Insights into the Potential Role of Gordonia alkanivorans Strains in Biotechnologies †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation Conditions
2.2. Genome Sequencing and Analysis
3. Results
3.1. Identifying the Strains
3.2. Physiological and Biochemical Characteristics of Strains
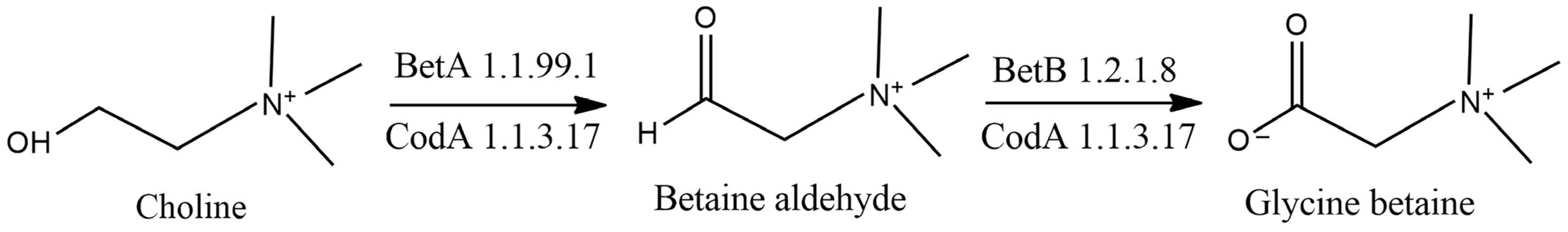
3.3. Osmoprotectant Metabolism
3.4. Pangenome Analysis
3.5. Features of the Genetic Organization of G. alkanivorans
3.5.1. Hydrocarbon Degradation Ability
3.5.2. Plasmids
3.5.3. Biosynthesis of Secondary Metabolites
- Terpene (SF2575 biosynthetic gene cluster: polyketide type II; carotenoid);
- Streptozotocin;
- Type I polyketide synthase;
- NRPS (non-ribosomal peptide synthetase);
- Non-alpha poly-amino acids (ε-Poly-L-lysine);
- Ectoine;
- Redox cofactor;
- NRPS-independent, IucA/IucC-like siderophores;
- Arylpolyene (primycin like most similar known cluster is found in all strains except for 134, 142, and 144, which do not exhibit similarity to this group).
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- LPSN—List of Prokaryotic Names with Standing in Nomenclature. Available online: https://lpsn.dsmz.de/ (accessed on 11 October 2023).
- Kim, H.; Dong, K.; Kim, J.; Lee, S. Characteristics of Crude Oil-degrading Bacteria Gordonia Iterans Isolated from Marine Coastal in Taean Sediment. MicrobiologyOpen 2019, 8, e00754. [Google Scholar] [CrossRef] [PubMed]
- Zargar, A.N.; Mishra, S.; Kumar, M.; Srivastava, P. Isolation and Chemical Characterization of the Biosurfactant Produced by Gordonia sp. IITR100. PLoS ONE 2022, 17, e0264202. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.A.; Stainsby, F.M.; Ward, A.C.; Goodfellow, M. Gordonia sinesedis sp. Nov., a Novel Soil Isolate. Antonie Van Leeuwenhoek 2003, 83, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.-C.; Xiong, L.; Poon, R.W.S.; Chen, J.H.K.; Leung, K.-W.; Lam, J.Y.W.; Wu, A.K.L.; Chan, J.F.W.; Lau, S.K.P.; Woo, P.C.Y. Gordonia hongkongensis sp. Nov., Isolated from Blood Culture and Peritoneal Dialysis Effluent of Patients in Hong Kong. Int. J. Syst. Evol. Microbiol. 2016, 66, 3942–3950. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, Z.; Chen, M.; Liu, Z. Gordonia crocea sp. Nov. Isolated from Wound Infection After Pacemaker Implantation: Case Report and Literature Review. IDR 2022, 15, 2915–2920. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-Q.; Ming, H.; Gonoi, T.; Chen, Y.; Cao, Y.; Wang, Y.-Y.; Cheng, J.; Koga, T.; Mikami, Y.; Li, W.-J. Gordonia iterans sp. Nov., Isolated from a Patient with Pneumonia. Int. J. Syst. Evol. Microbiol. 2014, 64, 3520–3525. [Google Scholar] [CrossRef]
- Sarkar, B.; Mandal, S. Gordonia sp. BSTG01 Isolated from Hevea Brasiliensis Plantation Efficiently Degrades Polyisoprene (Rubber). 3 Biotech 2021, 11, 508. [Google Scholar] [CrossRef]
- Lee, K.; Badaya, S.K.; Singh, R.; Lim, J.Y. Complete Genome Sequence of Gordonia sp. Strain JH63, Isolated from Human Skin. Microbiol. Resour. Announc. 2020, 9, e00059-20. [Google Scholar] [CrossRef]
- Brown, L.M.; Gunasekera, T.S.; Striebich, R.C.; Ruiz, O.N. Draft Genome Sequence of Gordonia sihwensis Strain 9, a Branched Alkane-Degrading Bacterium. Genome Announc. 2016, 4, e00622-16. [Google Scholar] [CrossRef]
- Kim, S.G.; Reyna, M.; Park, M.; Wang, S.X.; Foley, S.L. Genome Sequence of Gordonia alkaliphila Strain WW102, Isolated from Influent Wastewater at a Research Center with Multiple-Species Research Animal Facilities. Microbiol. Resour. Announc. 2023, 12, e00492-23. [Google Scholar] [CrossRef]
