Optimization of Production of Polyhydroxyalkanoates (PHAs) from Newly Isolated Ensifer sp. Strain HD34 by Response Surface Methodology
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Bacteria Isolation
2.2. Screening of Polyhydroxyalkanoate (PHA)-Producing Bacteria
2.2.1. Observation of Fluorescence Intensity on Agar
2.2.2. Determination of PHA Granules in Bacterial Cell
2.3. PHA Extraction and Determination
2.4. Identification of Selected Bacteria
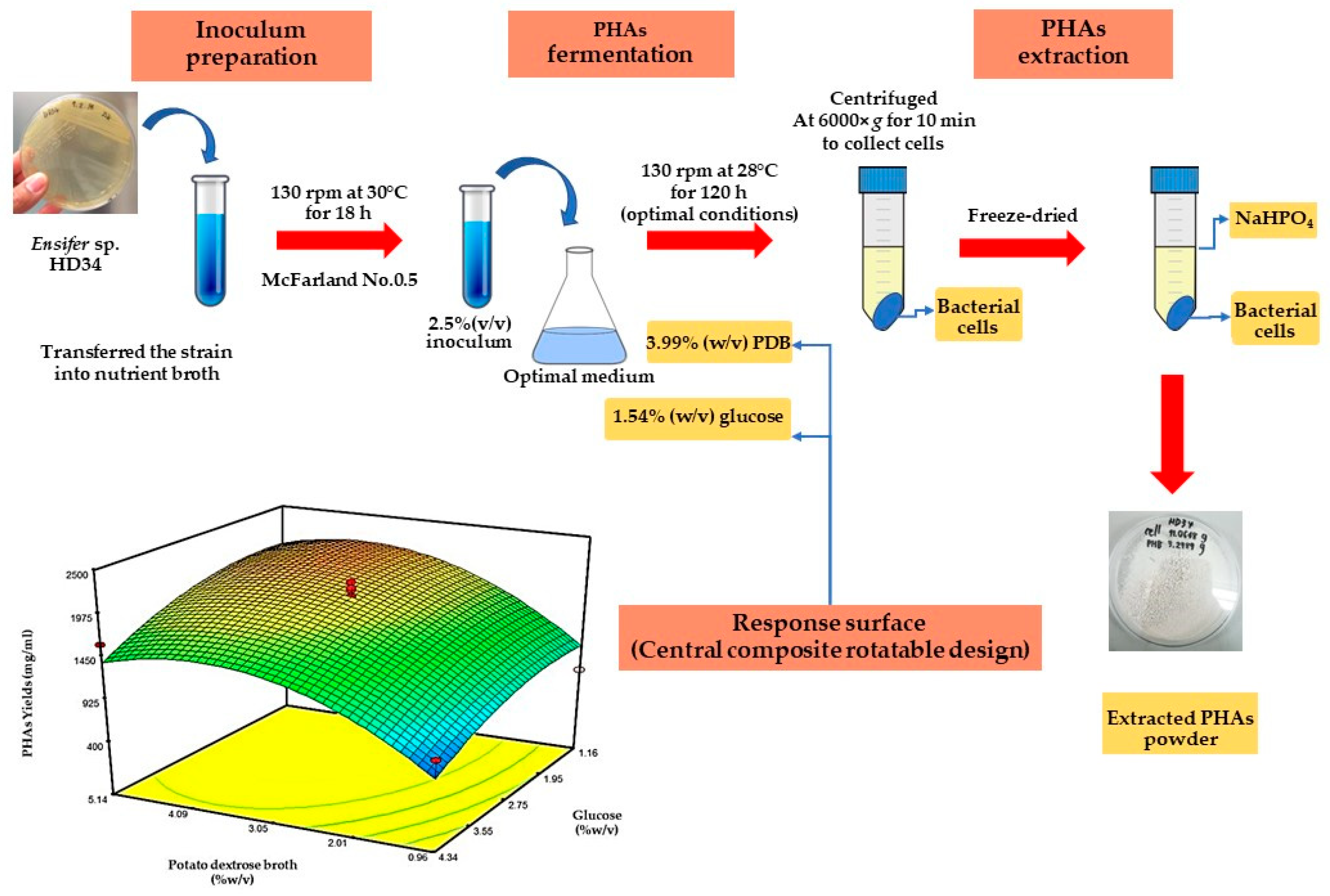
2.5. Optimization of PHA Production
2.5.1. Effect of Carbon Source
2.5.2. Effect of Production Medium
2.5.3. Central Composite Rotatable Design (CCRD)
2.5.4. One Factor at a Time Experiment for the Optimal Conditions of PHA Production
- Effect of incubation time.
- Effect of initial pH.
- Effect of inoculum size.
- Effect of temperature.
