Rhizosphere Bacterial Isolation from Indigenous Plants in Arid and Semi-Arid Algerian Soils: Implications for Plant Growth Enhancement
Abstract
:1. Introduction
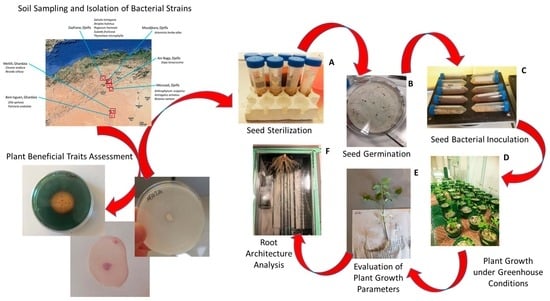
2. Materials and Methods
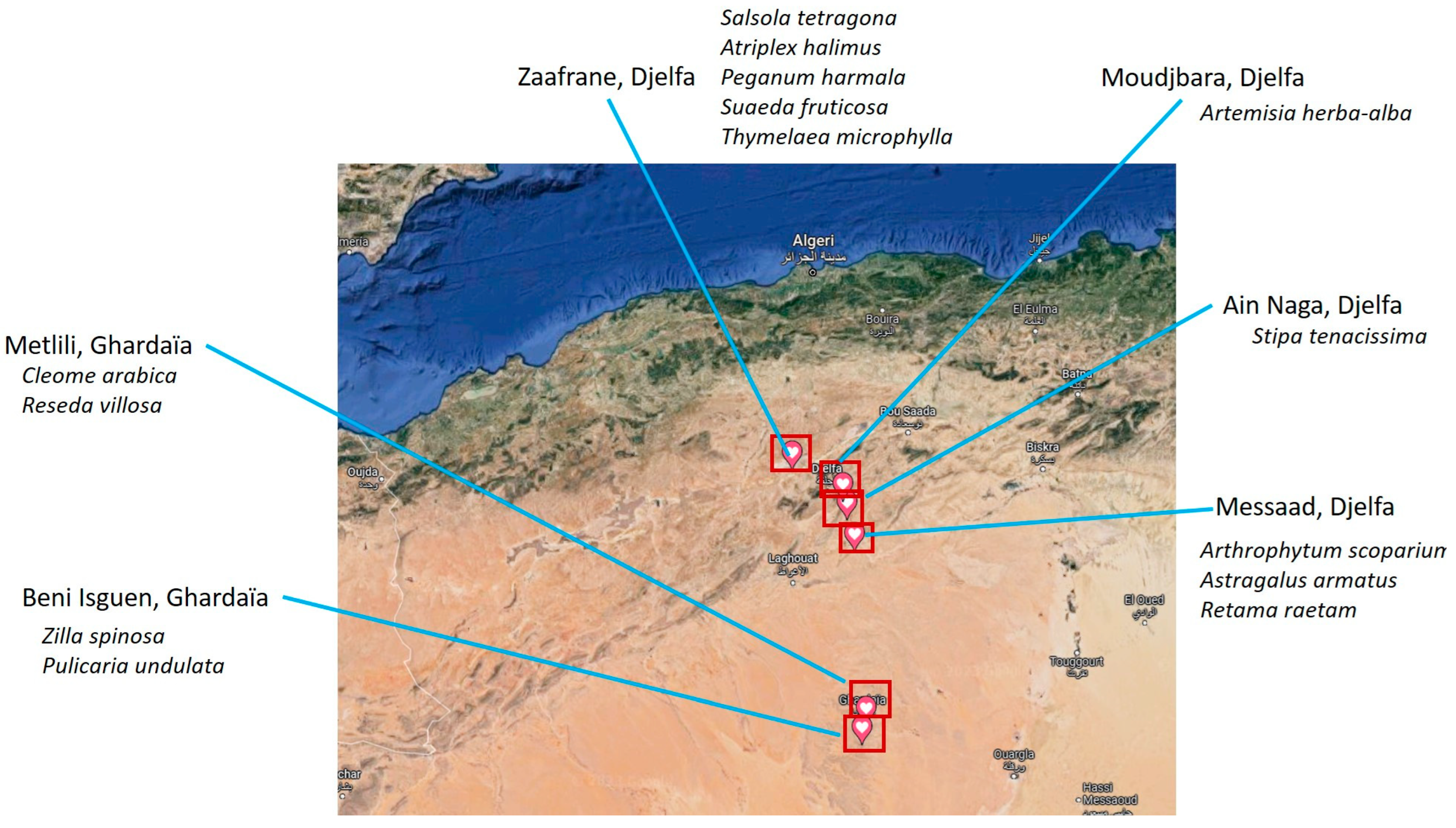
2.1. Soil Sampling
2.2. Isolation of Culturable Bacteria
2.3. Identification of Bacterial Strains
2.4. Growth Temperature Assessment and Evaluation of Salt Tolerance
2.5. Determination of Plant-Beneficial Physiological Traits
2.6. Seed Sterilization and Germination
2.7. Greenhouse Experiment
2.8. Statistical Analysis
3. Results
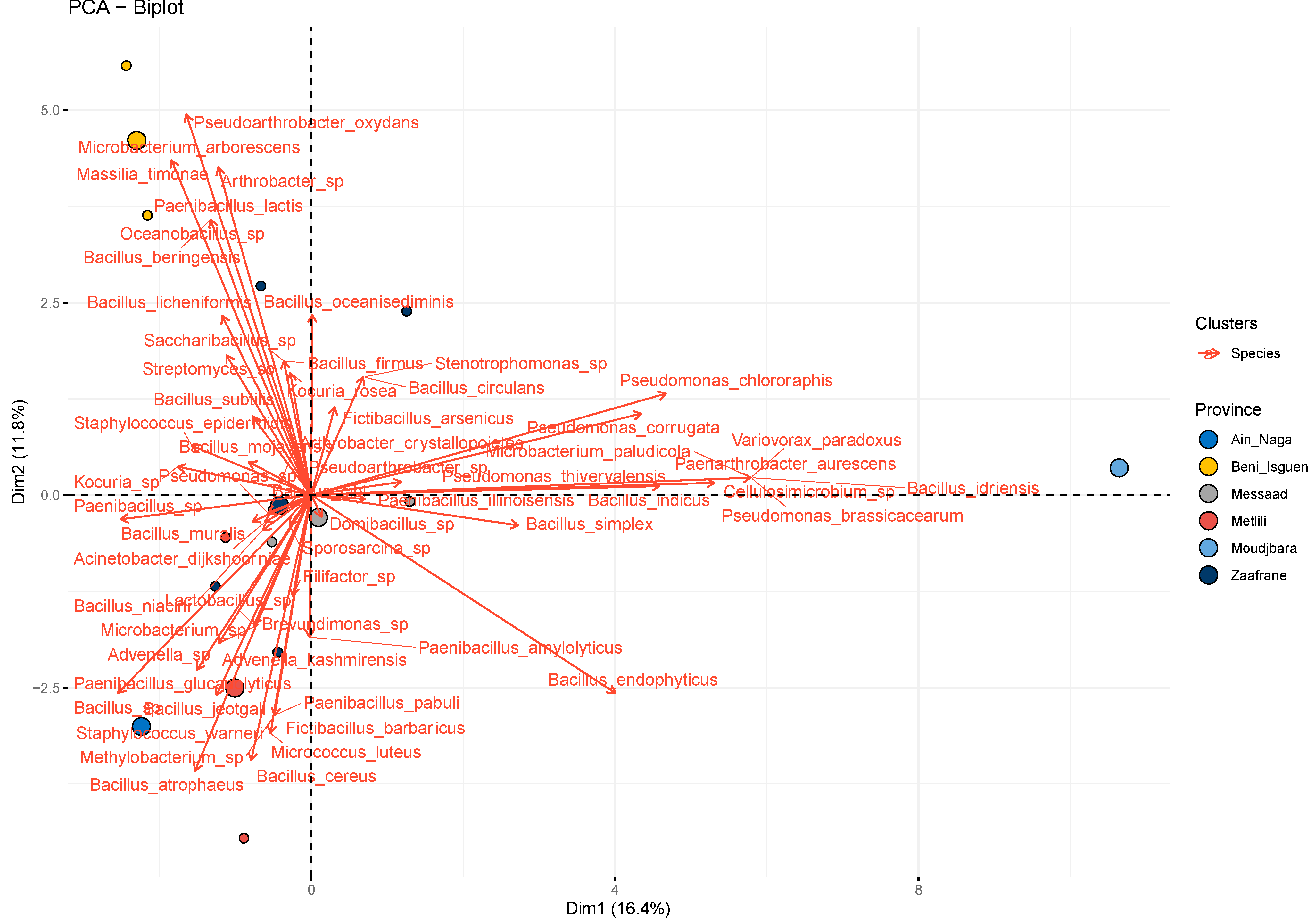
