scholar] [CrossRef] [PubMed]
- Klebe, G.; Abraham, U.; Mietzner, T. Molecular Similarity Indices in a Comparative Analysis (CoMSIA) of Drug Molecules to Correlate and Predict Their Biological Activity. J. Med. Chem. 1994, 37, 4130–4146. [Google Scholar] [CrossRef] [PubMed]
- Ståhle, L.; Wold, S. 6 Multivariate Data Analysis and Experimental Design in Biomedical Research. In Progress in Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 1988; Volume 25, pp. 291–338. ISBN 978-0-444-80965-0. [Google Scholar]
- Bush, B.L.; Nachbar, R.B. Sample-Distance Partial Least Squares: PLS Optimized for Many Variables, with Application to CoMFA. J. Comput. -Aided Mol. Des. 1993, 7, 587–619. [Google Scholar] [CrossRef] [PubMed]
- Rosipal, R.; Krämer, N. Overview and Recent Advances in Partial Least Squares. In Subspace, Latent Structure and Feature Selection; Lecture Notes in Computer Science; Saunders, C., Grobelnik, M., Gunn, S., Shawe-Taylor, J., Eds.; Springer: Berlin, Heidelberg, 2006; Volume 3940, pp. 34–51. ISBN 978-3-540-34137-6. [Google Scholar]
- Polat, E.; Gunay, S. A New Robust Partial Least Squares Regression Method Based on Multivariate MM-Estimators. Int. J. Math. Stat. 2017, 18, 82–99. [Google Scholar]
- Tropsha, A.; Gramatica, P.; Gombar, V. The Importance of Being Earnest: Validation Is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of Q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Baroni, M.; Clementi, S.; Cruciani, G.; Costantino, G.; Riganelli, D.; Oberrauch, E. Predictive Ability of Regression Models. Part II: Selection of the Best Predictive PLS Model. J. Chemom. 1992, 6, 347–356. [Google Scholar] [CrossRef]
- Daoui, O.; Elkhattabi, S.; Chtita, S.; Elkhalabi, R.; Zgou, H.; Benjelloun, A.T. QSAR, Molecular Docking and ADMET Properties in Silico Studies of Novel 4,5,6,7-Tetrahydrobenzo[D]-Thiazol-2-Yl Derivatives Derived from Dimedone as Potent Anti-Tumor Agents through Inhibition of C-Met Receptor Tyrosine Kinase. Heliyon 2021, 7, e07463. [Google Scholar] [CrossRef]
- Ebenezer, O.; Damoyi, N.; Jordaan, M.A.; Shapi, M. Unveiling of Pyrimidindinones as Potential Anti-Norovirus Agents—A Pharmacoinformatic-Based Approach. Molecules 2022, 27, 380. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, S. Molecular Docking, Drug Likeness, and ADMET Analyses of Passiflora Compounds as P-Glycoprotein (P-Gp) Inhibitor for the Treatment of Cancer. Curr. Pharmacol. Rep. 2020, 6, 429–440. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of Drug Absorption Using Multivariate Statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef] [PubMed]
- Egan, W.J.; Lauri, G. Prediction of Intestinal Permeability. Adv. Drug Deliv. Rev. 2002, 54, 273–289. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple Selection Criteria for Drug-like Chemical Matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Gomes, D.; Silvestre, S.; Duarte, A.P.; Venuti, A.; Soares, C.P.; Passarinha, L.; Sousa, Â. In Silico Approaches: A Way to Unveil Novel Therapeutic Drugs for Cervical Cancer Management. Pharmaceuticals 2021, 14, 741. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- PkCSM. Available online: http://biosig.unimelb.edu.au/pkcsm/run_example? (accessed on 8 March 2022).
