1. Introduction
Plant-based meat analogues are designed to provide an alternative to traditional meat products and mimic the taste, texture, and appearance of meat. These products have gained popularity due to concerns about the environmental impact of meat production, as well as health and ethical considerations. Soy leghemoglobin (LegH) is a small 16 kDa holoprotein (i.e., a protein plus a heme cofactor) found in soy plants that has a similar structure to heme in animal meat (a molecule found in animal blood and muscle tissue, responsible for the red color of meat and carrying oxygen in the blood). It is sometimes used as a meat flavoring and colorant in plant-based meat alternatives to create a more authentic meat-like taste, texture, and a reddish hue [
1,
2].
The Impossible Burger is a plant-based burger made by Impossible Foods Inc. (Redwood City, CA, USA), a company that specializes in developing and producing meat alternatives. The burger is made using a combination of plant-based ingredients, including soy and potato protein, coconut and sunflower oil, cellulose-based culinary binder, water, and the secret ingredient—soy leghemoglobin [
1]. The LegH molecule, expressed in recombinant yeast
Pichia pastoris, is what gives the burger its meat-like taste and aroma [
3,
4]. The Impossible Burger has gained popularity due to its close resemblance to traditional beef burgers in terms of taste, texture, and appearance. It is also marketed as a more sustainable and ethical alternative to beef, as it requires less land, water, and other resources to produce.
LegH is a naturally occurring molecule found in animal tissue and certain microorganisms. However, assessing its safety becomes crucial when produced through genetic engineering for use in food products. Extensive studies on LegH as a food ingredient indicate minimal toxicological or allergic concerns [
4,
5,
6], leading regulatory agencies in the United States and other countries to deem it safe for consumption [
7,
8]. However, approval in the EU and UK is still pending.
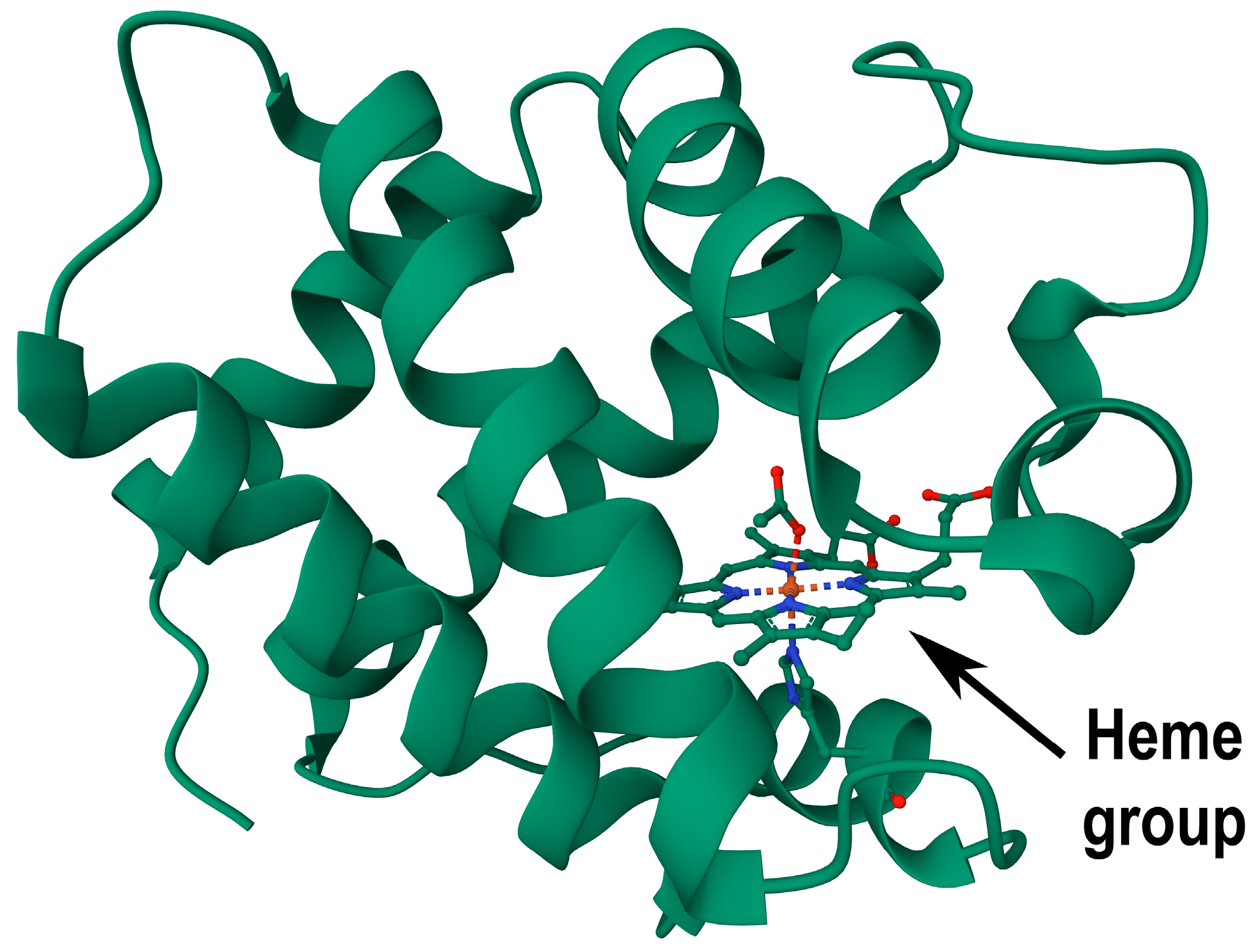
LegH, a single-unit hemoprotein present in leguminous plant root nodules, shares a three-dimensional structure akin to myoglobin, a hemoprotein in mammalian muscles. Its protein structure, mainly comprising alpha helices that form a stable framework, features eight helices creating a distinct pocket for heme binding [
9]. In contrast to mammalian globin with four subunits, plant LegH comprises a single monomeric unit [
10]. See
Figure 1 for the structure of a soybean LegH molecule.
Several microorganisms have successfully produced recombinant soy LegH. Recently, Shao et al. developed a high-yield secretion system for functional LegH expression using a
P. pastoris yeast strain, through gene dosage optimization and heme pathway consolidation. These strategies increased LegH secretion by more than 83-fold, resulting in a maximum titer of 3.5 g/L, which is the highest ever reported for a secretory production of not only LegH, but also all heme-containing proteins [
13]. Xue et al. reported the production of several heme proteins in the yeast