3.1. Screening of the Isolated Bacterial Strains
3.2. Bacterial Culturable Fraction Profiling
3.3. Plant Growth-Promoting Trait Characterization
3.4. Effects on Seed Germination
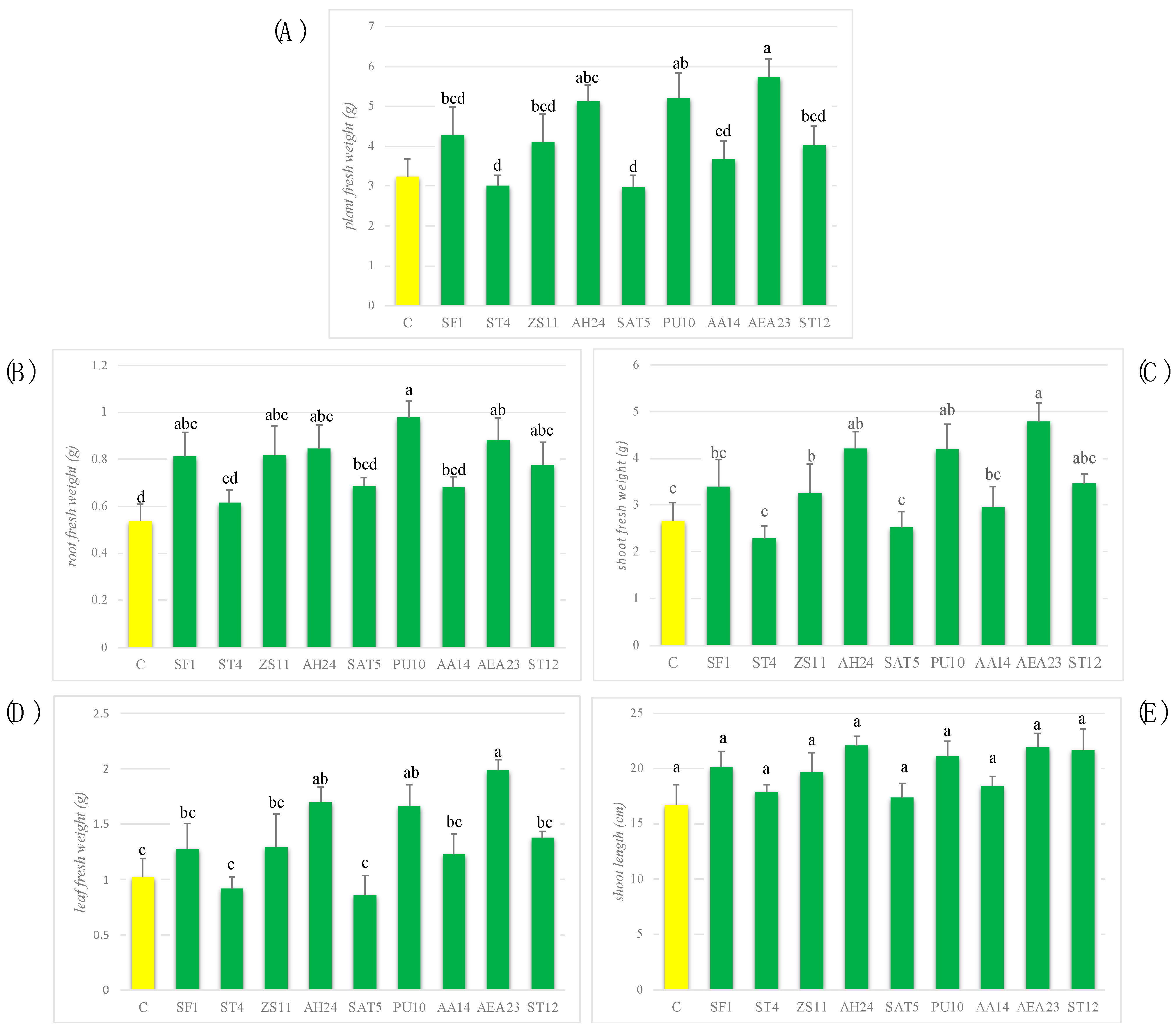
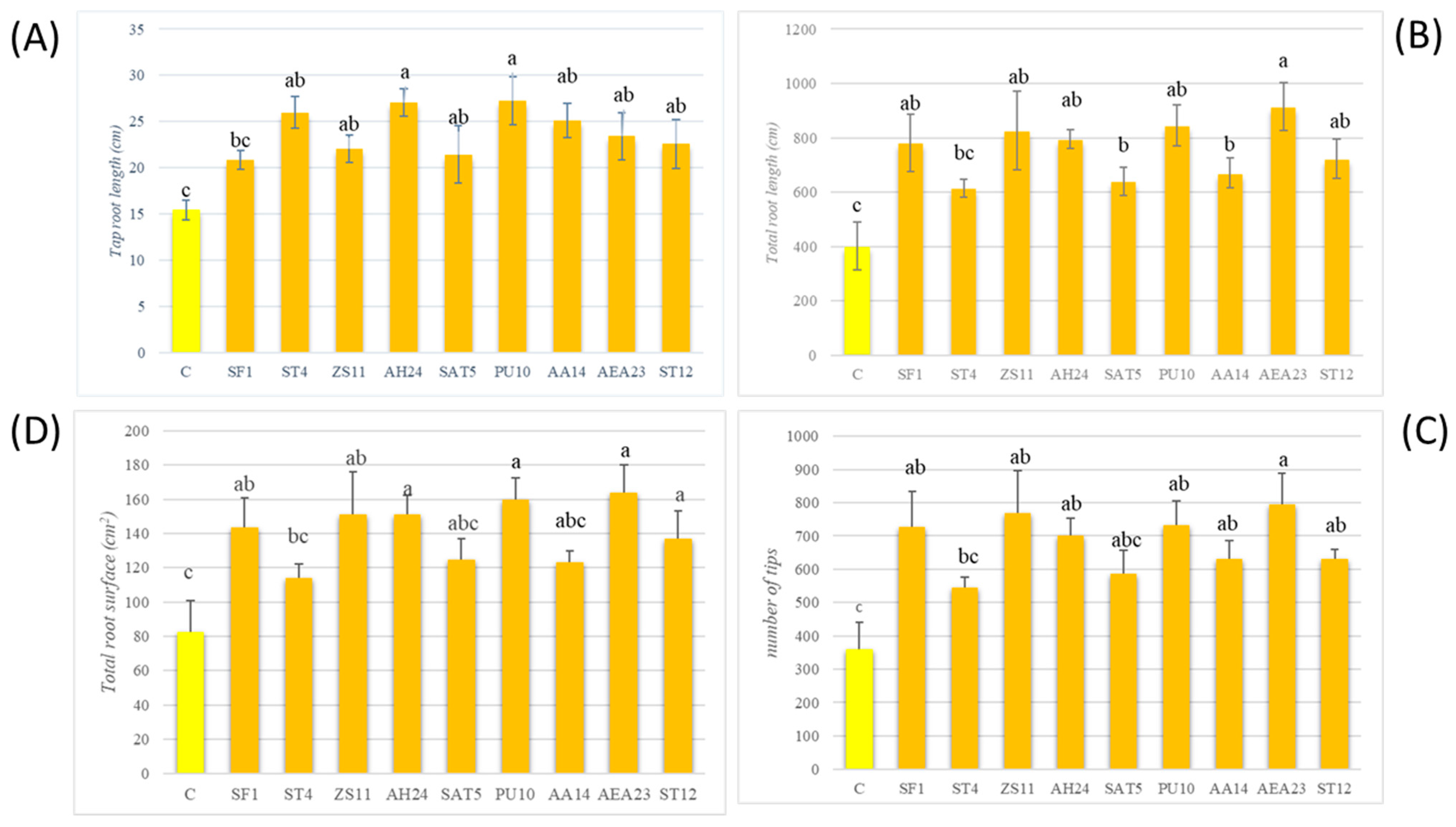
3.5. Selection of Possible PGP Bacterial Strains and Their Effects on Plant Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sellam, N.; Viñolas, A.; Zouggaghe, F.; Moulaï, R. Assessment of the physico-chemical and biological quality of surface waters in arid and semi-arid regions of Algeria (North-Africa). Bull. Soc. Zool. Fr. 2019, 144, 157–178. [Google Scholar]