- Serrano, A.; Imbernón, B.; Pérez-Sánchez, H.; Cecilia, J.M.; Bueno-Crespo, A.; Abellán, J.L. QN-Docking: An Innovative Molecular Docking Methodology Based on Q-Networks. Appl. Soft Comput. 2020, 96, 106678. [Google Scholar] [CrossRef]
- Berger, M.L.; Maciejewska, D.; Vanden Eynde, J.J.; Mottamal, M.; Żabiński, J.; Kaźmierczak, P.; Rezler, M.; Jarak, I.; Piantanida, I.; Karminski-Zamola, G.; et al. Pentamidine Analogs as Inhibitors of [3H]MK-801 and [3H]Ifenprodil Binding to Rat Brain NMDA Receptors. Bioorganic Med. Chem. 2015, 23, 4489–4500. [Google Scholar] [CrossRef]
- Kouranov, A.; Xie, L.; de la Cruz, J.; Chen, L.; Westbrook, J.; Bourne, P.E.; Berman, H.M. The RCSB PDB Information Portal for Structural Genomics. Nucleic Acids Research 2006, 34, D302–D305. [Google Scholar] [CrossRef]
- Signals Lead Discovery—Chemical Analytics—PerkinElmer Informatics; PerkinElmer: Waltham, MA, USA, 2022.
- Norgan, A.P.; Coffman, P.K.; Kocher, J.-P.A.; Katzmann, D.J.; Sosa, C.P. Multilevel Parallelization of AutoDock 4.2. J. Cheminform. 2011, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA Discovery Studio-BIOVIA-Dassault Systèmes®. Available online: https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (accessed on 17 November 2021).
- El Khatabi, K.; El-mernissi, R.; Aanouz, I.; Ajana, M.A.; Lakhlifi, T.; Shahinozzaman, M.; Bouachrine, M. Benzimidazole Derivatives in Identifying Novel Acetylcholinesterase Inhibitors: A Combination of 3D-QSAR, Docking and Molecular Dynamics Simulation. Phys. Chem. Res. 2022, 10, 237–249. [Google Scholar] [CrossRef]
- Hadni, H.; Elhallaoui, M. 3D-QSAR, Docking and ADMET Properties of Aurone Analogues as Antimalarial Agents. Heliyon 2020, 6, e03580. [Google Scholar] [CrossRef] [PubMed]
- El Fadili, M.; Er-Rajy, M.; Kara, M.; Assouguem, A.; Belhassan, A.; Alotaibi, A.; Mrabti, N.N.; Fidan, H.; Ullah, R.; Ercisli, S.; et al. QSAR, ADMET In Silico Pharmacokinetics, Molecular Docking and Molecular Dynamics Studies of Novel Bicyclo (Aryl Methyl) Benzamides as Potent GlyT1 Inhibitors for the Treatment of Schizophrenia. Pharmaceuticals 2022, 15, 670. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. ILOGP: A Simple, Robust, and Efficient Description of n -Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef]
- Williams, K. Ifenprodil, a Novel NMDA Receptor Antagonist: Site and Mechanism of Action. Curr. Drug Targets 2001, 2, 285–298. [Google Scholar] [CrossRef]
- Adamski, A.; Kruszka, D.; Dutkiewicz, Z.; Kubicki, M.; Gorczyński, A.; Patroniak, V. Novel Family of Fused Tricyclic [1,4]Diazepines: Design, Synthesis, Crystal Structures and Molecular Docking Studies. Tetrahedron 2017, 73, 3377–3386. [Google Scholar] [CrossRef]
- Zentrum Für Bioinformatik: Universität Hamburg-Proteins Plus Server. Available online: https://proteins.plus/ (accessed on 8 March 2022).