2.5.5. Time-Course Culture under Optimal Medium and Conditions for PHA Production
2.6. Characterization of PHAs
2.6.1. 1H Nuclear Magnetic Resonance (NMR)
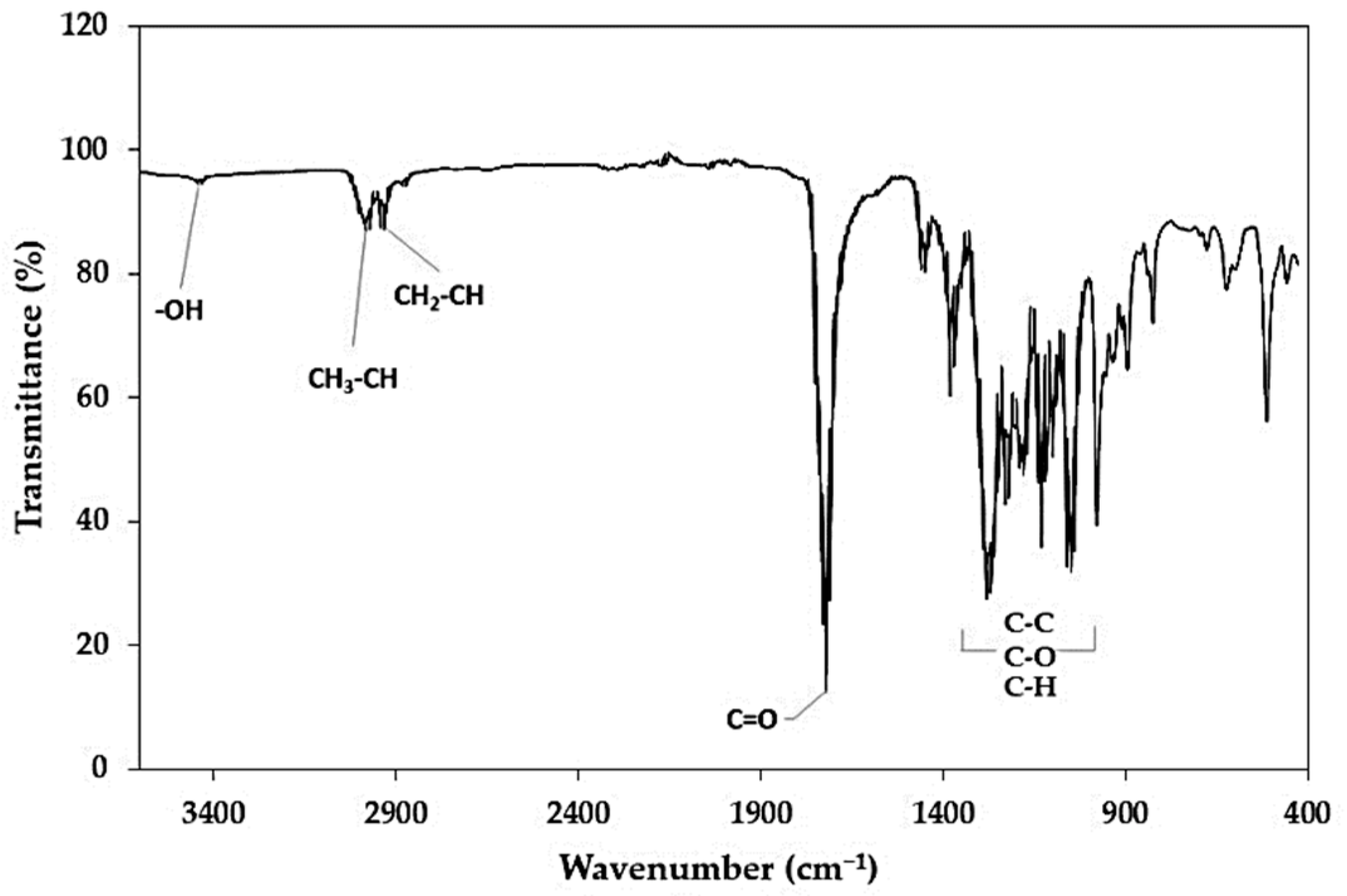
2.6.2. Fourier-Transform Infrared Spectroscopy (FTIR)
2.6.3. X-ray Diffraction (XRD)
2.6.4. Thermogravimetric Analysis (TGA)
2.6.5. Differential Scanning Calorimetry (DSC)
2.7. Statistical Analysis
3. Results
3.1. Sample Collection and Isolation of Bacteria
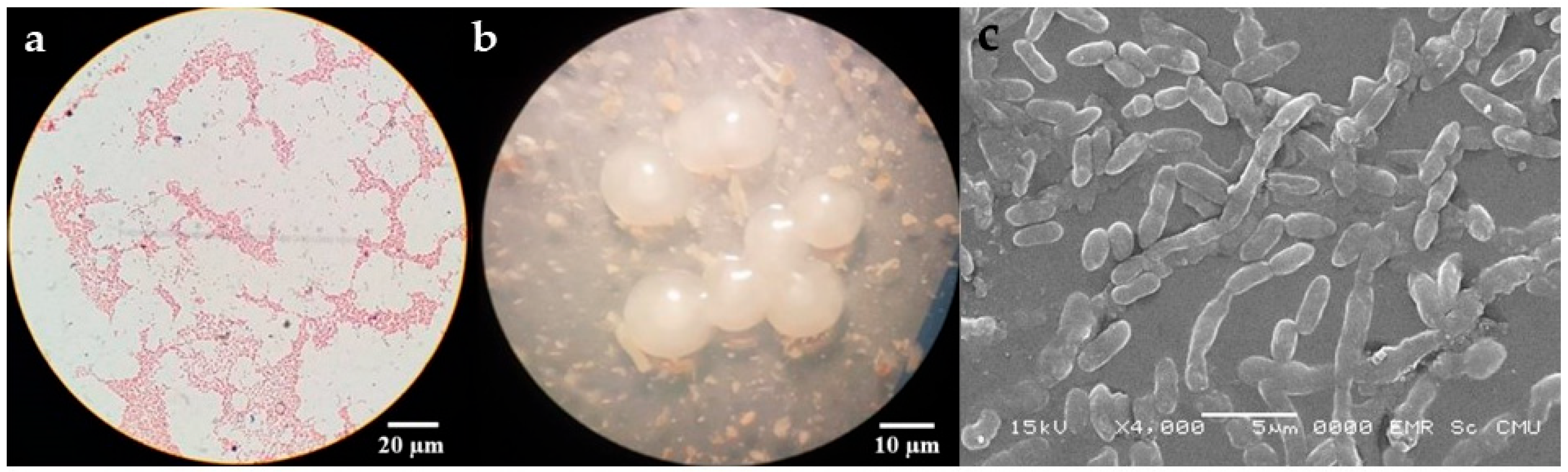
3.2. Screening of Polyhydroxyalkanoate (PHA)-Producing Bacteria
3.3. Identification of Selected Bacteria