- Menasria, T.; Aguilera, M.; Hocine, H.; Benammar, L.; Ayachi, A.; Bachir, A.S.; Dekak, A.; Monteoliva-Sánchez, M. Diversity and bioprospecting of extremely halophilic archaea isolated from Algerian arid and semi-arid wetland ecosystems for halophilic-active hydrolytic enzymes. Microbiol. Res. 2018, 207, 289–298. [Google Scholar] [CrossRef]
- Meddour, R.; Sahar, O.; Jury, J. New analysis of the endemic vascular plants of Algeria, their diversity, distribution pattern and conservation status. Willdenowia 2023, 53, 25–43. [Google Scholar] [CrossRef]
- Hemmami, H.; Seghir, B.B.; Zeghoud, S.; Ben Amor, I.; Kouadri, I.; Rebiai, A.; Zaater, A.; Messaoudi, M.; Benchikha, N.; Sawicka, B.; et al. Desert endemic plants in Algeria: A review on traditional uses, phytochemistry, polyphenolic compounds and pharmacological activities. Molecules 2023, 28, 1834. [Google Scholar] [CrossRef] [PubMed]
- Médail, F.; Quézel, P. Hot-Spots analysis for conservation of plant biodiversity in the Mediterranean basin. Ann. Mo. Bot. Gard. 1997, 84, 112–127. [Google Scholar] [CrossRef]
- EEA—European Environment Agency. Mapping Desertification in Europe. 2020. Available online: https://www.eea.europa.eu/themes/soil/desertification/mapping-desertification-in-europe (accessed on 2 July 2023).