Saccharomyces cerevisiae. An engineered
S. cerevisiae strain produced a titer of 108.2 mg/L soybean LegH [
14]. Recombinant LegH production has also been reported in bacterial
Escherichia coli cells [
11,
15]. Jones et al. managed to achieve a yield of 20 mg/L pure product, after the optimization of the growth conditions in shake flasks [
16].
Over the last two to three decades, the
P. pastoris expression system has proven its efficacy in generating a diverse range of recombinant proteins for both research and industrial purposes. This methylotrophic yeast stands out as an excellent choice for expressing foreign proteins, thanks to its straightforward genetic manipulation, high-frequency DNA transformation, functional complementation cloning, robust intra- and extracellular protein expression capabilities, and proficiency in executing complex higher eukaryotic protein modifications such as glycosylation, disulfide bond formation, and proteolytic processing. Additionally, the low levels of native secreted proteins simplify the purification of the expressed recombinant proteins. When considering economic factors like high cell growth in minimal medium, prolonged process stability, and the availability of potent genetic techniques,
P. pastoris undeniably emerges as the preferred system for heterologous protein expression [
17,
18].
The typical
P. pastoris two-stage cultivation process is described in Invitrogen’s “
Pichia Fermentation Process Guidelines” [
19]. This procedure comprises growing
P. pastoris cells in a minimal medium, first using glycerol as a growth substrate until a suitable biomass concentration is reached, then inducing product biosynthesis by switching the substrate feed to methanol. Recent developments indicate a shift away from conventional protocols, embracing a more conceptual approach that enables the customization of process-specific strategies based on the unique attributes of the product/genetic construct and the equipment in the bioreactor [
20].
Invitrogen Co.’s basal salt medium (BSM) stands out as the frequently employed minimal medium for achieving high cell density fermentation of the methylotrophic yeast P. pastoris. Despite its status as a standard medium, it may not be universally optimal and is known to exhibit certain drawbacks, including an unbalanced composition, precipitate formation, and issues related to ionic strength [
21]. To circumvent the aforementioned problems, optimization of the BSM medium components is often carried out; however, this can be time and labor intensive. Therefore, opting for a previously developed medium may be preferable, as several formulations have been reported. For example, the FM22 medium by Stratton et al. [
22] or the D’Anjou medium [
23]. More recent minimal medium formulations include the rich defined medium (RDM) by Matthews et al. [
24] and the MBSM medium by Pais-Chanfrau et al. [
25]. Several authors have also reported that, reducing the salt concentrations of BSM may prove beneficial for recombinant product synthesis, while having little to no effect on cell growth [
26,
27].