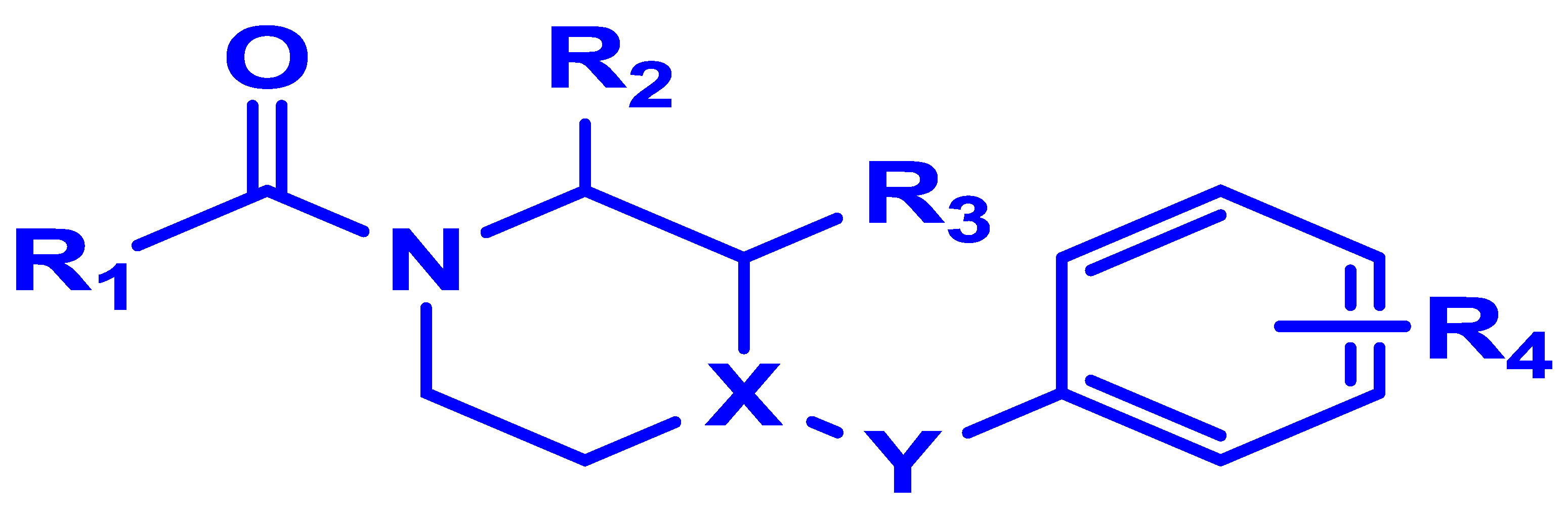




| Compounds | R1 | R2 | R3 | R4 | X | Y | pIC50 (bind) |
|---|---|---|---|---|---|---|---|
| 1 | 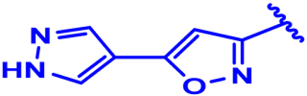 | H | H | H | CH | CH2 | 7.1 |
| 2 | 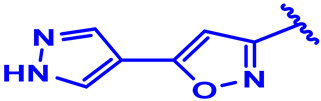 | H | H | 4-F | CH | CH2 | 7.1 |
| 3 | 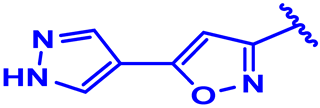 | H | H | 4-Cl | CH | CH2 | 7.2 |
| 4 | 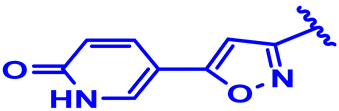 | H | H | H | CH | CH2 | 7.2 |
| 5 | 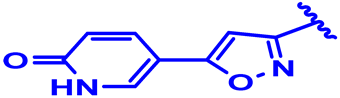 | H | H | 4-F | CH | CH2 | 7.3 |
| 6 | 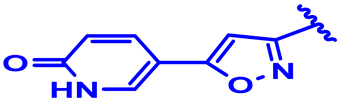 | H | H | 4-Cl | CH | CH2 | 7.6 |
| 7 |  | H | H | 4-F | CH | CH2 | 7.5 |
| 8 | 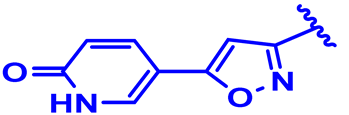 | H | H | 4-Cl | N | CH2 | 6 |
| 9 * | 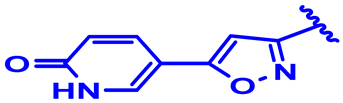 | H | H | 4-Cl | N | None | 6 |
| 10 | 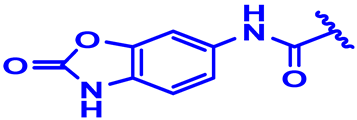 | H | H | 4-Cl | CH | CH2 | 7.5 |
| 11 | 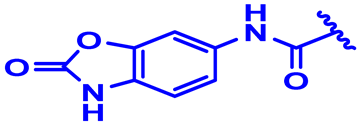 | H | H | 4-Cl | N | CH2 | 6.3 |
| 12 * |  | H | H | 4-Cl | N | None | 7.8 |
| 13 | 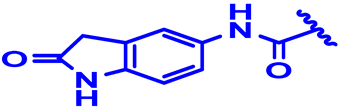 | H | H | 4-Cl | N | None | 6 |
| 14 | 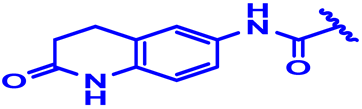 | H | H | 4-Cl | N | None | 6 |
| 15 |  | H | H | H | N | None | 6 |
| 16 * | 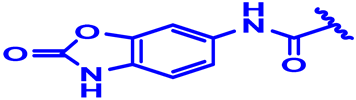 | H | H | 4-F | N | None | 6.9 |
| 17 * |  | H | H | 4-CF3 | N | None | 7.6 |
| 18 | 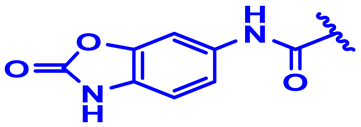 | H | H | 4-OCH3 | N | None | 6.6 |
| 19 | 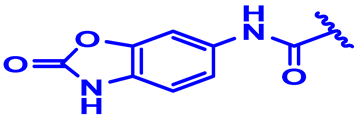 | H | H | 3.4-2F | N | None | 7.7 |
| 20 | 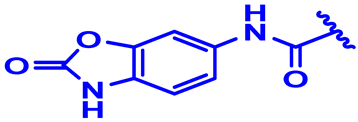 | H | H | 3.5-2F | N | None | 6.8 |
| 21 |  | H | H | 3-Cl.4-F | N | None | 7.5 |
| 22 | 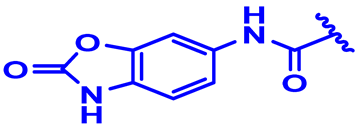 | H | H | 3-F.4-Cl | N | None | 7.9 |
| 23 |  | H | H | 3-F.4-CF3 | N | None | 7.8 |
| 24 * | 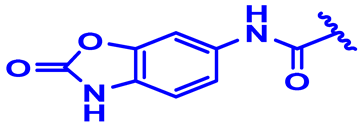 | H | H | 3-F.4-OCH3 | N | None | 7.8 |
| 25 |  | CH3 backward | H | 4-Cl | N | None | 7.5 |