- Ferreira, C.S.S.; Seifollahi-Aghmiuni, S.; Destouni, G.; Ghajarnia, N.; Kalantari, Z. Soil degradation in the European Mediterranean region: Processes, status and consequences. Sci. Total Environ. 2022, 805, 150106. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Recent advances in bacterial amelioration of plant drought and salt stress. Biology 2022, 11, 437. [Google Scholar] [CrossRef]
- Khan, N.; Ali, S.; Shahid, M.A.; Mustafa, A.; Sayyed, R.Z.; Curá, J.A. Insights into the Interactions among Roots, Rhizosphere, and Rhizobacteria for Improving Plant Growth and Tolerance to Abiotic Stresses: A Review. Cells 2021, 10, 1551. [Google Scholar] [CrossRef]
- Shahid, M.; Singh, U.B.; Khan, M.S.; Singh, P.; Kumar, R.; Singh, R.N.; Kumar, A.; Singh, H.V. Bacterial ACC deaminase: Insights into enzymology, biochemistry, genetics, and potential role in amelioration of environmental stress in crop plants. Front. Microbiol. 2023, 14, 1132770. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Santoyo, G.; Yadav, A.N.; Babalola, O.O. Efforts towards overcoming drought stress in crops: Revisiting the mechanisms employed by plant growth-promoting bacteria. Front Microbiol. 2022, 13, 962427. [Google Scholar] [CrossRef]
- Seleiman, M.F.; Al-Suhaibani, N.; Ali, N.; Akmal, M.; Alotaibi, M.; Refay, Y.; Dindaroglu, T.; Abdul-Wajid, H.H.; Battaglia, M.L. Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants 2021, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Trovato, M.; Brini, F.; Mseddi, K.; Rhizopoulou, S.; Jones, M.A. A holistic and sustainable approach linked to drought tolerance of Mediterranean crops. Front. Plant Sci. 2023, 14, 1167376. [Google Scholar] [CrossRef] [PubMed]
- Foo, J.L.; Ling, H.; Lee, Y.S.; Chang, M.W. Microbiome engineering: Current applications and its future. Biotechnol. Adv. 2018, 36, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Bokhari, A.; Essack, M.; Lafi, F.F.; Andres-Barrao, C.; Jalal, R.; Alamoudi, S.; Razali, R.; Alzubaidy, H.; Shah, K.H.; Siddique, S.; et al. Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Sci. Rep. 2019, 9, 18154. [Google Scholar] [CrossRef]
- Alsharif, W.; Saad, M.M.; Hirt, H. Desert microbes for boosting sustainable agriculture in extreme environments. Front. Microbiol. 2020, 11, 1666. [Google Scholar] [CrossRef]
- Eida, A.A.; Ziegler, M.; Lafi, F.F.; Michell, C.T.; Voolstra, C.R.; Hirt, H.; Saad, M.M. Desert plant bacteria reveal host influence and beneficial plant growth properties. PLoS ONE 2018, 13, e0208223. [Google Scholar] [CrossRef]
- Bona, E.; Massa, N.; Toumatia, O.; Novello, G.; Cesaro, P.; Todeschini, V.; Boatti, L.; Mignone, F.; Titouah, H.; Zitouni, A.; et al. Climatic zone and soil properties determine the biodiversity of the soil bacterial communities associated to native plants from desert areas of North-Central Algeria. Microorganisms 2021, 9, 1359. [Google Scholar] [CrossRef]
- Massa, N.; Cesaro, P.; Todeschini, V.; Capraro, J.; Scarafoni, A.; Cantamessa, S.; Copetta, A.; Anastasia, F.; Gamalero, E.; Lingua, G.; et al. Selected autochthonous Rhizobia, applied in combination with AM Fungi, improve seed quality of common bean cultivated in reduced fertilization condition. Appl. Soil Ecol. 2020, 148, 103507. [Google Scholar] [CrossRef]