The use of complex cultivation medium (a nutrient-rich medium that contains a variety of undefined components such as yeast extract, peptone), can sometimes produce better results than the minimal (defined) medium. However, the use of complex medium can make it difficult to control and optimize the growth conditions, result in batch-to-batch variability, and is generally much more expensive. The buffered glycerol complex medium (BMGY) is often employed in
P. pastoris cultivations and is the go-to complex medium [
24,
26].
When analyzing the formulation of a
P. pastoris cultivation medium, the study conducted by Wegner often serves as a benchmark [
28]. In this study, the optimal ranges of important elements for cell growth (P, K, Mg, Ca, S, Fe, Zn, Cu, Mn) were determined experimentally in a continuous fermentation.
Artificial neural networks (ANNs) are computational algorithms designed to emulate the structure of biological brain networks, enabling the estimation and prediction of bioprocess variables using real-time sensor data, offering the capability to model intricate nonlinear systems without intricate model equations, although they necessitate substantial historical process data for precise network training and establishing the connections between input and output parameters [
29]. While ANNs are typically efficient and easy to deploy with strong performance, their drawback lies in their lack of interpretability, leading to a restricted acquisition of process knowledge. Despite this limitation, ANNs have demonstrated success in predicting the behavior of diverse fermentation systems, prompting their utilization in bioprocess control applications. Recent applications of ANN models in cell biomass estimation, encompass regulating specific growth rate [
30], optimizing cell biomass [
31,
32,
33], and estimating [
33,
34] or tracking a predefined substrate concentration trajectory [
35].
In this study, we explored the expression of recombinant LegH in P. pastoris using various documented cultivation media (BSM, BMGY, FM22, D’Anjou, BSM/2, RDM) and employed different feeding strategies (µ-stat and mixed feed with sorbitol). Generated process data were used to establish and train a novel artificial neural network-based soft sensor for cell biomass estimation, utilizing only standard bioreactor measurements (stirrer speed, dissolved oxygen, O2 enrichment, base feed, glycerol feed, methanol feed, and reactor volume).
2. Materials and Methods
2.1. Construction of an Expression Vector and Selection of Clones
An artificial gene with P. pastoris optimized codons encoding LegH sequence (GenBank Acc. NP_001235248.2) was designed by GenScript and synthesized by BioCat GmbH (Heidelberg, Germany). This gene was subsequently incorporated into the pPICZC vector (Invitrogen) through EcoRI and NotI restriction sites. The resulting plasmid underwent linearization with PmeI and was introduced into the P. pastoris X-33 strain through electroporation. Mut+ transformants were successfully obtained on agarized YPD plates containing 800 µg/mL zeocin, and the selected clones were further cultivated analytically in flasks using the rich BMGY medium with methanol induction over three days to identify the most efficient producer.
2.2. Experimental Conditions
A recombinant P. pastoris X-33 Mut+ strain was used for the cultivation processes. The bioreactor vessel was filled with distilled water and subjected to sterilization at 121 °C for 30 min. Simultaneously, the cultivation media and glycerol fed-batch solutions underwent separate autoclaving under identical conditions. The trace element, vitamin, and methanol fed-batch solutions were sterilized through filtration using a 0.2 µm filter.
The fermentations were carried out in a 5 L bench-top fermenter (Bioreactors.net, EDF-5.4/BIO-4, Riga, Latvia) with a working volume ranging from 2 to 4 L, as illustrated in
Figure 2. The pH levels were monitored using a calibrated pH sensor probe (Hamilton, EasyFerm Bio, Bonaduz, Switzerland) and adjusted to 5.0 ± 0.1 before initiating cultivation, maintaining the set value throughout fermentation using a 28% NH
4OH solution. Temperature control was set at 30.0 ± 0.1 °C, regulated by a temperature sensor and adjustments to the vessel jacket temperature. Dissolved oxygen (DO) levels were measured with a DO probe (Hamilton, Oxyferm Bio, Bonaduz, Switzerland) and kept above 30 ± 5% by modulating stirrer speed (200–1000 RPM) in Cascade 1 or enriching the inlet air with pure O
2 in Cascade 2. A consistent flow of air or an air/oxygen mixture at 3.0 slpm was maintained throughout the process. A condenser was employed to condense moisture from outlet gases, and antifoam 204 (Sigma, St. Louis, MO, USA) was added when needed to manage excessive foam formation. Substrate feed solutions were pumped using a high-precision peristaltic pump (Longer-Pump, BT100–2J, Baoding, China). A turbidity probe (Optek, ASD19- EB-01, Kitzingen, Germany) was employed in Experiments 1 and 9. Sensor signal was converted to wet cell biomass (g/L), according to a previously established correlation [
36].
