Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview
Abstract
:1. Introduction
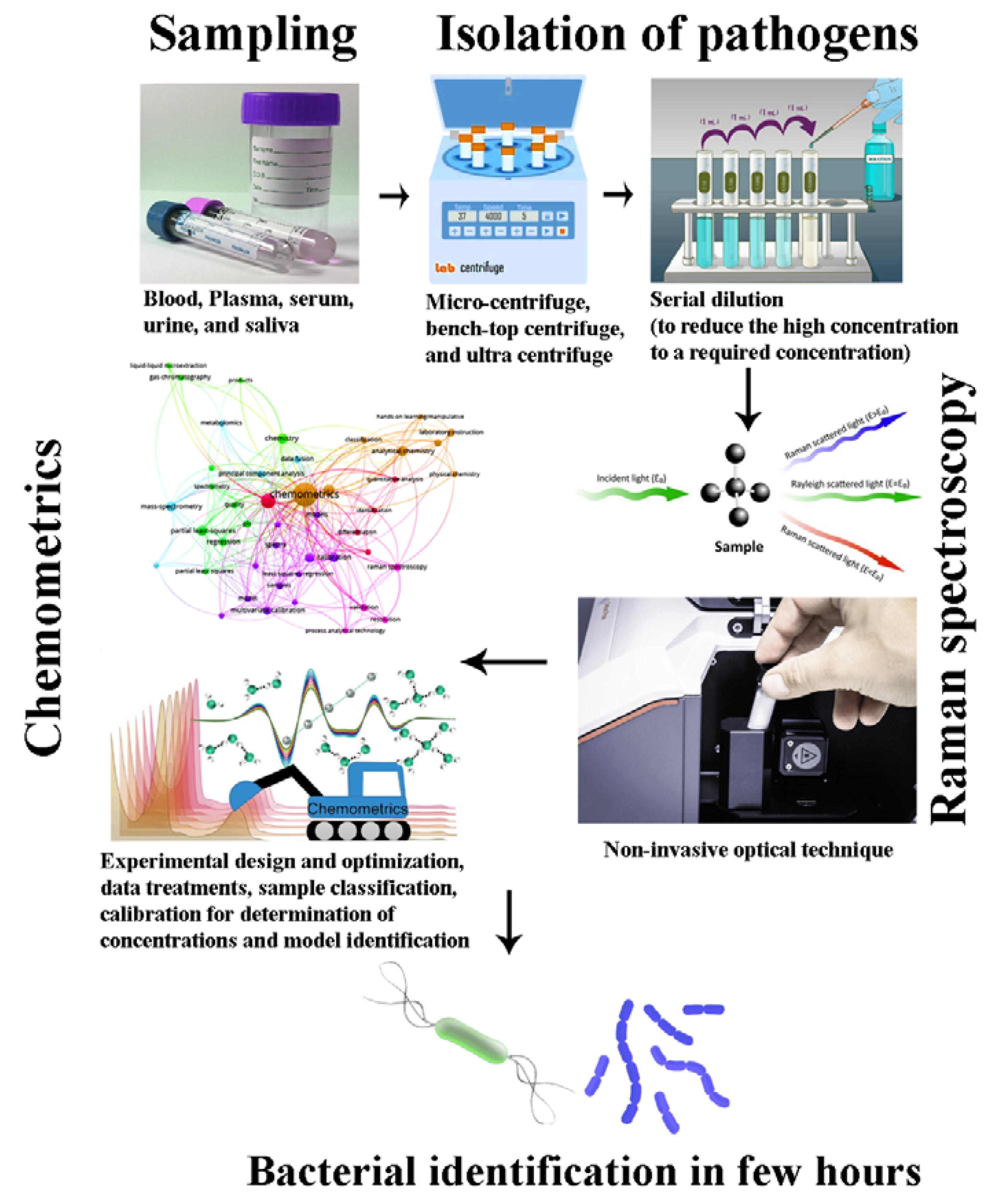
2. Spectroscopic Methods for the Identification and Characterization of Microbial Pathogens
2.1. Wavelength-Based Microbial Growth Using Spectroscopic Analysis
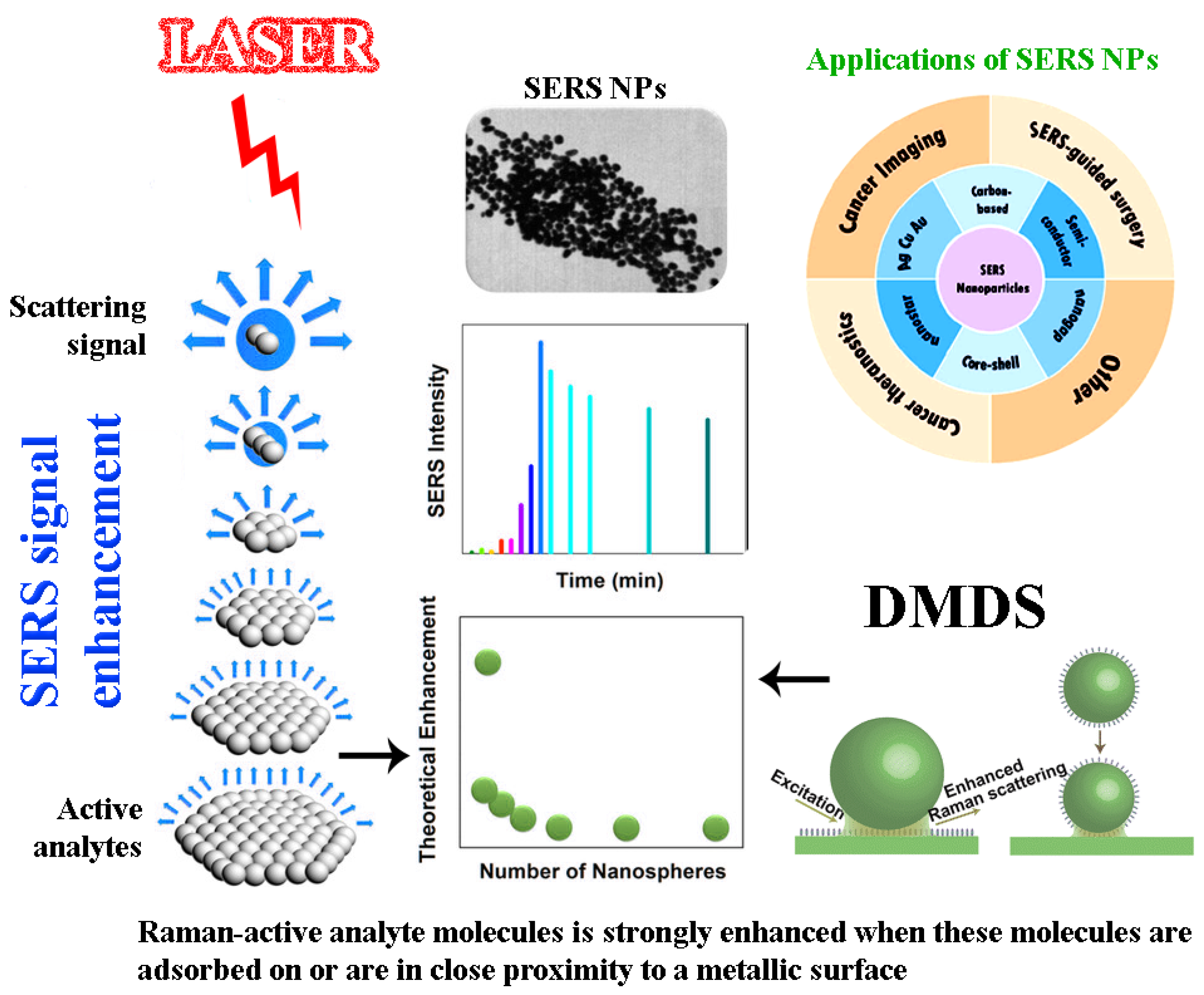
2.2. Surface-Enhanced Raman Spectroscopy (SERS)
2.3. Fourier Transform Infrared Spectroscopy (FTIR)
2.4. Electrochemical Impedance Spectroscopy (EIS)
2.5. MALDI-TOF/TOF Tandem Mass Spectrometry
2.6. Near-Infrared Spectroscopy (NIRS) and Chemometrics
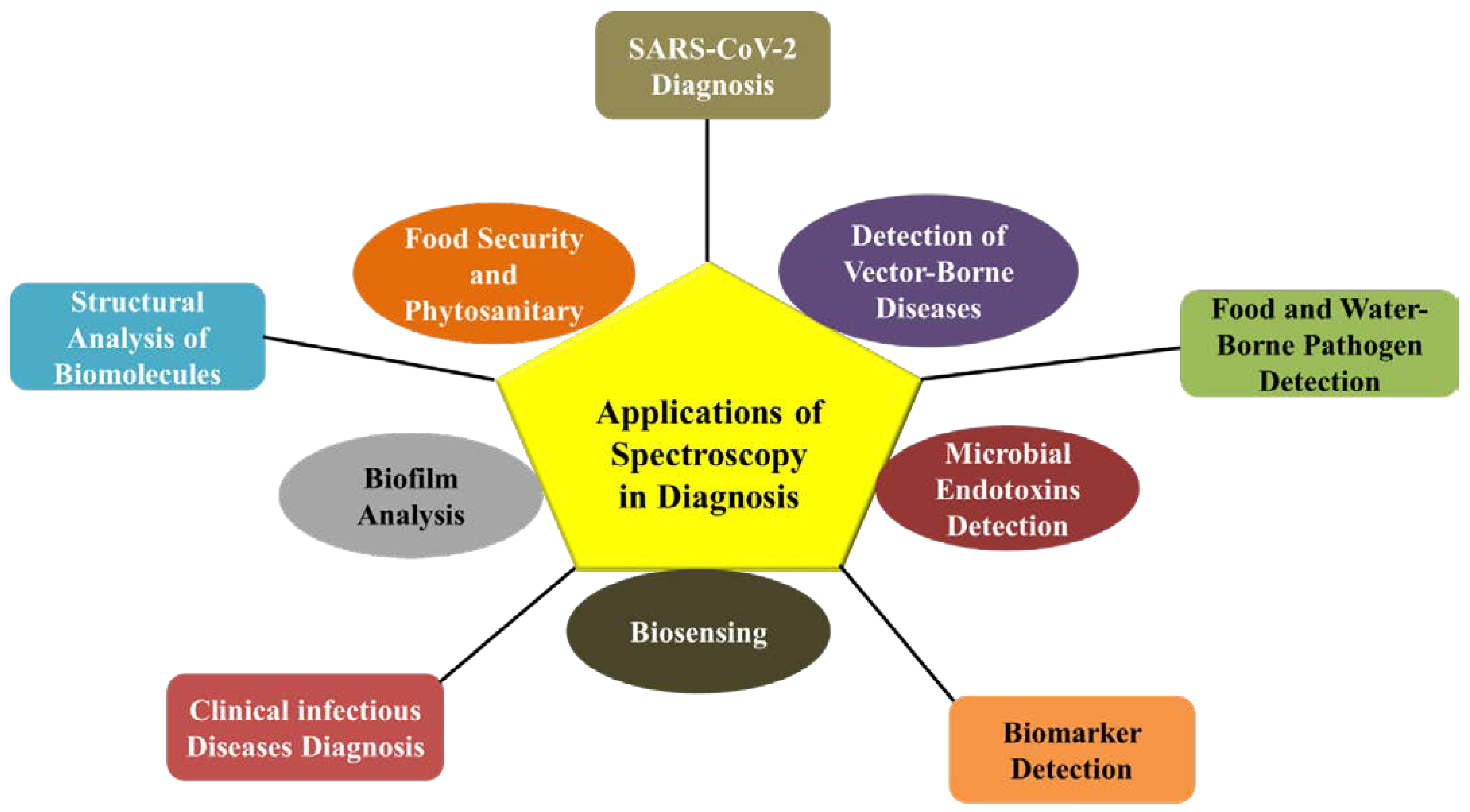
3. Applications of Spectroscopy in Diagnostics
3.1. Epidemiology
3.2. Diagnosis of Clinical Infectious and Vector-Borne Diseases
3.3. Food- and Waterborne Pathogen Detection
3.4. Antibiotic Resistance and Virulence Factors
3.5. SARS-CoV-2 Diagnosis
3.6. Microbial Endotoxins/Biomarker Detection
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign: International guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013, 39, 165–228. [Google Scholar] [CrossRef] [PubMed]