| 26 | 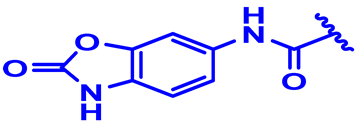 | CH3 forward | H | 4-Cl | N | None | 6 |
| 27 | 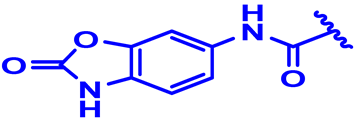 | H | CH3 backward | 4-Cl | N | None | 7.2 |
| 28 | 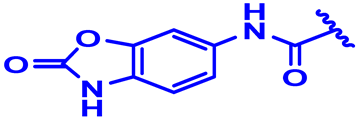 | H | CH3 forward | 4-Cl | N | None | 6.2 |
| 29 * |  | CH3 backward | H | 3.4-2F | N | None | 7.3 |
| 30 | 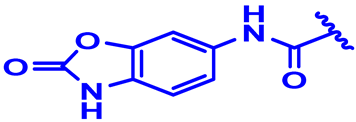 | H | CH3 backward | 3.4-2F | N | None | 7.2 |
| 31 | 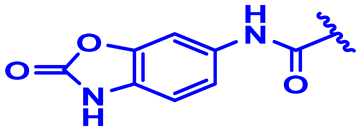 | CH3 backward | H | 3-F.4-Cl | N | None | 7.9 |
| 32 |  | H | CH3 backward | 3-F.4-Cl | N | None | 7.3 |
| Q2 | R2 | SEE | F | ONC | R2pr | Fractions | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Steric | Electrostatic | Hydrophobic | Donor | Acceptor | |||||||
| CoMFA | 0.540 | 0.980 | 0.110 | 124.198 | 7 | 0.613 | 0.857 | 0.143 | - | - | - |
| CoMSIA/SEA | 0.667 | 0.903 | 0.228 | 37.421 | 5 | 0.420 | 0.304 | 0.524 | - | - | 0.172 |
| CoMSIA/SEH | 0.592 | 0.821 | 0.296 | 33.700 | 3 | 0.110 | 0.141 | 0.221 | 0.638 | - | - |
| CoMSIA/SED | 0.704 | 0.908 | 0.218 | 51.754 | 4 | 0.465 | 0.399 | 0.601 | - | 0 | - |
| CoMSIA/SHD | 0.537 | 0.755 | 0.347 | 22.618 | 3 | 0.131 | 0.204 | - | 0.796 | 0 | - |
| CoMSIA/SHA | 0.568 | 0.763 | 0.341 | 23.641 | 3 | 0.015 | 0.161 | - | 0.678 | - | 0.161 |
| CoMSIA/SDA | 0.320 | 0.564 | 0.462 | 9.505 | 3 | 0.522 | 0.5 | - | - | 0 | 0.5 |
| CoMSIA/EHA | 0.617 | 0.817 | 0.300 | 32.718 | 3 | 0.018 | - | 0.209 | 0.634 | - | 0.157 |
| CoMSIA/EHD | 0.579 | 0.756 | 0.338 | 35.624 | 2 | 0.010 | - | 0.211 | 0.789 | 0 | - |
| CoMSIA/EDA | 0.665 | 0.916 | 0.238 | 19.365 | 9 | 0.701 | - | 0.627 | - | 0 | 0.373 |
| CoMSIA/HDA | 0.546 | 0.762 | 0.342 | 23.449 | 3 | 0.025 | - | - | 0.801 | 0 | 0.199 |
| CoMSIA/EHDA | 0.617 | 0.817 | 0.300 | 32.718 | 3 | 0.018 | - | 0.209 | 0.634 | 0 | 0.157 |
| CoMSIA/SHDA | 0.568 | 0.763 | 0.341 | 23.641 | 3 | 0.015 | 0.161 | - | 0.678 | 0 | 0.161 |
| CoMSIA/SEDA | 0.667 | 0.903 | 0.228 | 37.421 | 5 | 0.420 | 0.304 | 0.524 | - | 0 | 0.172 |