The cultivation commenced with a glycerol batch phase. After 18–24 h, once the batch glycerol was exhausted, indicated by a sudden DO spike, a glycerol fed-batch solution was introduced into the reactor at a rate of 0.61 mL/min for 4 h or until reaching an optical density of 100–120. Subsequently, a brief feeding pause of 10–30 min allowed cells to consume any residual glycerol. Following this, the substrate feed transitioned to methanol, supplied to the reactor at a rate of 0.12 mL/min for 5 h, followed by 0.24 mL/min for 2 h, and finally 0.36 mL/min for the remainder of the cultivation.
In the mixed feed cultivation (Experiment 10), a mixture of methanol/sorbitol at a ratio of 0.5 C-mol/0.5 C-mol was used, according to Niu et al. [
37]. The feed rate profile was the same as in previous cultivations; however, after the stirrer (Cascade 1) reached 1000 RPM, DO-stat feeding (Cascade 2) was activated instead of O
2 enrichment.
2.3. Cultivation Media
In order to evaluate the effect that the cultivation medium has on recombinant LegH biosynthesis in yeast
P. pastoris, several reported minimal media formulations were selected. Namely, Invitrogen’s BSM [
19], FM22 medium reported by Stratton et al. [
22], D’Anjou medium [
23], BSM with the salt concentration reduced by half (denoted as BSM/2) [
26,
27], and the RDM without the addition of lipids reported by Matthews et al. [
24]. In order to compare the performance of minimal and complex media, one experiment was carried out in BMGY medium. The compositions of the previously mentioned media are shown in
Table A1 (
Appendix A).
2.4. Downstream Processing of LegH
A total of 7.0 g of wet cells were resuspended in 35 mL of lysis buffer (20 mM Tris 8.0, 100 mM NaCl) and disrupted by French press (3 × 10,000 psi). The suspension was then centrifuged for 30 min at 18,500× g and the supernatant was buffer exchanged to 20 mM Tris 8.0 on XK26/20 column packed with 60 mL of Sephadex G-25 at 5 mL/min. Proteins were then loaded onto XK16/20 column packed with 20 mL of Sepharose Q HP equilibrated with 20 mM Tris 8.0. Bound proteins were eluted with a salt gradient using 20 mM Tris 8.0, 1 M NaCl at 3 mL/min. Finally, fractions containing the target protein were loaded onto XK16/70 column packed with 120 mL of Superdex 200 in PBS at 1 mL/min. All the columns were purchased from Cytiva. The first two processes were operated by Akta Pure 25, while the third was processed by Akta Prime Plus.
2.5. Analytical Measurements
Cell growth was monitored through offline measurements of wet cell weight (WCW), determined gravimetrically. Biomass samples were placed in pre-weighed Eppendorf® tubes and centrifuged at 15,500× g for 3 min. Subsequently, the supernatant was discarded, and the cells were resuspended in distilled water before undergoing another round of centrifugation. The liquid phase was discarded, and the remaining wet cell biomass was then weighed.
Protein samples collected during cultivation underwent analysis through sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), employing a 5% stacking and 15% separating polyacrylamide gel (PAAG), following established protocols. To visualize the distinct protein bands, the gels were stained with 0.4% Coomassie Brilliant Blue G-250 dye.
LegH amount was estimated by Coomassie-stained PAAG, using protein concentration standards. Relative proportions of target protein outcome after purification were calculated by measuring peak squares after size-exclusion chromatography.
2.6. ANN-Based Cell Biomass Soft Sensor Development
The ANN-based cell biomass soft sensor was developed in MATLAB R2021b, using the Neural Net Fitting toolbox. The cell biomass dataset (12,631 entries) generated from turbidity sensor measurements in Experiments 1 and 9, were used as the response variables. Corresponding recorded process data of stirrer speed (RPM), dissolved oxygen (%), O
2 enrichment (%), pumped base (mL), glycerol feed (mL), methanol feed (mL), and reactor volume (L) were used as the predictor variables. Then, 70% of data were used for neural network training, 15% for testing, and 15% for validation. A two-layer feedforward network with 10 sigmoid hidden neurons and 1 linear output neuron, schematically illustrated in
Figure 3, was selected for training.