- Woo, H.L.; DeAngelis, K.M.; Teshima, H.; Davenport, K.; Daligault, H.; Erkkila, T.; Goodwin, L.; Gu, W.; Lo, C.-C.; Munk, C.; et al. High-Quality Draft Genome Sequences of Four Lignocellulose-Degrading Bacteria Isolated from Puerto Rican Forest Soil: Gordonia sp., Paenibacillus sp., Variovorax sp., and Vogesella sp. Genome Announc. 2017, 5, e00300-17. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Suárez, J.; Díaz, L.; Melo-Bolivar, J.; Villamil, L. Draft Genome Sequence Data of Gordonia hongkongensis Strain EUFUS-Z928 Isolated from the Octocoral Eunicea Fusca. Data Brief 2022, 42, 108076. [Google Scholar] [CrossRef] [PubMed]
- Domingos, D.F.; Dellagnezze, B.M.; Greenfield, P.; Reyes, L.R.; Melo, I.S.; Midgley, D.J.; Oliveira, V.M. Draft Genome Sequence of the Biosurfactant-Producing Bacterium Gordonia amicalis Strain CCMA-559, Isolated from Petroleum-Impacted Sediment. Genome Announc. 2013, 1, e00894-13. [Google Scholar] [CrossRef]
- Badariotti, E.H.; Beyrne, E.; Tejerina, M.; Raymond, M.L.; Soria, N.W. Draft Genome Sequence of Gordonia sp. Strain Campus, a Bacterium Isolated from Diesel-Contaminated Soil with Potential Use in Phytoremediation Systems. Microbiol. Resour. Announc. 2022, 11, e00139-22. [Google Scholar] [CrossRef] [PubMed]
- Istvan, P.; Ronen, Z. Draft Genome Sequence of Gordonia sp. Strain YY1, Isolated from an Explosive-Contaminated Environment. Microbiol. Resour. Announc. 2020, 9, e00070-20. [Google Scholar] [CrossRef] [PubMed]
- Brandão, P.F.B.; Maldonado, L.A.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Gordonia namibiensis sp. Nov., a Novel Nitrile Metabolising Actinomycete Recovered from an African Sand. Syst. Appl. Microbiol. 2001, 24, 510–515. [Google Scholar] [CrossRef]
- Durrell, K.; Prins, A.; Le Roes-Hill, M. Draft Genome Sequence of Gordonia lacunae BS2 T. Genome Announc. 2017, 5, e00959-17. [Google Scholar] [CrossRef]
- Kummer, C.; Schumann, P.; Stackebrandt, E. Gordonia alkanivorans sp. Nov., Isolated from Tar-Contaminated Soil. Int. J. Syst. Evol. Microbiol. 1999, 49, 1513–1522. [Google Scholar] [CrossRef]
- Delegan, Y.; Valentovich, L.; Vetrova, A.; Frantsuzova, E.; Kocharovskaya, Y. Complete Genome Sequence of Gordonia sp. 135, a Promising Dibenzothiophene- and Hydrocarbon-Degrading Strain. Microbiol. Resour. Announc. 2020, 9, e01450-19. [Google Scholar] [CrossRef]
- Frantsuzova, E.; Solomentsev, V.; Vetrova, A.; Travkin, V.; Solyanikova, I.; Delegan, Y. Complete Genome Sequence of Gordonia polyisoprenivorans 135, a Promising Degrader of Aromatic Compounds. Microbiol. Resour. Announc. 2023, 12, e00058-23. [Google Scholar] [CrossRef]
- Xie, Y.; Zhou, S.; Xu, Y.; Wu, W.; Xia, W.; Zhang, R.; Huang, D.; Huang, X. Gordonia Mangrovi sp. Nov., a Novel Actinobacterium Isolated from Mangrove Soil in Hainan. Int. J. Syst. Evol. Microbiol. 2020, 70, 4537–4543. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lyu, L.; Hu, Z.; Yu, Z.; Shao, Z. Gordonia tangerina sp. Nov., Isolated from Seawater. Int. J. Syst. Evol. Microbiol. 2022, 72, 005632. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Maser, E.; Zhang, T. Genomic Analysis of Gordonia polyisoprenivorans Strain R9, a Highly Effective 17 Beta-Estradiol- and Steroid-Degrading Bacterium. Chem. Biol. Interact. 2021, 350, 109685. [Google Scholar] [CrossRef] [PubMed]
- Loh, W.L.C.; Huang, K.-C.; Ng, H.S.; Lan, J.C.-W. Exploring the Fermentation Characteristics of a Newly Isolated Marine Bacteria Strain, Gordonia terrae TWRH01 for Carotenoids Production. J. Biosci. Bioeng. 2020, 130, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Pansomsuay, R.; Duangupama, T.; Pittayakhajonwut, P.; Intaraudom, C.; Suriyachadkun, C.; He, Y.-W.; Tanasupawat, S.; Thawai, C. Gordonia aquimaris sp. Nov., a Novel Marine Actinobacterium Isolated from Seawater in the Upper Gulf of Thailand. Int. J. Syst. Evol. Microbiol. 2023, 73, 005804. [Google Scholar] [CrossRef]