| CoMSIA/SEHA | 0.620 | 0.808 | 0.307 | 30.844 | 3 | 0.007 | 0.122 | 0.172 | 0.568 | - | 0.138 |
| CoMSIA/SEHD | 0.592 | 0.821 | 0.296 | 33.700 | 3 | 0.110 | 0.141 | 0.221 | 0.638 | 0 | - |
| CoMSIA/SEHDA | 0.620 | 0.808 | 0.307 | 30.844 | 3 | 0.007 | 0.122 | 0.172 | 0.568 | 0 | 0.138 |
| Compounds Number | Observed pIC50 | Predicted pIC50 of CoMFA | Predicted pIC50 of CoMSIA/EDA |
|---|---|---|---|
| 9 * | 6 | 8.2 | 6.7 |
| 12 * | 7.8 | 9.6 | 7.3 |
| 16 * | 6.9 | 9.7 | 7.4 |
| 17 * | 7.6 | 10.0 | 7.4 |
| 24 * | 7.8 | 10.2 | 7.4 |
| 29 * | 7.3 | 10.6 | 7.5 |
| Physical and Chemical Properties | Lipinski Violations | Veber Violations | Egan Violations | Ghose Violations | Muegge Violations | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligands number | Molecular weight (g/mol) | Molar refractive index | Rotatable bonds Number | Log P (Octanol/Water) | Hydrogen bond acceptors | H-bond donors Number | |||||
| Threshold | ≤500 | 40 ≤ MR ≤ 130 | <10 | <5 | ≤10 | <5 | Yes/No | Yes/No | Yes/No | Yes/No | Yes/No |
| L1 | 336.39 | 97.50 | 5 | 2.26 | 4 | 1 | Yes | Yes | Yes | Yes | Yes |
| L2 | 354.38 | 97.46 | 5 | 2.41 | 5 | 1 | Yes | Yes | Yes | Yes | Yes |
| L3 | 370.83 | 102.51 | 5 | 2.53 | 4 | 1 | Yes | Yes | Yes | Yes | Yes |
| Ifenprodil | 323.43 | 101.49 | 5 | 3.47 | 3 | 1 | Yes | Yes | Yes | Yes | Yes |
| Absorption | Distribution | Metabolism | Excretion | Toxicity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligands number | Intestinal Absorption (human) | VDss (human) | BBB permeability | CNS permeability | Substrate | Inhibitor | Total Clearance | AMES toxicity | |||||
| CYP | |||||||||||||
| 2D6 | 3A4 | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | |||||||
| Numeric (% Absorbed) | Numeric (Log L/kg) | Numeric (Log BB) | Numeric (Log PS) | Categorical (Yes/No) | Numeric (Log mL/min/kg) | Categorical (Yes/No) | |||||||