- Novello, G.; Gamalero, E.; Massa, N.; Cesaro, P.; Lingua, G.; Todeschini, V.; Caramaschi, A.; Favero, F.; Corà, D.; Manfredi, M.; et al. Proteome and Physiological Characterization of Halotolerant Nodule Endophytes: The Case of Rahnella aquatilis and Serratia plymuthica. Microorganisms 2022, 10, 890. [Google Scholar] [CrossRef]
- De Brito, A.M.; Gagne, S.; Antoun, H. Effect of compost on rhizosphere microflora of the tomato and on the incidence of plant growth-promoting rhizobacteria. Appl. Environ. Microbiol. 1995, 61, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, Y.; Teng, D.; Zhang, X.; He, X.; Zhang, Y.; Lv, G. Rhizobacterial communities of five co-occurring desert halophytes. PeerJ 2018, 6, e5508. [Google Scholar] [CrossRef]
- Belov, A.A.; Cheptsov, V.S.; Vorobyova, E.A. Soil bacterial communities of Sahara and Gibson deserts: Physiological and taxonomical characteristics. AIMS Microbiol. 2018, 4, 685–710. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Bacillus: A Biological Tool for Crop Improvement through Bio-Molecular Changes in Adverse Environments. Front Physiol. 2017, 8, 667. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Tabassum, B.; Fathi Abd Allah, E. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for Plant Growth Promotion and Stress Resilience: What Have We Learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef]
- Lyu, D.; Msimbira, L.A.; Nazari, M.; Antar, M.; Pagé, A.; Shah, A.; Monjezi, N.; Zajonc, J.; Tanney, C.A.S.; Backer, R.; et al. The Coevolution of Plants and Microbes Underpins Sustainable Agriculture. Microorganisms 2021, 9, 1036. [Google Scholar] [CrossRef]
- Fields, B.; Friman, V.P. Microbial eco-evolutionary dynamics in the plant rhizosphere. Curr. Opin. Microbiol. 2022, 68, 102153. [Google Scholar] [CrossRef]
- Wang, P.; Ding, L.; Zou, C.; Zhang, Y.; Wang, M. Rhizosphere element circling, multifunctionality, aboveground productivity and trade-offs are better predicted by rhizosphere rare taxa. Front. Plant Sci. 2022, 13, 985574. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed]
- Numan, M.; Bashir, S.; Khan, Y.A.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L. Exploration of plant growth-promoting bacteria as a strategy for improving plant growth and tolerance against abiotic stress. In Plant-Microbe Interactions in Agro-Ecological Perspectives: Volume 2: Microbial Interactions and Agro-Ecological Impacts; Springer: Singapore, 2020; pp. 355–372. [Google Scholar]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Manda, R.; Gupta, S.; Kumar, S.; Kumar, V. Isolation and characterization of osmotolerant bacteria from thar desert of western Rajasthan (India). Rev. Biol. Trop. 2013, 61, 1551–1562. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Singh, A.; Shukla, P.; Singh, S. Plant growth-promoting rhizobacteria: Mechanisms and applications in agriculture. In Plant Microbiome: Stress Response and Developmental Functions; Springer: Berlin/Heidelberg, Germany, 2021; pp. 47–74. [Google Scholar]
- Aanniz, T.; Ouadghiri, M.; Melloul, M.; Swings, J.; Elfahime, E.; Ibijbijen, J.; Ismaili, M.; Amar, M. Thermophilic bacteria in Moroccan hot springs, salt marshes and desert soils. Braz. J. Microbiol. 2015, 46, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Kearl, J.; McNary, C.; Lowman, J.S.; Mei, C.; Aanderud, Z.T.; Smith, S.T.; West, J.; Colton, E.; Hamson, M.; Nielsen, B.L. Salt-tolerant halophyte rhizosphere bacteria stimulate growth of Alfalfa in salty soil. Front. Microbiol. 2019, 10, 1849. [Google Scholar] [CrossRef]