3.4. Optimization of PHA Production
3.5. One Factor at a Time Experiment for the Optimal Conditions of PHA Production
3.5.1. Effect of Incubation Time
3.5.2. Effect of Initial pH
3.5.3. Effect of Inoculum Size
3.5.4. Effect of Temperature
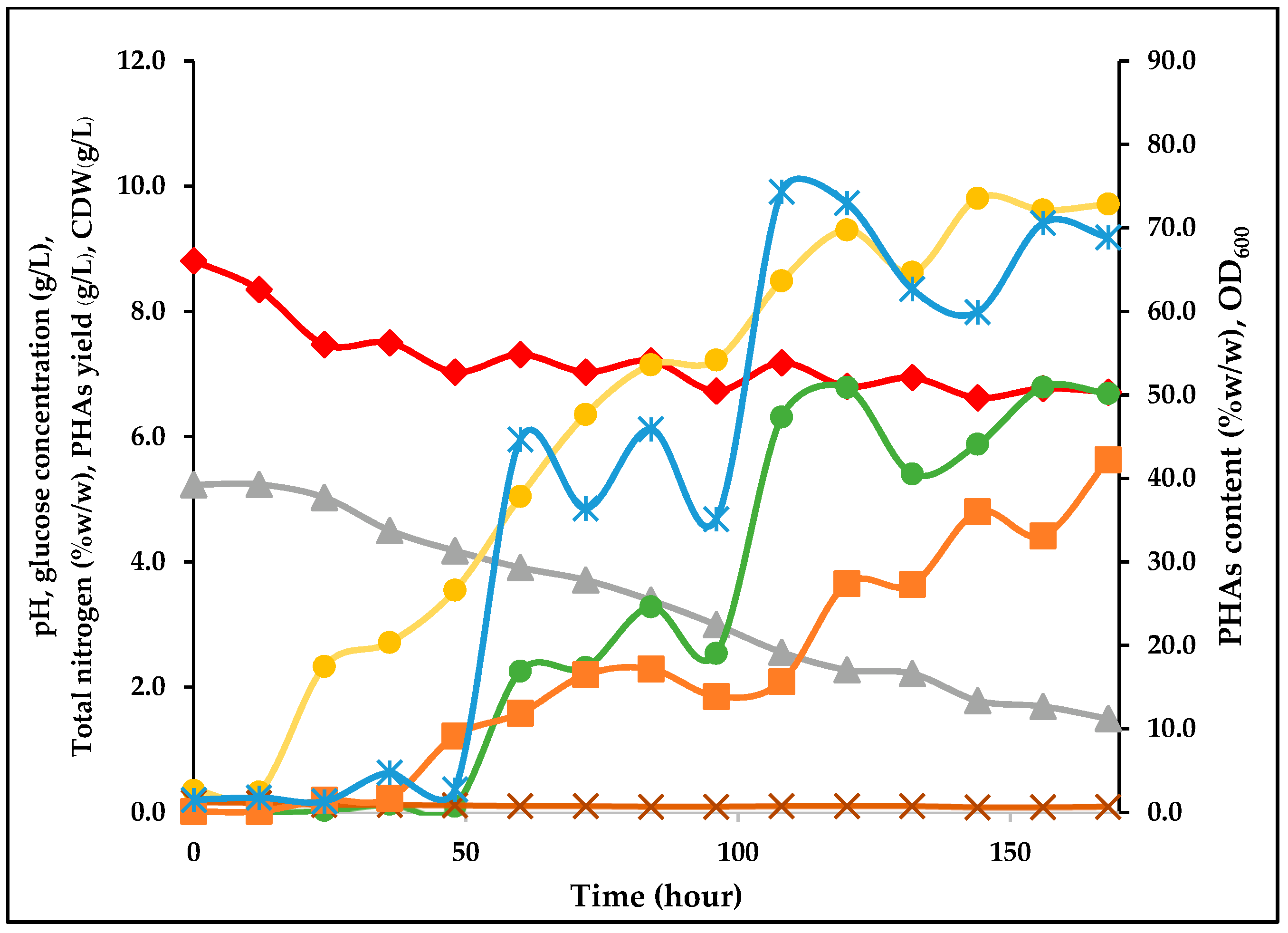
3.6. Time-Course Culture under Optimal Medium and Conditions for PHA Production
3.7. Characterization of PHAs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Plastics—The Facts 2021. Available online: https://plasticseurope.org/knowledge-hub/plastics-the-facts-2021/ (accessed on 5 April 2022).