| L1 | 94.525 | 0.191 | −0.965 | −2.361 | Yes | Yes | Yes | Yes | Yes | No | Yes | 0.264 | Not toxic |
| L2 | 89.981 | 0.346 | −0.176 | −2.53 | No | Yes | Yes | Yes | Yes | No | No | 0.197 | Not toxic |
| L3 | 89.079 | 0.475 | −1.138 | −2.376 | No | Yes | Yes | Yes | Yes | No | No | 0.156 | Not toxic |
| Ifenprodil | 92.417 | 1.23 | 0.046 | −1.079 | Yes | Yes | Yes | No | No | Yes | No | 0.993 | Not toxic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El fadili, M.; Er-rajy, M.; Imtara, H.; Kara, M.; Zarougui, S.; Altwaijry, N.; Al kamaly, O.; Al Sfouk, A.; Elhallaoui, M. 3D-QSAR, ADME-Tox In Silico Prediction and Molecular Docking Studies for Modeling the Analgesic Activity against Neuropathic Pain of Novel NR2B-Selective NMDA Receptor Antagonists. Processes 2022, 10, 1462. https://doi.org/10.3390/pr10081462
El fadili M, Er-rajy M, Imtara H, Kara M, Zarougui S, Altwaijry N, Al kamaly O, Al Sfouk A, Elhallaoui M. 3D-QSAR, ADME-Tox In Silico Prediction and Molecular Docking Studies for Modeling the Analgesic Activity against Neuropathic Pain of Novel NR2B-Selective NMDA Receptor Antagonists. Processes. 2022; 10(8):1462. https://doi.org/10.3390/pr10081462
Chicago/Turabian StyleEl fadili, Mohamed, Mohammed Er-rajy, Hamada Imtara, Mohammed Kara, Sara Zarougui, Najla Altwaijry, Omkulthom Al kamaly, Aisha Al Sfouk, and Menana Elhallaoui. 2022. "3D-QSAR, ADME-Tox In Silico Prediction and Molecular Docking Studies for Modeling the Analgesic Activity against Neuropathic Pain of Novel NR2B-Selective NMDA Receptor Antagonists" Processes 10, no. 8: 1462. https://doi.org/10.3390/pr10081462
APA StyleEl fadili, M., Er-rajy, M., Imtara, H., Kara, M., Zarougui, S., Altwaijry, N., Al kamaly, O., Al Sfouk, A., & Elhallaoui, M. (2022). 3D-QSAR, ADME-Tox In Silico Prediction and Molecular Docking Studies for Modeling the Analgesic Activity against Neuropathic Pain of Novel NR2B-Selective NMDA Receptor Antagonists. Processes, 10(8), 1462. https://doi.org/10.3390/pr10081462