The network was trained using the Levenberg–Marquard training algorithm until 6 consecutive validation checks were failed. The model was then exported to MATLAB workspace and used for cell biomass concentration estimation.
Cultivation parameters are often influenced by external factors or signal noise; hence, signal filtering methods are popular in bioengineering. To reduce the sudden jumps and noise of the developed biomass soft sensor, a Savitzky–Golay filter was used with an order of 1 and frame length of 29. This filter significantly reduced sudden signal jumps, noise, and improved the overall performance of the sensor.
3. Results
3.1. Clone Selection
To select the best producer cells, eight zeocin-resistant clones were cultivated in flasks with rich BMGY medium, and LegH synthesis level was assessed three days post methanol induction. A product of predicted molecular mass was detectable for all clones compared (
Figure 4A). Although LegH synthesis level is well detectable in all cases, it varies from clone to clone; however, the best production can be noted in clone No 3. Henceforth, this producer strain was selected for further investigation.
Flask experiments were also used to investigate the optimal LegH expression temperature. Inducing biosynthesis at a lowered temperature is a popular strategy to improve protein yield in some cases; therefore, two induction temperatures—24 and 30 °C were investigated. The Coomassie-stained PAAG from these experiments can be seen in (
Figure 4B). Thicker LegH bands can be noted at an expression temperature of 30 °C; hence, this temperature was used during induction in consequent bioreactor experiments.
3.2. Cultivation Experiments
In order to establish a standardized bioreactor process, we cultured the chosen producer cells in Invitrogen’s classical BSM medium five days after induction, and then monitored LegH synthesis levels at various time intervals using SDS–PAGE. For comparison, cultivation in complex BMGY medium was also carried out, according to the same protocol, in order to compare the productivity between minimal and complex media. The cultivation process parameters during these experiments are presented in
Figure 5 and the LegH accumulation dynamics in post induction samples, visualized by Coomassie-stained PAAG, are shown in
Figure 6.
According to the PAAG from Experiment 1 in
Figure 6, it can be noted that the thickness of LegH band increases in the 7 h, 24 h, and 48 h samples post methanol induction. In the remaining samples, the increase is insignificant and difficult to observe. After performing sample purification, a LegH concentration of 1.56 mg/g wet cells is reached after 48 h on methanol. Although, the maxima of synthesis (1.62 mg/g) was reached on the fifth day of post methanol induction (120 h sample), the increase in specific product yield was only gradual. Therefore, the time point of 48 h after methanol induction was used in further experiments to compare the efficiency of LegH biosynthesis in different reported cultivation media.
The cultivation in rich BMGY medium was carried out, according to the same Invitrogen cultivation protocol and continued for 48 h after methanol induction. In this cultivation, an even higher yield of LegH—1.77 mg/g wet cells was achieved, indicating that the cultivation medium might have a significant effect on LegH productivity. Yet, employing complex (rich) medium in cultivations does present notable drawbacks, particularly on an industrial scale. These include diminished batch-to-batch repeatability stemming from variations in component composition, increased costs, and challenges in product purification. Additionally, the inclusion of meat peptone in the medium formulation raises ethical concerns, given LegH’s primary use in vegan nutrition. Given the well-documented cultivation of P. pastoris on minimal media, our focus shifted exclusively to investigating minimal media.
3.3. Purification
Purification of LegH was processed in three steps. In the first step, excess salt was removed and a pH of 8.0 was established. In the second step, protein was attached to an anion-exchange matrix and eluted by increasing the amount of salt, resulting in removal of major contaminants (
Figure 7A). Then, 1 mL of four major fractions, corresponding to the LegH peak, were taken for further purification and analysis. For final polishing, the four fractions were merged and the protein was passed through a size-exclusion chromatography column, which indicated that the majority of the protein is eluted in a monomeric state according to its molecular weight (
Figure 7B). Moreover, Superdex column peak fractions, in contrast to the anion Q HP fractions, represented at least 90% pure LegH protein (
Figure 7C), which allowed to further use the square of this peak for quantification of target protein and comparison of different cultivation processes. Attachment of the heme group to LegH is proven by the characteristic reddish color of peak protein fractions from the Superdex column (
Figure 7D).