- Braunegg, G.; Lefebvre, G.; Genser, K.F. Polyhydroxyalkanoates, biopolyesters from renewable resources: Physiological and engineering aspects. J. Biotechnol. 1998, 65, 127–161. [Google Scholar] [CrossRef]
- Lemoigne, M. Products of dehydration and of polymerization of β-hydroxybutyric acid. Bull. Soc. Chem. Biol. 1926, 8, 770–782. [Google Scholar]
- Suwannasing, W.; Imai, T.; Kaewkannetra, P. Cost-effective defined medium for the production of polyhydroxyalkanoates using agricultural raw materials. Bioresour. Technol. 2015, 194, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Trainer, M.A.; Charles, T.C. The role of PHB metabolism in the symbiosis of rhizobia with legumes. Appl. Microbiol. Biotechnol. 2006, 71, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Belal, E.B. Production of poly-β-hydroxybutyric acid (PHB) by Rhizobium elti and Pseudomonas stutzeri. Curr. Res. J. Biol. Sci. 2013, 5, 273–284. [Google Scholar] [CrossRef]
- Heng, K.S.; Hatti-Kaul, R.; Adam, F.; Fukui, T.; Sudesh, K. Conversion of rice husks to polyhydroxyalkanoates (PHA) via a three-step process: Optimized alkaline pretreatment, enzymatic hydrolysis, and biosynthesis by Burkholderia cepacia USM (JCM 15050). J. Chem. Technol. Biotechnol. 2017, 92, 100–108. [Google Scholar] [CrossRef]
- Steinbüchel, A. Polyhydroxyalkanoic acids. In Biomaterials; Springer: Berlin/Heidelberg, Germany, 1991; pp. 123–213. [Google Scholar]
- Bugnicourt, E.; Cinelli, P.; Lazzeri, A.; Alvarez, V.A. Polyhydroxyalkanoate (PHA): Review of synthesis, characteristics, processing and potential applications in packaging. Express Polym. Lett. 2014, 8, 791–808. [Google Scholar] [CrossRef]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A. Recent advances and challenges towards sustainable polyhydroxyalkanoate (PHA) production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef]
- Mukheem, A.; Shahabuddin, S.; Khan, A.A.; Hossain, M.M.; Jasni, A.H.; Sridewi, N. Bio-plastic Polyhydroxyalkanoate (PHA): Applications in Modern Medicine. In Bioplastics for Sustainable Development; Springer: Berlin/Heidelberg, Germany, 2021; pp. 231–257. [Google Scholar]
- Mukheem, A.; Shahabuddin, S.; Akbar, N.; Miskon, A.; Muhamad Sarih, N.; Sudesh, K.; Ahmed Khan, N.; Saidur, R.; Sridewi, N. Boron nitride doped polyhydroxyalkanoate/chitosan nanocomposite for antibacterial and biological applications. Nanomaterials 2019, 9, 645. [Google Scholar] [CrossRef]
- Yetilmezsoy, K.; Demirel, S.; Vanderbei, R.J. Response surface modeling of Pb (II) removal from aqueous solution by Pistacia vera L.: Box–Behnken experimental design. J. Hazard. Mater. 2009, 171, 551–562. [Google Scholar] [CrossRef]
- Naik, P.; Ranade, V. Mathematical Modelling as a Tool to Optimize PHA Production by Massilia spp. J. Adv. Biol. Biotechnol. 2022, 23, 19–29. [Google Scholar] [CrossRef]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Repaske, R.; Repaske, A. Quantitative requirements for exponential growth of Alcaligenes eutrophus. Appl. Environ. Microbiol. 1976, 32, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Spiekermann, P.; Rehm, B.H.; Kalscheuer, R.; Baumeister, D.; Steinbüchel, A. A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch. Microbiol. 1999, 171, 73–80. [Google Scholar] [CrossRef]
- Lageveen, R.G.; Huisman, G.W.; Preusting, H.; Ketelaar, P.; Eggink, G.; Witholt, B. Formation of polyesters by Pseudomonas oleovorans: Effect of substrates on formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 1988, 54, 2924–2932. [Google Scholar] [CrossRef]
- Penkhrue, W.; Khanongnuch, C.; Masaki, K.; Pathom-Aree, W.; Punyodom, W.; Lumyong, S. Isolation and screening of biopolymer-degrading microorganisms from northern Thailand. World J. Microbiol. Biotechnol. 2015, 31, 1431–1442. [Google Scholar] [CrossRef]