- Delgado-García, M.; Contreras-Ramos, S.M.; Rodríguez, J.A.; Mateos-Díaz, J.C.; Aguilar, C.N.; Camacho-Ruíz, R.M. Isolation of halophilic bacteria associated with saline and alkaline-sodic soils by culture dependent approach. Heliyon 2018, 4, e00954. [Google Scholar] [CrossRef]
- Bachran, M.; Kluge, S.; Lopez-Fernandez, M.; Cherkouk, A. Microbial diversity in an arid, naturally saline environment. Microb. Ecol. 2019, 78, 494–505. [Google Scholar] [CrossRef]
- Dehimi, K.; Speciale, A.; Saija, A.; Dahamna, S.; Raciti, R.; Cimino, F.; Cristani, M. Antioxidant and Anti-inflammatory Properties of Algerian Thymelaea microphylla Coss. and Dur. Extracts. Pharmacogn. Mag. 2016, 12, 203–210. [Google Scholar]
- Li, Y.; Wu, X.; Chen, T.; Wang, W.; Liu, G.; Zhang, W.; Li, S.; Wang, M.; Zhao, C.; Zhou, H.; et al. Plant phenotypic traits eventually shape its microbiota: A common garden test. Front. Microbiol. 2018, 9, 2479. [Google Scholar] [CrossRef]
- Contreras, M.J.; Leal, K.; Bruna, P.; Nuñez-Montero, K.; Goméz-Espinoza, O.; Santos, A.; Bravo, L.; Valenzuela, B.; Solis, F.; Gahona, G.; et al. Commonalities between the Atacama Desert and Antarctica rhizosphere microbial communities. Front. Microbiol. 2023, 14, 1197399. [Google Scholar] [CrossRef]
- Yang, G.; Jiang, L.; Li, W.; Li, E.; Lv, G. Structural Characteristics and Assembly Mechanisms of Soil Microbial Communities under Water–Salt Gradients in Arid Regions. Microorganisms 2023, 11, 1060. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, W.; Huang, T.; Yang, Y. Similarities and differences in the rhizosphere biota among different ephemeral desert plants in Gurbantünggüt Desert. Environ. Sci. Eur. 2023, 35, 18. [Google Scholar] [CrossRef]
- Yang, Z.W.; Salam, N.; Mohany, M.; Chinnathambi, A.; Alharbi, S.A.; Xiao, M.; Hozzein, W.N.; Li, W.J. Microbacterium album sp. nov. and Microbacterium deserti sp. nov., two halotolerant actinobacteria isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 217–222. [Google Scholar] [CrossRef]
- Park, H.Y.; Kim, K.K.; Jin, L.; Lee, S.T. Microbacterium paludicola sp. nov., a novel xylanolytic bacterium isolated from swamp forest. Int. J. Syst. Evol. Microbiol. 2006, 56 Pt 3, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Takeuchi, M.; Banno, I. Reclassification of “Flavobacterium arborescens” (Frankland and Frankland) Bergey et al. in the genus Microbacterium (Orla-Jensen) Collins et al., as Microbacterium arborescens comb. nov., nom. rev. Curr. Microbiol. 1984, 11, 281–284. [Google Scholar] [CrossRef]
- Gamalero, E.; Martinotti, M.G.; Trotta, A.; Lemanceau, P.; Berta, G. Morphogenetic modifications induced by Pseudomonas fluorescens A6RI and Glomus mosseae BEG12 in the root system of tomato differ according to plant growth conditions. New Phytol. 2002, 155, 293–300. [Google Scholar] [CrossRef]
- Santoyo, G.; Gamalero, E.; Glick, B.R. Mycorrhizal-Bacterial amelioration of plant abiotic and biotic stress. Front. Sustain. Food Syst. 2021, 57, 672881. [Google Scholar] [CrossRef]
- Mao, W.; Wu, Y.; Li, F.; Tang, W.; Gong, W.; Han, X.; White, J.F.; Ji, X.; Li, H. Seed Endophytes and Their Roles in Host Plant Stress Resistance. J. Soil Sci. Plant Nutr. 2023, 23, 2927–2937. [Google Scholar] [CrossRef]
- De Andrade, L.A.; Santos, C.H.B.; Frezarin, E.T.; Sales, L.R.; Rigobelo, E.C. Plant Growth-Promoting Rhizobacteria for Sustainable Agricultural Production. Microorganisms 2023, 11, 1088. [Google Scholar] [CrossRef]