- Kwon, S.J.; Choi, Y.J.; Kim, J.M.; Lee, P.C. Complete Genome Sequence of the Carotenoid-Producing Strain Gordonia ajoucoccus A2. Microbiol. Resour. Announc. 2020, 9, e00662-20. [Google Scholar] [CrossRef]
- Kim, Y.S.; Roh, S.G.; Kim, S.B. Gordonia insulae sp. Nov., Isolated from an Island Soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 2079–2083. [Google Scholar] [CrossRef]
- Kotani, T.; Yamamoto, T.; Yurimoto, H.; Sakai, Y.; Kato, N. PropaneMonooxygenase and NAD+-Dependent Secondary AlcoholDehydrogenase in Propane Metabolism by Gordonia sp. Strain TY-5. J. Bacteriol. 2003, 185, 7120–7128. [Google Scholar] [CrossRef]
- Arenskötter, M.; Bröker, D.; Steinbüchel, A. Biology of the Metabolically Diverse Genus Gordonia. Appl. Environ. Microbiol. 2004, 70, 3195–3204. [Google Scholar] [CrossRef]
- Shen, F.-T.; Lu, H.-L.; Lin, J.-L.; Huang, W.-S.; Arun, A.B.; Young, C.-C. Phylogenetic Analysis of Members of the Metabolically Diverse Genus Gordonia Based on Proteins Encoding the gyrB Gene. Res. Microbiol. 2006, 157, 367–375. [Google Scholar] [CrossRef]
- Shen, F.-T.; Young, C.-C. Rapid Detection and Identification of the Metabolically Diverse Genus Gordonia by 16S rRNA-Gene-Targeted Genus-Specific Primers. FEMS Microbiol. Lett. 2005, 250, 221–227. [Google Scholar] [CrossRef]
- Lienkamp, A.C.; Burnik, J.; Heine, T.; Hofmann, E.; Tischler, D. Characterization of the Glutathione S-Transferases Involved in Styrene Degradation in Gordonia rubripertincta CWB2. Microbiol. Spectr. 2021, 9, e00474-21. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, J.; Liu, Y.; Wu, X. Biological Process of Alkane Degradation by Gordonia sihwaniensis. ACS Omega 2022, 7, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Sowani, H.; Kulkarni, M.; Zinjarde, S. Harnessing the Catabolic Versatility of Gordonia Species for Detoxifying Pollutants. Biotechnol. Adv. 2019, 37, 382–402. [Google Scholar] [CrossRef] [PubMed]
- Mai, Z.; Wang, L.; Li, Q.; Sun, Y.; Zhang, S. Biodegradation and Metabolic Pathway of Phenanthrene by a Newly Isolated Bacterium Gordonia sp. SCSIO19801. Biochem. Biophys. Res. Commun. 2021, 585, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Weng, L.; Chen, D.; Hu, H.; Jia, Y.; Zhou, J.L. Bioremediation of PAEs-Contaminated Saline Soil: The Application of a Marine Bacterial Strain Isolated from Mangrove Sediment. Mar. Pollut. Bull. 2023, 192, 115071. [Google Scholar] [CrossRef]
- Hu, T.; Yang, C.; Hou, Z.; Liu, T.; Mei, X.; Zheng, L.; Zhong, W. Phthalate Esters Metabolic Strain Gordonia sp. GZ-YC7, a Potential Soil Degrader for High Concentration Di-(2-Ethylhexyl) Phthalate. Microorganisms 2022, 10, 641. [Google Scholar] [CrossRef]
- Dhar, R.; Basu, S.; Bhattacharyya, M.; Dutta, T.K. Evaluation of Distinct Molecular Architectures and Coordinated Regulation of the Catabolic Pathways of Oestrogenic Dioctyl Phthalate Isomers in Gordonia sp. Microbiology 2023, 169, 001353. [Google Scholar] [CrossRef]
- Wang, H.; Guan, F.; Zhu, Y.; Pan, Y.; Liu, Q.; Liu, Q.; He, W.; Gong, D.; Tian, J.; Han, D. Biofilm Formation Promoted Biodegradation of Polyethylene in Gordonia polyisoprenivorans B251 Isolated from Bacterial Enrichment Acclimated by Hexadecane for Two Years. Chemosphere 2023, 344, 140383. [Google Scholar] [CrossRef]
- Kalita, M.; Chutia, M.; Jha, D.K.; Subrahmanyam, G. Mechanistic Understanding of Gordonia sp. in Biodesulfurization of Organosulfur Compounds. Curr. Microbiol. 2022, 79, 82. [Google Scholar] [CrossRef]