- Penkhrue, W.; Jendrossek, D.; Khanongnuch, C.; Pathom-Aree, W.; Aizawa, T.; Behrens, R.L.; Lumyong, S. Response surface method for polyhydroxybutyrate (PHB) bioplastic accumulation in Bacillus drentensis BP17 using pineapple peel. PLoS ONE 2020, 15, e0230443. [Google Scholar] [CrossRef]
- Valappil, S.P.; Misra, S.K.; Boccaccini, A.R.; Keshavarz, T.; Bucke, C.; Roy, I. Large-scale production and efficient recovery of PHB with desirable material properties, from the newly characterised Bacillus cereus SPV. J. Biotechnol. 2007, 132, 251–258. [Google Scholar] [CrossRef]
- Dhangdhariya, J.H.; Dubey, S.; Trivedi, H.B.; Pancha, I.; Bhatt, J.K.; Dave, B.P.; Mishra, S. Polyhydroxyalkanoate from marine Bacillus megaterium using CSMCRI’s Dry Sea Mix as a novel growth medium. Int. J. Biol. Macromol. 2015, 76, 254–261. [Google Scholar] [CrossRef]
- Hassan, M.; Bakhiet, E.; Hussein, H.; Ali, S. Statistical optimization studies for polyhydroxybutyrate (PHB) production by novel Bacillus subtilis using agricultural and industrial wastes. Int. J. Environ. Sci. Technol. 2019, 16, 3497–3512. [Google Scholar] [CrossRef]
- Naheed, N.; Jamil, N. Optimization of biodegradable plastic production on sugar cane molasses in Enterobacter sp. SEL2. Braz. J. Microbiol. 2014, 45, 417–426. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Singh, G.; Mittal, A.; Kumari, A.; Goel, V.; Aggarwal, N.K.; Yadav, A. Optimization of poly-B-hydroxybutyrate production from Bacillus species. Eur. J. Biol. Sci. 2011, 3, 112–116. [Google Scholar]
- Brenner, D.J.; Krieg, N.R.; Staley, J.T.; Garrity, G. Bergey’s Manual® of Systematic Bacteriology: Volume Two the Proteobacteria Part C: The Alpha-, Beta-, Delta-, and Epsilonproteobacteria; Springer: Berlin, Germany, 2005. [Google Scholar]
- Wang, Y.C.; Wang, F.; Hou, B.C.; Wang, E.T.; Chen, W.F.; Sui, X.H.; Chen, W.X.; Li, Y.; Zhang, Y.B. Proposal of Ensifer psoraleae sp. nov., Ensifer sesbaniae sp. nov., Ensifer morelense comb. nov. and Ensifer americanum comb. nov. Syst. Appl. Microbiol. 2013, 36, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Khamkong, T.; Penkhrue, W.; Lumyong, S. Effect of acid pretreatment agricultural wastes on Polyhydroxyalkanoates (PHAs) production by Ensifer sp. HD34. In Proceedings of the International Conference of Agriculture and Natural Resources, Bangkok, Thailand, 26–28 April 2018. [Google Scholar]
- Ratledge, C.; Kristiansen, B. Basic Biotechnology; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar]
- Jung, Y.-M.; Lee, Y.-H. Utilization of oxidative pressure for enhanced production of poly-β-hydroxybutyrate and poly (3-hydroxybutyrate-3-hydroxyvalerate) in Ralstonia eutropha. J. Biosci. Bioeng. 2000, 90, 266–270. [Google Scholar] [CrossRef]
- Haas, R.; Jin, B.; Zepf, F.T. Production of poly (3-hydroxybutyrate) from waste potato starch. Biosci. Biotechnol. Biochem. 2008, 72, 253–256. [Google Scholar] [CrossRef]
- Gomez, J.; Rodrigues, M.; Alli, R.; Torres, B.; Netto, C.; Oliveira, M.; Da Silva, L. Evaluation of soil gram-negative bacteria yielding polyhydroxyalkanoic acids from carbohydrates and propionic acid. Appl. Microbiol. Biotechnol. 1996, 45, 785–791. [Google Scholar] [CrossRef]
- Lasemi, Z.; Darzi, G.N.; Baei, M.S. Media optimization for poly (β-hydroxybutyrate) production using Azotobacter beijerinckii. Int. J. Polym. Mater. Polym. Biomater. 2013, 62, 265–269. [Google Scholar] [CrossRef]
- Wendlandt, K.-D.; Jechorek, M.; Helm, J.; Stottmeister, U. Production of PHB with a high molecular mass from methane. Polym. Degrad. Stab. 1998, 59, 191–194. [Google Scholar] [CrossRef]