| Number of Species (S) | Simpson’s Dominance (D) | Simpson’s Biodiversity (1-D) | Simpson’s Biodiversity (1/D) | Shannon’s (H’) | Evenness EH = H/ln(S) | |
|---|---|---|---|---|---|---|
| CA | 11 | 0.240 | 0.760 | 4.172 | 0.833 | 0.347 |
| AH | 11 | 0.128 | 0.872 | 7.806 | 0.961 | 0.401 |
| PU | 13 | 0.116 | 0.884 | 8.647 | 1.030 | 0.402 |
| RV | 13 | 0.129 | 0.871 | 7.737 | 1.014 | 0.395 |
| ZS | 14 | 0.088 | 0.912 | 11.308 | 1.100 | 0.417 |
| AHA | 14 | 0.088 | 0.912 | 11.308 | 1.100 | 0.417 |
| AS | 10 | 0.164 | 0.836 | 6.080 | 0.876 | 0.380 |
| AA | 8 | 0.191 | 0.809 | 5.232 | 0.799 | 0.384 |
| PH | 13 | 0.095 | 0.905 | 10.526 | 1.068 | 0.416 |
| RR | 14 | 0.091 | 0.909 | 10.939 | 1.096 | 0.415 |
| SF | 12 | 0.107 | 0.893 | 9.308 | 1.021 | 0.411 |
| SAT | 10 | 0.130 | 0.870 | 7.681 | 0.938 | 0.407 |
| ST | 14 | 0.140 | 0.860 | 7.118 | 1.019 | 0.386 |
| TM | 6 | 0.225 | 0.775 | 4.446 | 0.708 | 0.395 |
| Bacterial strain | Growth at | MIC NaCl% | IAA | Siderophores | DCP | TCP | |||
|---|---|---|---|---|---|---|---|---|---|
| 4 °C | 28 °C | 37 °C | 45 °C | ||||||
| SF1 | + | + | + | + | 14.4 | − | + | − | + |
| ST4 | + | + | + | + | 28.8 | + | − | + | + |
| ZS11 | − | + | + | + | 14.4 | − | + | + | + |
| AH24 | − | + | + | + | 14.4 | + | − | − | + |
| SAT5 | − | + | + | − | 14.4 | − | − | + | + |
| PU10 | + | + | + | + | 14.4 | − | − | + | + |
| AA14 | + | + | + | + | 14.4 | + | − | − | + |
| AEA23 | − | + | + | + | 14.4 | + | − | − | − |
| ST12 | − | + | + | + | 28.8 | + | + | − | + |
| Treatments | Root Dry Weight (g) | Shoot Dry Weight (g) | Leaf Dry Weight (g) |
|---|---|---|---|
| C | 0.06 ± 0.01 d | 0.20 ± 0.04 c | 0.16 ± 0.03 c |
| SF1 | 0.08 ± 0.01 abcd | 0.26 ± 0.05 abc | 0.19 ± 0.03 bc |
| ST4 | 0.06 ± 0.01 cd | 0.17 ± 0.02 c | 0.14 ± 0.01 c |
| ZS11 | 0.09 ± 0.01 abcd | 0.25 ± 0.04 abc | 0.18 ± 0.03 bc |
| AH24 | 0.09 ± 0.01 abcd | 0.31 ± 0.03 ab | 0.24 ± 0.02 ab |
| SAT5 | 0.08 ± 0.01 bcd | 0.22 ± 0.03 bc | 0.16 ± 0.01 c |
| PU10 | 0.11 ± 0.01 a | 0.30 ± 0.04 ab | 0.24 ± 0.02 ab |
| AA14 | 0.07 ± 0.00 cd | 0.22 ± 0.03 bc | 0.18 ± 0.03 bc |
| AEA23 | 0.10 ± 0.01 ab | 0.33 ± 0.04 a | 0.27 ± 0.02 a |
| ST12 | 0.09 ± 0.01 abc | 0.26 ± 0.02 abc | 0.21 ± 0.01 abc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novello, G.; Bona, E.; Toumatia, O.; Vuolo, F.; Bouras, N.; Titouah, H.; Zitouni, A.; Gorrasi, S.; Massa, N.; Cesaro, P.; et al. Rhizosphere Bacterial Isolation from Indigenous Plants in Arid and Semi-Arid Algerian Soils: Implications for Plant Growth Enhancement. Processes 2023, 11, 2907. https://doi.org/10.3390/pr11102907
Novello G, Bona E, Toumatia O, Vuolo F, Bouras N, Titouah H, Zitouni A, Gorrasi S, Massa N, Cesaro P, et al. Rhizosphere Bacterial Isolation from Indigenous Plants in Arid and Semi-Arid Algerian Soils: Implications for Plant Growth Enhancement. Processes. 2023; 11(10):2907. https://doi.org/10.3390/pr11102907
Chicago/Turabian StyleNovello, Giorgia, Elisa Bona, Omrane Toumatia, Francesco Vuolo, Noureddine Bouras, Houda Titouah, Abdelghani Zitouni, Susanna Gorrasi, Nadia Massa, Patrizia Cesaro, and et al. 2023. "Rhizosphere Bacterial Isolation from Indigenous Plants in Arid and Semi-Arid Algerian Soils: Implications for Plant Growth Enhancement" Processes 11, no. 10: 2907. https://doi.org/10.3390/pr11102907