3.4. Reported Cultivation Medium Evaluation
The choice of cultivation medium holds considerable importance in bioprocess development. To explore whether the yield of LegH is impacted by the cultivation media employed, cultivations were conducted in reported FM22, D’Anjou, BSM/2, and RDM media under uniform conditions, following the Invitrogen protocol. The cultivation parameter dynamics during these cultivation experiments are shown in
Figure A1 (
Appendix A).
The purified LegH results from the six cultivation experiments are compiled in
Table 1 and visualized in
Figure 8. The results indicate that the highest LegH productivity was achieved in BMGY medium; however, a slightly lower, but similar yield was noted in BSM. Out of the reported media formulations, the best performance was shown by BSM/2 medium (BSM salt concentrations reduced two times). However, the yields in the reported media were almost two times lower than in BSM or BMGY.
3.5. Experiments to Improve LegH Expression
Since experiments with different cultivation media did not achieve an increased LegH yield, we decided to investigate, whether supplementing the BSM medium with 1 g/L glycine (Experiment 7) or a vitamin solution (Experiment 8) would have an effect on product yield. Additionally, a µ-stat feeding profile (Experiment 9) and mixed substrate (0.5 C-mol methanol/0.5 C-mol sorbitol) feed (Experiment 10) were investigated.
Glycine is an amino acid involved in the heme biosynthesis pathway. In a recent paper, we also hypothesized that upregulating the C1 metabolism pathway in mitochondria to increase glycine synthesis is necessary for improved heme biosynthesis [
38]. Vitamin addition to cultivation media has been shown to improve recombinant product yield in some cases [
24,
39]. To investigate the effect of vitamin addition, we supplemented the BSM medium with the vitamin solution used in the RDM medium formulation.
Inducing recombinant product biosynthesis by methanol mixed feed induction with sorbitol, is a popular strategy to improve recombinant protein yields. To investigate the effect that a mixed substrate feed has on LegH production, a cultivation process was carried out.
According to the popular Luedeking–Piret model, the protein production rate has an empirical relationship with the cell growth rate. There are many reports in the literature of a positive correlation between the specific cell growth rate (µ) and specific target protein production rate (q). To investigate, whether this correlation is also true for LegH, we conducted an experiment, in which we attempted to control the specific cell growth rate of P. pastoris, by manipulating the substrate (methanol) feed during the induction phase.
The cultivation parameters from the aforementioned experiments are presented in
Figure A2 (
Appendix A) and LegH yields in
Table 2.
The BSM medium supplementation with 1 g/L glycine did not yield a positive effect on the LegH synthesis level. The achieved LegH yield of 1.05 mg/g WCW was slightly lower than in Experiment 1; however, a higher cell concentration was achieved.
In the experiment where BSM was enriched with vitamins, we observed a reduced lag phase and a quicker adjustment to methanol uptake. These effects can be attributed to the presence of crucial vitamins that facilitate yeast metabolism. However, no increase in LegH productivity could be noted in this experiment.
In Experiment 9, we attempted to control the specific cell growth rate (µ) at 0.02 h−1 post methanol induction, by varying the methanol feed rate using a PID algorithm-based controller. Soon after initiating µ-stat control, it was noted that the increased feed rate caused a significant increase in metabolic heat production, as the fermentation temperature began to rise. The bioreactor cooling system was unable to maintain the temperature at 30 °C; therefore, the maximum feed rate was adjusted so that the temperature would not exceed 32 °C, which can be detrimental to recombinant protein biosynthesis. Although this restriction led to a lower average specific growth rate of 0.015 h−1, it was still higher than in the typical BSM process (approx. 0.006–0.008 h−1). Based on the findings, no enhancement in LegH productivity was observed in this experiment.
Finally, an experiment (Experiment 10) with mixed substrate induction was carried out. Methanol solution was supplemented with sorbitol at a ratio of 0.5 C-mol methanol/0.5 C-mol sorbitol. Induction was carried out, according to the Invitrogen protocol. This experiment revealed a rapid adjustment to methanol uptake, a well-known occurrence in mixed substrate induction with sorbitol. Although, a higher cell biomass concentration could be noted at the end of the process, the specific LegH productivity was reduced, perhaps due to the lower fraction of methanol in the feed solution.