- Naitam, M.G.; Tomar, G.S.; Pushpad, U.; Singh, S.; Kaushik, R. Halophilic bacteria mediated poly-β-hydroxybutyrate production using paddy straw as a substrate. Bioresour. Technol. Rep. 2022, 17, 100915. [Google Scholar] [CrossRef]
- Jin, Y.-X.; Shi, L.-H.; Kawata, Y. Metabolomics-based component profiling of Halomonas sp. KM-1 during different growth phases in poly (3-hydroxybutyrate) production. Bioresour. Technol. 2013, 140, 73–79. [Google Scholar] [CrossRef]
- Lathwal, P.; Nehra, K.; Singh, M.; Jamdagni, P.; Rana, J.S. Optimization of culture parameters for maximum polyhydroxybutyrate production by selected bacterial strains isolated from rhizospheric soils. Pol. J. Microbiol. 2015, 64, 7. [Google Scholar] [CrossRef]
- Zihayat, B.; Shakibaie, M.; Sabouri-Shahrbabak, S.; Doostmohammadi, M.; Ameri, A.; Adeli-Sardou, M.; Forootanfar, H. Medium optimization for polyhydroxyalkanoate production by Pseudomonas pseudoalcaligenes strain Te using D-optimal design. Biocatal. Agric. Biotechnol. 2019, 18, 101001. [Google Scholar] [CrossRef]
- Thapa, C.; Shakya, P.; Shrestha, R.; Pal, S.; Manandhar, P. Isolation of polyhydroxybutyrate (PHB) producing bacteria, optimization of culture conditions for PHB production, extraction and characterization of PHB. Nepal J. Biotechnol. 2018, 6, 62–68. [Google Scholar] [CrossRef]
- Bora, L. Polyhydroxybutyrate accumulation in Bacillus megaterium and optimization of process parameters using response surface methodology. J. Polym. Environ. 2013, 21, 415–420. [Google Scholar] [CrossRef]
- Nishida, M.; Tanaka, T.; Hayakawa, Y.; Nishida, M. Solid-state nuclear magnetic resonance (NMR) and nuclear magnetic relaxation time analyses of molecular mobility and compatibility of plasticized polyhydroxyalkanoates (PHA) copolymers. Polymers 2018, 10, 506. [Google Scholar] [CrossRef]
- Saratale, R.G.; Cho, S.K.; Saratale, G.D.; Ghodake, G.S.; Bharagava, R.N.; Kim, D.S.; Nair, S.; Shin, H.S. Efficient bioconversion of sugarcane bagasse into polyhydroxybutyrate (PHB) by Lysinibacillus sp. and its characterization. Bioresour. Technol. 2021, 324, 124673. [Google Scholar] [CrossRef]
- Mascarenhas, J.; Aruna, K. Production and Optimization of Polyhydroxyalkaonoate Obtained from Bacillus megaterium JHA. J. Appl. Biotechnol. Rep. 2021, 8, 346–360. [Google Scholar]
- Radwan, W.H.; Abu Hussein, S.H.; Nassef, M.A.-M.; Selim, S.M. Optimization of Poly-β-Hydroxybutyrate (PHB) Production by an Egyption strain of Rhizobium fabae F44 using Response Surface Methodology. Arab Univ. J. Agric. Sci. 2019, 27, 1383–1397. [Google Scholar] [CrossRef]
- Panda, B.; Jain, P.; Sharma, L.; Mallick, N. Optimization of cultural and nutritional conditions for accumulation of poly-β-hydroxybutyrate in Synechocystis sp. PCC 6803. Bioresour. Technol. 2006, 97, 1296–1301. [Google Scholar] [CrossRef]
- Liangqi, Z.; Jingfan, X.; Tao, F.; Haibin, W. Synthesis of poly (3-hydroxybutyrate-co-3-hydroxyoctanoate) by a Sinorhizobium fredii strain. Lett. Appl. Microbiol. 2006, 42, 344–349. [Google Scholar] [CrossRef]
- Hashemi, B.M.; Nakhaei, M.M. Biodegradable Plastics from Sinorhizobium meliloti as Plastic Compatible with the Environment and Human Health. Biol. J. Microorg. 2016, 4, 9. [Google Scholar]
- Balakrishna Pillai, A.; Jaya Kumar, A.; Kumarapillai, H. Enhanced production of poly (3-hydroxybutyrate) in recombinant Escherichia coli and EDTA–microwave-assisted cell lysis for polymer recovery. AMB Express 2018, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Bhuwal, A.K.; Singh, G.; Aggarwal, N.K.; Goyal, V.; Yadav, A. Poly-β-hydroxybutyrate production and management of cardboard industry effluent by new Bacillus sp. NA10. Bioresour. Bioprocess. 2014, 1, 9. [Google Scholar] [CrossRef]