- Shin, K.-C.; Lee, H.-J.; Oh, D.-K. Substrate Specificity of β-Glucosidase from Gordonia terrae for Ginsenosides and Its Application in the Production of Ginsenosides Rg3, Rg2, and Rh1 from Ginseng Root Extract. J. Biosci. Bioeng. 2015, 119, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, J.; Li, C.; Zheng, M.; He, Z.; Zou, Y.; Xiong, H.; Xu, B.; Xiang, W.; Tang, J. Characterization and Bioactive Potential of Carotenoid Lutein from Gordonia rubripertncta GH-1 Isolated from Traditional Pixian Douban. Foods 2022, 11, 3649. [Google Scholar] [CrossRef] [PubMed]
- Nahurira, R.; Wang, J.; Yan, Y.; Jia, Y.; Fan, S.; Khokhar, I.; Eltoukhy, A. In Silico Genome Analysis Reveals the Metabolic Versatility and Biotechnology Potential of a Halotorelant Phthalic Acid Esters Degrading Gordonia alkanivorans Strain YC-RL2. AMB Expr. 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Fusconi, R.; Maria Nascimento Assunção, R.; De Moura Guimarães, R.; Rodrigues Filho, G.; Eduardo Da Hora Machado, A. Exopolysaccharide Produced by Gordonia polyisoprenivorans CCT 7137 in GYM Commercial Medium and Sugarcane Molasses Alternative Medium: FT-IR Study and Emulsifying Activity. Carbohydr. Polym. 2010, 79, 403–408. [Google Scholar] [CrossRef]
- Nazina, T.N.; Sokolova, D.S.; Grigor’ian, A.A.; Xue, Y.F.; Beliaev, S.S.; Ivanov, M.V. Production of oil-processing compounds by microorganisms from the Daqing oil field, China. Mikrobiologiia 2003, 72, 206–211. [Google Scholar]
- Delegan, Y.; Sargsyan, A.; Hovhannisyan, N.; Babayan, B.; Petrikov, K.; Vainstein, M. Analysis of Genome Sequence and Trehalose Lipid Production Peculiarities of the Thermotolerant Gordonia Strain. J. Basic Microbiol. 2020, 60, 14–21. [Google Scholar] [CrossRef]
- Mikolasch, A.; Hammer, E.; Schauer, F. Synthesis of Imidazol-2-Yl Amino Acids by Using Cells from Alkane-Oxidizing Bacteria. Appl. Environ. Microbiol. 2003, 69, 1670–1679. [Google Scholar] [CrossRef]
- Franzetti, A.; Caredda, P.; Ruggeri, C.; Colla, P.L.; Tamburini, E.; Papacchini, M.; Bestetti, G. Potential Applications of Surface Active Compounds by Gordonia sp. Strain BS29 in Soil Remediation Technologies. Chemosphere 2009, 75, 801–807. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, M.; Liu, H.; Yu, T.; Guo, P.; Liu, W.; Jin, X. Antimicrobial Compounds Were Isolated from the Secondary Metabolites of Gordonia, a Resident of Intestinal Tract of Periplaneta americana. AMB Expr. 2021, 11, 111. [Google Scholar] [CrossRef]
- Claverías, F.P.; Undabarrena, A.; González, M.; Seeger, M.; Cámara, B. Culturable Diversity and Antimicrobial Activity of Actinobacteria from Marine Sediments in Valparaíso Bay, Chile. Front. Microbiol. 2015, 6, 737. [Google Scholar] [CrossRef]
- Liu, W.; Li, E.; Liu, L.; Tian, F.; Luo, X.; Cai, Y.; Wang, J.; Jin, X. Antifungal Activity of Compounds from Gordonia sp. WA8-44 Isolated from the Gut of Periplaneta americana and Molecular Docking Studies. Heliyon 2023, 9, e17777. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Marett, L.; Hughen, R.W.; Flores, M.; Forteza, I.; Ammon, M.A.; Concepcion, G.P.; Espino, S.; Olivera, B.M.; Rosenberg, G.; et al. Neuroactive Diol and Acyloin Metabolites from Cone Snail-Associated Bacteria. Bioorg. Med. Chem. Lett. 2013, 23, 4867–4869. [Google Scholar] [CrossRef] [PubMed]
- Graffius, S.; Garzón, J.F.G.; Zehl, M.; Pjevac, P.; Kirkegaard, R.; Flieder, M.; Loy, A.; Rattei, T.; Ostrovsky, A.; Zotchev, S.B. Secondary Metabolite Production Potential in a Microbiome of the Freshwater Sponge Spongilla lacustris. Microbiol. Spectr. 2023, 11, e04353-22. [Google Scholar] [CrossRef]