3.6. ANN-Based Cell Biomass Soft Sensor
In Experiments 1 and 9, we employed an in situ turbidity probe to monitor the real-time growth of P. pastoris cell biomass. This monitoring process produced a substantial dataset, comprising 12,631 entries. This dataset served as the foundation for developing a neural network-based soft sensor for estimating cell biomass. To ensure that no additional expensive sensors are necessary, we exclusively utilized parameters directly measured by the bioreactor system, which included stirrer speed, dissolved oxygen, oxygen enrichment, base pump, feed pump, and reactor volume.
The datasets from previous experiments were used to test and validate the created ANN model. The model was used to calculate WCW values, based on the input data recorded in the experiments. These values were then compared to their corresponding experimentally measured WCW values to determine the model accuracy. The dataset generated by the ANN-based soft sensor and experimental measurements is illustrated in
Figure 9.
As we can see, the developed soft sensor is able to accurately describe cell biomass dynamics in the selected cultivations. A good fit can be noted in almost all experiments.
In Experiment 4, the sensor fails to follow the biomass trajectory, as it overestimates the biomass concentration post induction. This most likely can be explained by the D’Anjou medium used in this experiment, as it significantly differs from other cultivation media. Also, significantly lower cell biomass measurements were registered in the particular cultivation experiment. Taking this into consideration, we can surmise that the developed soft sensor is not entirely applicable for cultivations in these media.
In Experiment 10, it is observed that at the beginning of methanol induction, the soft sensor tends to overestimate the cell biomass concentration. This could be attributed to the influence of sorbitol when co-fed with methanol. The transition phase to methanol uptake is expedited, and cell metabolism resumes more rapidly than usual. Consequently, the sensor overestimates the presumed adapting cell biomass concentration.
To investigate the performance of the ANN-based soft sensor, RMSE and NRMSE values for each experiment and overall accuracy are compiled in
Table 3.
The overall precision of the developed ANN-based soft sensor for cell biomass estimation, is evaluated at ± 13.36 g/L WCW or 3.72%. Considering that the cultivation medium can be a significant factor in this case (e.g., Experiment 4), and that eight different media were employed in 10 performed cultivations, it speaks to the robustness of the developed sensor. Another factor that must be considered, is that the sensor does not use any additional sensor signals (e.g., CO2 measurement), which, although, may reduce sensor accuracy, does not require the purchase of additional sensor systems. Overall, the sensor accuracy can be deemed as sufficient for application in recombinant P. pastoris cultivations.
4. Discussion
In this research, we investigated several reported cultivation media for recombinant LegH production with the yeast
P. pastoris. For improved results interpretation, we estimated the elemental composition of each cultivation medium and compared the respective concentrations with the so-called Wegner ranges (
Table A2,
Appendix A).
The highest LegH yield was achieved in rich BMGY medium. Rich medium is known for improved cell growth, as the cells do not need to synthesize all of the required metabolic intermediates, as is the case in minimal media. Frequently, this impact can result in enhanced yields of recombinant proteins, as the Luedeking–Piret model suggests that, in numerous instances, the cell growth rate can be directly related to recombinant protein production. However, there are several drawbacks to using a complex cultivation medium, for example, increased costs, composition variability, and hindered purification. Also, it should be noted that the use of meat peptone in BMGY formulation for the production of a product mainly used as a vegan food supplement could be considered controversial. Substituting meat peptone with, for example, soy peptone could be a viable alternative; however, the changes in ingredient composition can not only result in reduced product yields, but potentially necessitate adjustments in the cultivation process itself.
The second best result was achieved, when cultivation was carried out in standard BSM medium, as the yields were comparable to those achieved in BMGY medium. As the elemental composition of BSM shows, it contains, per Wegner, all of the necessary elements for
P. pastoris growth, most of them—even in excess of the preferred range. This, in part, accounts for the precipitation problems noted in BSM, as well as the increased osmotic pressure that is considered as a stress factor on the cells [
40]. However, as a reduced salt concentration (Experiment 5 (BSM/2)) did not result in an improved LegH yield, elevated osmotic pressure is probably not severely hindering LegH expression.
Considering the reported media formulations (FM22, D’Anjou, and RDM), an underwhelming LegH yield was achieved. The FM22 and RDM media are fairly similar to BSM; however, both have lower elemental concentrations in most cases. RDM is also supplemented with a mixture of vitamins suitable for yeast cultivation. Although, some salt precipitation was noted, when preparing these media, it was not on the same scale as with BSM. The D’Anjou medium has the least salts of any other media tested; hence, no precipitation was noted. The low LegH yield achieved in these media is somewhat perplexing. Considering the elemental compositions of the cultivation media, perhaps some of the excess elements in BSM amounted to an increased LegH expression.