- Shamala, T.; Divyashree, M.; Davis, R.; Kumari, K.; Vijayendra, S.; Raj, B. Production and characterization of bacterial polyhydroxyalkanoate copolymers and evaluation of their blends by fourier transform infrared spectroscopy and scanning electron microscopy. Indian J. Microbiol. 2009, 49, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Fatma, T. Cyanobacterial polyhydroxybutyrate (PHB): Screening, optimization and characterization. PLoS ONE 2016, 11, e0158168. [Google Scholar] [CrossRef]
- López, J.A.; Naranjo, J.M.; Higuita, J.C.; Cubitto, M.A.; Cardona, C.A.; Villar, M.A. Biosynthesis of PHB from a new isolated Bacillus megaterium strain: Outlook on future developments with endospore forming bacteria. Biotechnol. Bioprocess Eng. 2012, 17, 250–258. [Google Scholar] [CrossRef]
- López-Cuellar, M.; Alba-Flores, J.; Rodríguez, J.G.; Pérez-Guevara, F. Production of polyhydroxyalkanoates (PHAs) with canola oil as carbon source. Int. J. Biol. Macromol. 2011, 48, 74–80. [Google Scholar] [CrossRef]
- Oliveira, L.M.; Araújo, E.S.; Guedes, S.M. Gamma irradiation effects on poly (hydroxybutyrate). Polym. Degrad. Stab. 2006, 91, 2157–2162. [Google Scholar] [CrossRef]
- Pradhan, S.; Borah, A.J.; Poddar, M.K.; Dikshit, P.K.; Rohidas, L.; Moholkar, V.S. Microbial production, ultrasound-assisted extraction and characterization of biopolymer polyhydroxybutyrate (PHB) from terrestrial (P. hysterophorus) and aquatic (E. crassipes) invasive weeds. Bioresour. Technol. 2017, 242, 304–310. [Google Scholar] [CrossRef]
- Florez, J.P.; Fazeli, M.; Simão, R.A. Preparation and characterization of thermoplastic starch composite reinforced by plasma-treated poly (hydroxybutyrate) PHB. Int. J. Biol. Macromol. 2019, 123, 609–621. [Google Scholar] [CrossRef]
- Grassie, N.; Murray, E.; Holmes, P. The thermal degradation of poly (-(D)-β-hydroxybutyric acid): Part 2—Changes in molecular weight. Polym. Degrad. Stab. 1984, 6, 95–103. [Google Scholar] [CrossRef]
- Shrivastav, A.; Mishra, S.K.; Mishra, S. Polyhydroxyalkanoate (PHA) synthesis by Spirulina subsalsa from Gujarat coast of India. Int. J. Biol. Macromol. 2010, 46, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.N.; Lee, M.Y.; Park, W.H. Thermal stabilization of poly (3-hydroxybutyrate) by poly (glycidyl methacrylate). J. Appl. Polym. Sci. 2002, 83, 2945–2952. [Google Scholar] [CrossRef]







| Soil Sources | Number of Isolates | Number of PHA-Positive Isolates |
|---|---|---|
| 7 soil samples from Phayao Province | 64 | 10 |
| 3 soil samples from Chiang Rai Province | 11 | 2 |
| 30 soil samples from Chiang Mai Province | 266 | 91 |
| Total 40 | 341 | 103 |
| Isolate | Amount of PHA Granules per Cell | PHA Content (%) |
|---|---|---|
| EP07 | 5 | 0 |
| IK01 | 3 | 3.89 |
| HD11 | 3 | 0 |
| HD22 | 4 | 0 |
| HD33 | 5 | 0 |
| HD34 | 3 | 26.79 |
| HD103 | 3 | 0 |
| HD501 | 5 | 1.96 |
| HD701 | 7 | 1.71 |
| HD121 | 4 | 0 |
| HD221 | 4 | 1.66 |
| HD224 | 2 | 0 |
| HD324 | 2 | 2.36 |
| HD523 | 7 | 0 |
| HD722 | 4 | 1.82 |
| Characteristics | Results |
|---|---|
| Gram stain | negative |
| Indole production (IND) | − |
| Acetoin production (VP) | + |
| Citrate utilization (CIT) | − |
| H2S production (H2S) | − |
| Fermentation/oxidation: | |
| D-Glucose (GLU) | + |
| D-Mannitol (MAN) | + |
| Inositol (INO) | + |
| D-Sorbitol (SOR) | + |
| L-Rhamnose (RHA) | + |
| D-Sucrose (SAC) | + |
| D-Melibiose (MEL) | + |
| Amygdalin (AMY) | + |
| L-Arabinose (ARA) | + |
| Dextrin | + |
| D-Melezitose | + |
| Starch hydrolysis | − |
| NO2 production (NO2) | − |