- Silva, T.P.; Paixão, S.M.; Fernandes, A.S.; Roseiro, J.C.; Alves, L. New Insights on Carotenoid Production by Gordonia alkanivorans Strain 1B. In Physiology; María Martínez-Espinosa, R., Ed.; IntechOpen: London, UK, 2022; Volume 16, ISBN 978-1-80355-423-5. [Google Scholar]
- Yang, Y.; Zhang, W.; Zhang, Z.; Yang, T.; Xu, Z.; Zhang, C.; Guo, B.; Lu, W. Efficient Bioremediation of Petroleum-Contaminated Soil by Immobilized Bacterial Agent of Gordonia alkanivorans W33. Bioengineering 2023, 10, 561. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, M.; Paixão, S.M.; Silva, T.P.; Alves, L. On the Road to Cost-Effective Fossil Fuel Desulfurization by Gordonia alkanivorans Strain 1B. RSC Adv. 2019, 9, 25405–25413. [Google Scholar] [CrossRef] [PubMed]
- Alves, L.; Marques, S.; Matos, J.; Tenreiro, R.; Gírio, F.M. Dibenzothiophene Desulfurization by Gordonia alkanivorans Strain 1B Using Recycled Paper Sludge Hydrolyzate. Chemosphere 2008, 70, 967–973. [Google Scholar] [CrossRef]
- Jaishankar, J.; Singh, P.; Srivastava, P. Draft Genome Sequence of a Biodesulfurizing Bacterium, Gordonia sp. Strain IITR100. Genome Announc. 2017, 5, e00230-17. [Google Scholar] [CrossRef]
- Delegan, Y.; Kocharovskaya, Y.; Frantsuzova, E.; Streletskii, R.; Vetrova, A. Characterization and Genomic Analysis of Gordonia alkanivorans 135, a Promising Dibenzothiophene-Degrading Strain. Biotechnol. Rep. 2021, 29, e00591. [Google Scholar] [CrossRef]
- Frantsuzova, E.; Delegan, Y.; Bogun, A.; Sokolova, D.; Nazina, T. Comparative Genomic Analysis of the Hydrocarbon-Oxidizing Dibenzothiophene-Desulfurizing Gordonia Strains. Microorganisms 2022, 11, 4. [Google Scholar] [CrossRef]
- Zhou, X.; Tang, J.; Wang, S.; Zhang, Y.; Ye, H.; Zhang, Q.; Xiang, W.; Cai, T.; Zeng, C. Whole Genome Sequencing and Transcriptomics-Based Characterization of a Novel β-Cypermethrin-Degrading Gordonia alkanivorans GH-1 Isolated from Fermented Foods. Chemosphere 2023, 320, 138017. [Google Scholar] [CrossRef]
- Bertani, G. Studies on Lysogenesis I: The Mode of Phage Liberation by Lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.; Lim, J.; Kwon, S.; Chun, J. A Large-Scale Evaluation of Algorithms to Calculate Average Nucleotide Identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Orata, F.D.; Meier-Kolthoff, J.P.; Sauvageau, D.; Stein, L.Y. Phylogenomic Analysis of the Gammaproteobacterial Methanotrophs (Order Methylococcales) Calls for the Reclassification of Members at the Genus and Species Levels. Front. Microbiol. 2018, 9, 3162. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Lu, F.; Luo, Y.; Bie, L.; Xu, L.; Wang, Y. OrthoVenn3: An Integrated Platform for Exploring and Visualizing Orthologous Data across Genomes. Nucleic Acids Res. 2023, 51, W397–W403. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Nazarova, E.A.; Kiryanova, T.D.; Egorova, D.O. Diversity of the Gene of Benzoate Dioxygenase in Bacterial Associations Isolated from Long Term Organochlorine-Contaminated Soils. Ecol. Genet. 2019, 17, 13–22. [Google Scholar] [CrossRef]
- Field, J.A.; Sierra-Alvarez, R. Microbial Transformation of Chlorinated Benzoates. Rev. Environ. Sci. Biotechnol. 2008, 7, 191–210. [Google Scholar] [CrossRef]
- Leewis, M.-C.; Uhlik, O.; Leigh, M.B. Synergistic Processing of Biphenyl and Benzoate: Carbon Flow Through the Bacterial Community in Polychlorinated-Biphenyl-Contaminated Soil. Sci. Rep. 2016, 6, 22145. [Google Scholar] [CrossRef]
- UniProt: Coda—Choline Oxidase. Available online: https://www.uniprot.org/uniprotkb/Q7X2H8/entry (accessed on 12 October 2023).