The investigated addition of glycine or vitamin solution to BSM, did not yield any significant improvement to LegH yield. In both experiments, a lower specific productivity was achieved, even though a higher cell biomass concentration was recorded. We also noted that the glycine addition promoted excessive salt precipitation in this experiment. The addition of vitamins, did positively impact cell growth, as some authors have reported [
24]; however, it did not have a positive effect on LegH production.
As testing different cultivation media formulations did not result in an improved LegH yield, we decided to test two of the more popular P. pastoris feed strategies—µ-stat and mixed feed induction with sorbitol. Unfortunately, neither of these strategies produced improved results. In both cases, LegH yield was lower than in a standard BSM cultivation, according to the Invitrogen protocol.
Methanol acts both as a growth substrate and product synthesis inducer in
P. pastoris cultivations. Increased methanol feed (Experiment 9), however, did not amount to a higher LegH synthesis level, indicating that some bottleneck, probably in the heme biosynthesis pathway, may be present and consequently, limit LegH synthesis. Although
P. pastoris has been defined as a GRAS (Generally Regarded As Safe) microorganism, some concerns, regarding the toxicity of residual methanol may arise for recombinant product use in food applications and promote the consideration of other promoters for biosynthesis induction, such as the galactose-induced
LAC4 promoter in
Kluyveromyces lactis expression vector. However, these concerns are offset by the several studies that investigated the toxicity and allergenicity of LegH produced by Impossible foods Inc. and found no significant risks [
4,
5,
6].
Taking into consideration the results from the previous experiments, we can conclude that process-specific optimization strategies did not have a positive impact on LegH yield, as the best result was achieved in the “unoptimized” cultivation, according to the Invitrogen guidelines. The results suggest that LegH expression in this particular case is most likely not hindered by expression conditions, but for strain-specific reasons. Strain engineering of
P. pastoris is likely the key to improved LegH production, as clearly illustrated by the research of Shao et al. [
13]. In a recent article, we also developed a metabolic model for
P. pastoris LegH production, suggesting the reactions to up-/downregulate with the most potential for improved LegH production [
38].
An efficient purification procedure was developed to ensure a purity level of at least 90% for the expressed LegH. Although three chromatography columns are involved in this method, the overall process takes less than one day. No detectable losses of target protein were observed during purification. The quality of the purified LegH was confirmed by PAAG. The inclusion of the heme group to LegH is proven by the characteristic reddish color of peak protein fractions.
For this method, a 7.0 g wet cell portion was chosen, considering the volume limitations of the utilized French press for cell disruption. Theoretically, this press currently acts as the bottleneck for the purification method. Scaling up cell lysis would enable the expansion of the purification process and the utilization of larger chromatography columns, thereby purifying a significantly greater quantity of LegH. However, given that the primary focus of this research was to examine LegH production at the laboratory scale, we consider this purification method adequate for the stated objective.
Shao et al. used an Ni–NTA agarose column to purify and Amicon Ultra 3 K centrifugal filter units to desalt the LegH secreted in the culture medium [
13]. Both Ni agarose and the Amicon filters are very expensive. The filtration procedure in this case is also volume limited and time intensive. Overall, this method is not suitable for large-scale production. Impossible foods Inc., on the other hand, did not employ chromatography at all in the purification of their product [
7]. Insoluble material was removed by centrifugation and microfiltration. Then, ultrafiltration was used to concentrate the LegH, resulting in the end product purity of ~80%. Their approach was to identify all of the remaining contaminants in the product and to assess their toxicity and allergic properties. Although, this would be more time consuming and expensive at first, this approach is more suited to the large-scale commercial production of LegH.
Generated process data were used to establish and train a neural network model for cell biomass estimation. Several similar models have been previously reported [
41,
42]; however, the novelty of our approach is based on the absence of external sensor signals. Both reported examples utilize the CO
2 measurement signal, which requires an additional exhaust gas analyzer. However, our sensor is able to estimate cell biomass concentration, with sufficient precision, by only using standard real-time measurements by the bioreactor system itself.
The soft sensor can be used in cultivation processes in real time to estimate cell biomass concentration—one of the more important process parameters. Perhaps, it is most suited particularly for fed-batch fermentations, as feed rate profile calculation often requires precise and rapid biomass measurements. The inclusion of real-time cell biomass estimation can also benefit several advanced bioprocess control strategies, such as PID or model predictive (MPC) controllers [
29].