| N2 production (N2) | + |
| Nitrate reduction | − |
| Catalase | + |
| L-Phenylalaninase | + |
| β-Galactosidase (ONPG) | + |
| Arginine dihydrolase (ADH) | − |
| Lysine decarboxylase (LDC) | − |
| Ornithine decarboxylase (ODC) | − |
| Urease (URE) | + |
| Tryptophan deaminase (TDA) | + |
| Gelatinase (GEL) | − |
| Growth at NaCl (% w/v) | 0–3 |
| Growth at different pH | 6.0–10.0 |
| Characteristics | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Carbon source utilizations: | ||||
| Dextrin | w | + | + | − |
| D-Melezitose | + | + | + | + |
| Sodium acetate | − | + | + | w |
| Growth at conditions: | ||||
| 1% (w/v) NaCl | + | − | + | + |
| 2% (w/v) NaCl | + | + | + | − |
| Ampicillin (5 µg/mL) | + | + | + | + |
| L-Phenylalaninase activity | + | + | − | + |
| Catalase activity | + | + | + | − |
| Nitrate reduction | − | + | + | + |
| Growth in nutrient broth | + | + | + | + |
| Hydrolysis of starch | − | + | + | + |
| Nutrient Sources | PHAs Yield (g/L) |
|---|---|
| Carbon sources | |
| Glucose | 1.604 ± 0.109 a |
| Cellobiose | 0.394 ± 0.265 c |
| Fructose | 0.107 ± 0.037 c |
| Sucrose | 1.096 ± 0.426 ab |
| Galactose | 1.372 ± 0.322 a |
| Lactose | 1.239 ± 0.436 ab |
| Xylose | 0.461 ± 0.096 c |
| Xylan | 0.180 ± 0.068 c |
| Mannose | 0.537 ± 0.202 c |
| Arabinose | 0.618 ± 0.167 bc |
| Culture media | |
| Nutrient broth (NB) | 0.34 ± 0.013 d |
| Casein yeast magnesium broth (CYM) | N/A |
| Tryptic soy broth (TSB) | N/A |
| Glucose broth (GB) | 1.34 ± 0.119 b |
| Luria-Bertani broth (LB) | N/A |
| Peptone water (PW) | N/A |
| Potato dextrose broth (PDB) | 2.75 ± 0.274 a |
| Sabouraud dextrose broth (SDB) | 0.40 ± 0.041 d |
| Yeast extract peptone dextrose broth (YPD) | 0.38 ± 0.093 d |
| Yeast malt broth (YM) | 0.59 ± 0.179 cd |
| Peptone broth (PEP) | 0.68 ± 0.017 c |
| Nitrogen-deficient minimal medium (NDMM) | 1.17 ± 0.027 b |
| Source | Coefficient Estimate | Sum of Square | df | Mean Square | F-Value | p-Value Prob > F |
|---|---|---|---|---|---|---|
| Model | 3.576 × 10−6 | 5 | 7.152 × 10−5 | 13.96 | 0.0016 | |
| Xi-PDB | 422.58 | 1.429 × 10−6 | 1 | 1.429 × 10−6 | 27.88 | 0.0011 |
| Xj-Glucose | −173.41 | 2.406 × 10−5 | 1 | 2.406 × 10−5 | 4.70 | 0.0669 |
| XiXj | −28.57 | 3.266 × 10−3 | 1 | 3.266 | 0.064 | 0.8079 |
| Xi2 | −476.02 | 1.576 × 10−6 | 1 | 1.576 × 10−6 | 30.76 | 0.0009 |
| Xj2 | −277.08 | 5.341 × 10−5 | 1 | 5.341 × 10−5 | 10.42 | 0.0145 |
| Residual | 3.587 × 10−5 | 7 | 51,237.33 | |||
| Lack of fit | 2.205 × 10−5 | 3 | 73,493.37 | 2.13 | 0.2395 | |
| Pure Error | 1.382 × 10−5 | 4 | 34,545.30 | |||
| Cor total | 3.935 × 10−6 | 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khamkong, T.; Penkhrue, W.; Lumyong, S. Optimization of Production of Polyhydroxyalkanoates (PHAs) from Newly Isolated Ensifer sp. Strain HD34 by Response Surface Methodology. Processes 2022, 10, 1632. https://doi.org/10.3390/pr10081632
Khamkong T, Penkhrue W, Lumyong S. Optimization of Production of Polyhydroxyalkanoates (PHAs) from Newly Isolated Ensifer sp. Strain HD34 by Response Surface Methodology. Processes. 2022; 10(8):1632. https://doi.org/10.3390/pr10081632
Chicago/Turabian StyleKhamkong, Thitichaya, Watsana Penkhrue, and Saisamorn Lumyong. 2022. "Optimization of Production of Polyhydroxyalkanoates (PHAs) from Newly Isolated Ensifer sp. Strain HD34 by Response Surface Methodology" Processes 10, no. 8: 1632. https://doi.org/10.3390/pr10081632
APA StyleKhamkong, T., Penkhrue, W., & Lumyong, S. (2022). Optimization of Production of Polyhydroxyalkanoates (PHAs) from Newly Isolated Ensifer sp. Strain HD34 by Response Surface Methodology. Processes, 10(8), 1632. https://doi.org/10.3390/pr10081632