- Breisch, J.; Bendel, M.; Averhoff, B. The Choline Dehydrogenase BetA of Acinetobacter baumannii: A Flavoprotein Responsible for Osmotic Stress Protection. Environ. Microbiol. 2022, 24, 1052–1061. [Google Scholar] [CrossRef]
- Loll, P.J.; Upton, E.C.; Nahoum, V.; Economou, N.J.; Cocklin, S. The High Resolution Structure of Tyrocidine A Reveals an Amphipathic Dimer. Biochim. Biophys. Acta BBA Biomembr. 2014, 1838, 1199–1207. [Google Scholar] [CrossRef]
- Habe, H.; Chung, J.-S.; Lee, J.-H.; Kasuga, K.; Yoshida, T.; Nojiri, H.; Omori, T. Degradation of Chlorinated Dibenzofurans and Dibenzo-p-Dioxins by Two Types of Bacteria Having Angular Dioxygenases with Different Features. Appl. Environ. Microbiol. 2001, 67, 3610–3617. [Google Scholar] [CrossRef] [PubMed]
- Solyanikova, I.P.; Emelyanova, E.V.; Shumkova, E.S.; Egorova, D.O.; Korsakova, E.S.; Plotnikova, E.G.; Golovleva, L.A. Peculiarities of the Degradation of Benzoate and Its Chloro- and Hydroxy-Substituted Analogs by Actinobacteria. Int. Biodeterior. Biodegrad. 2015, 100, 155–164. [Google Scholar] [CrossRef]
- Feng, L.; Wang, W.; Cheng, J.; Ren, Y.; Zhao, G.; Gao, C.; Tang, Y.; Liu, X.; Han, W.; Peng, X.; et al. Genome and Proteome of Long-Chain Alkane Degrading Geobacillus thermodenitrificans NG80-2 Isolated from a Deep-Subsurface Oil Reservoir. Proc. Natl. Acad. Sci. USA 2007, 104, 5602–5607. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, M.; Hosseini, A. Streptozotocin. In Encyclopedia of Toxicology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 402–404. ISBN 978-0-12-386455-0. [Google Scholar]
- Prado-Alonso, L.; Pérez-Victoria, I.; Malmierca, M.G.; Montero, I.; Rioja-Blanco, E.; Martín, J.; Reyes, F.; Méndez, C.; Salas, J.A.; Olano, C. Colibrimycins, Novel Halogenated Hybrid Polyketide Synthase-Nonribosomal Peptide Synthetase (PKS-NRPS) Compounds Produced by Streptomyces sp. Strain CS147. Appl. Environ. Microbiol. 2022, 88, e01839-21. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Asano, D.; Sawamura, M.; In, Y.; Ishida, T.; Imoto, M. Ulbactins F and G, Polycyclic Thiazoline Derivatives with Tumor Cell Migration Inhibitory Activity from Brevibacillus sp. Org. Lett. 2016, 18, 1658–1661. [Google Scholar] [CrossRef]
- Melinda, Y.N.; Widada, J.; Wahyuningsih, T.D.; Febriansah, R.; Damayanti, E.; Mustofa, M. Metabologenomics Approach to the Discovery of Novel Compounds from Streptomyces sp. GMR22 as Anti-SARS-CoV-2 Drugs. Heliyon 2021, 7, e08308. [Google Scholar] [CrossRef]
- Park, S.R.; Tripathi, A.; Wu, J.; Schultz, P.J.; Yim, I.; McQuade, T.J.; Yu, F.; Arevang, C.-J.; Mensah, A.Y.; Tamayo-Castillo, G.; et al. Discovery of Cahuitamycins as Biofilm Inhibitors Derived from a Convergent Biosynthetic Pathway. Nat. Commun. 2016, 7, 10710. [Google Scholar] [CrossRef]
- Dose, B.; Niehs, S.P.; Scherlach, K.; Flórez, L.V.; Kaltenpoth, M.; Hertweck, C. Unexpected Bacterial Origin of the Antibiotic Icosalide: Two-Tailed Depsipeptide Assembly in Multifarious Burkholderia Symbionts. ACS Chem. Biol. 2018, 13, 2414–2420. [Google Scholar] [CrossRef]
- Jenner, M.; Jian, X.; Dashti, Y.; Masschelein, J.; Hobson, C.; Roberts, D.M.; Jones, C.; Harris, S.; Parkhill, J.; Raja, H.A.; et al. An Unusual Burkholderia gladioli Double Chain-Initiating Nonribosomal Peptide Synthetase Assembles Fungal Icosalide Antibiotics. Chem. Sci. 2019, 10, 5489–5494. [Google Scholar] [CrossRef]
- Dangi, A.; Pande, B.; Agrawal, S.; Sarkar, D.; Vamkudoth, K.R.; Marelli, U.K. Total Synthesis, Structure Elucidation and Expanded Bioactivity of Icosalide A: Effect of Lipophilicity and Ester to Amide Substitution on Its Bioactivity. Org. Biomol. Chem. 2023, 21, 5725–5731. [Google Scholar] [CrossRef]
- Koomsiri, W.; Inahashi, Y.; Leetanasaksakul, K.; Shiomi, K.; Takahashi, Y.; Omura, S.; Samborskyy, M.; Leadlay, P.F.; Wattana-Amorn, P.; Thamchaipenet, A.; et al. Sarpeptins A and B, Lipopeptides Produced by Streptomyces sp. KO-7888 Overexpressing a Specific SARP Regulator. J. Nat. Prod. 2019, 82, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jin, D.; Zhou, L.; Wu, L.; An, W.; Zhao, L. Draft Genome Sequence of Gordonia alkanivorans Strain CGMCC6845, a Halotolerant Hydrocarbon-Degrading Bacterium. Genome Announc. 2014, 2, e01274-13. [Google Scholar] [CrossRef] [PubMed]
- Goel, D.; Singh, A.K.; Yadav, V.; Babbar, S.B.; Murata, N.; Bansal, K.C. Transformation of Tomato with a Bacterial codA Gene Enhances Tolerance to Salt and Water Stresses. J. Plant Physiol. 2011, 168, 1286–1294. [Google Scholar] [CrossRef] [PubMed]



| Strain | Collection Number | Isolation Source | Previously Identified As |
|---|---|---|---|
| 96 | IEGM 96 | crude oil-contaminated soil, Ukraine | G. rubripertincta |
| 129 | IEGM 129 | crude oil-contaminated soil, Ivano-Frankovsk, Ukraine | G. rubripertincta |
| 132 | IEGM 132 | crude oil-contaminated soil, Ivano-Frankovsk, Ukraine | G. rubripertincta |
| 133 | IEGM 133 | crude oil-contaminated soil, Ivano-Frankovsk, Ukraine | G. rubripertincta |
| 134 | IEGM 134 | crude oil-contaminated soil, oilfield, Lvov, Ukraine | G. rubripertincta |
| 142 | IEGM 142 | crude oil-contaminated soil, Ukraine | G. rubripertincta |
| 144 | IEGM 144 | crude oil-contaminated soil, Lvov, Ukraine | G. terrae |
| 12 | - | oil-polluted soil, Moscow, Russia | G. alkanivorans |
| 152 | - | oil-polluted soil, Moscow, Russia | G. alkanivorans |
| Strain | Read Pairs before Filtration | Read Pairs after Filtration | Read Pairs after Filtration (%) |
|---|---|---|---|
| 96 | 2,962,813 | 2,835,521 | 95.70 |
| 129 | 3,147,953 | 3,003,848 | 95.42 |
| 132 | 4,354,967 | 4,162,088 | 95.57 |
| 133 | 5,192,473 | 5,019,737 | 96.67 |
| 134 | 5,815,565 | 5,558,107 | 95.57 |
| 142 | 3,330,918 | 3,171,608 | 95.22 |
| 144 | 5,382,881 | 5,150,561 | 95.68 |
| 12 | 9,985,963 | 8,692,673 | 87.05 |
| 152 | 6,985,286 | 4,928,247 | 70.55 |
| Strain | ANI Value with the Type Strain of G. rubripertincta, % | DDH Value with the Type Strain of G. rubripertincta, % | ANI Value with the Type Strain of G. alkanivorans, % | DDH Value with the Type Strain of G. alkanivorans, % | Taxonomic Position of the Strain |
|---|---|---|---|---|---|
| 96 | 92.58 | 76.20 | 98.45 | 89.70 | G. alkanivorans |
| 129 | 92.45 | 76.00 | 98.42 | 88.60 | G. alkanivorans |
| 132 | 92.40 | 76.30 | 98.42 | 88.60 | G. alkanivorans |
| 133 | 92.40 | 76.10 | 98.37 | 88.50 | G. alkanivorans |
| 134 | 92.25 | 76.80 | 98.23 | 87.60 | G. alkanivorans |
| 142 | 92.54 | 76.30 | 98.23 | 91.00 | G. alkanivorans |
| 144 | 92.42 | 66.80 | 98.63 | 78.10 | G. alkanivorans |
| 12 | 92.38 | 76.20 | 98.40 | 90.10 | G. alkanivorans |
| 152 | 92.48 | 76.60 | 98.34 | 88.00 | G. alkanivorans |
| Strain | Alkanes C10–C16 | Alkanes C18–C20 | Benzoate | Phenol | Naphthalene | Catechol | DBT |
|---|---|---|---|---|---|---|---|
| 96 | ++ | + | + | - | - | - | ± |
| 129 | + | + | + | - | - | - | - |
| 132 | + | + | + | - | - | - | ± |
| 133 | + | + | + | - | - | - | - |
| 134 | ++ | ++ | + | - | - | - | - |
| 142 | + | + | + | - | - | - | - |
| 144 | + | + | + | - | - | - | - |
| 12 | ++ | + | + | - | - | - | ± |
| 152 | ++ | + | + | - | - | - | - |
| 135 | ++ | ++ | + | - | - | - | + |
| Strain | betA/codA | betB | betT | opuA | opuBD | opuC |
|---|---|---|---|---|---|---|
| 96 | 1 | 4 | 1 | 1 | 2 | 2 |
| 129 | - | 5 | 1 | 1 | 3 | 2 |
| 132 | 1 | 3 | 1 | 1 | 2 | 2 |
| 133 | 1 | 3 | 1 | 1 | 2 | 2 |
| 134 | 1 | 3 | 1 | 1 | 2 | 2 |
| 142 | 1 | 4 | 1 | 1 | 2 | 2 |
| 144 | 1 | 3 | 1 | 1 | 2 | 2 |
| 12 | 1 | 4 | 1 | 1 | 2 | 2 |
| 152 | 1 | 3 | 1 | 1 | 2 | 2 |
| Strain | Concentration of NaCl | |||
|---|---|---|---|---|
| 1% | 3% | 7% | 10% | |
| 96 | + | + | + | + |
| 129 | + | ± | ± | - |
| 132 | + | + | - | - |
| 133 | + | + | ± | ± |
| 134 | + | + | - | - |
| 142 | + | + | - | - |
| 144 | + | ± | ± | - |
| 12 | + | + | - | - |
| 152 | + | + | ± | - |
| Strain | Proteins | Clusters | Singletons |
|---|---|---|---|
| 96 | 4446 | 4318 | 61 |
| 129 | 4380 | 4252 | 22 |
| 132 | 4521 | 4404 | 1 |
| 133 | 4520 | 4399 | 4 |
| 134 | 4587 | 4312 | 60 |
| 142 | 4530 | 4329 | 63 |
| 144 | 5390 | 4506 | 315 |
| 12 | 4480 | 4522 | 25 |
| 152 | 4421 | 4183 | 88 |
| NBRC 16433 | 4445 | 4155 | 109 |
| Strain | CYP153 |
|---|---|
| 96 | 2 |
| 129 | 1 |
| 132 | 3 |
| 133 | 3 |
| 134 | 2 |
| 142 | 2 |
| 144 | 3 |
| 12 | 2 |
| 152 | 2 |
| Strain | Most Similar Known Cluster | Similarity |
|---|---|---|
| 96 | sarpeptin | 25% |
| 129 | ulbactin echoside | 28% 11% |
| 144 | cahuitamycin | 12% |
| 12 | leucomycin | 3% |
| 152 | icosalide | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frantsuzova, E.; Bogun, A.; Shishkina, L.; Vetrova, A.; Solyanikova, I.; Delegan, Y. Insights into the Potential Role of Gordonia alkanivorans Strains in Biotechnologies. Processes 2023, 11, 3184. https://doi.org/10.3390/pr11113184
Frantsuzova E, Bogun A, Shishkina L, Vetrova A, Solyanikova I, Delegan Y. Insights into the Potential Role of Gordonia alkanivorans Strains in Biotechnologies. Processes. 2023; 11(11):3184. https://doi.org/10.3390/pr11113184
Chicago/Turabian StyleFrantsuzova, Ekaterina, Alexander Bogun, Lidiya Shishkina, Anna Vetrova, Inna Solyanikova, and Yanina Delegan. 2023. "Insights into the Potential Role of Gordonia alkanivorans Strains in Biotechnologies" Processes 11, no. 11: 3184. https://doi.org/10.3390/pr11113184