- American Thoracic Society, Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenstein, B.I. New molecular techniques for microbial epidemiology and the diagnosis of infectious diseases. J. Infect. Dis. 1990, 161, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.J.; van Zanten, E.; Akkerboom, V.; Wisselink, G.; van Slochteren, K.; de Boer, R.F.; Hendrix, R.; Friedrich, A.W.; Rossen, J.W.; Kooistra-Smid, A. Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification-increased discrimination of closely related species. Sci. Rep. 2017, 7, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Ko, J.; Park, S.G.; Lee, S.; Wang, X.; Mun, C.; Kim, S.; Kim, D.H.; Choo, J. Culture-free detection of bacterial pathogens on plasmonic nanopillar arrays using rapid Raman mapping. ACS Appl. Mater. Interfaces 2018, 10, 6831–6840. [Google Scholar] [CrossRef]
- Shrivastava, S.; Lee, W.I.; Lee, N.E. Culture-free, highly sensitive, quantitative detection of bacteria from minimally processed samples using fluorescence imaging by smartphone. Biosens. Bioelectron. 2018, 109, 90–97. [Google Scholar] [CrossRef]
- Wang, J.C.; Tung, Y.C.; Ichiki, K.; Sakamoto, H.; Yang, T.H.; Suye, S.I.; Chuang, H.S. Culture-free detection of methicillin-resistant Staphylococcus aureus by using self-driving diffusometric DNA nanosensors. Biosens. Bioelectron. 2020, 148, 111817. [Google Scholar] [CrossRef]
- Fenollar, F.; Raoult, D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int. J. Antimicrob. Agents 2007, 30, 7–15. [Google Scholar] [CrossRef]
- Şen Karaman, D.; Ercan, U.K.; Bakay, E.; Topaloğlu, N.; Rosenholm, J.M. Evolving technologies and strategies for combating antibacterial resistance in the advent of the postantibiotic era. Adv. Funct. Mater. 2020, 30, 1908783. [Google Scholar] [CrossRef]
- Ashton, L.; Lau, K.; Winder, C.L.; Goodacre, R. Raman spectroscopy: Lighting up the future of microbial identification. Future Microbiol. 2011, 6, 991–997. [Google Scholar] [CrossRef] [Green Version]
- Kotanen, C.N.; Martinez, L.; Alvarez, R.; Simecek, J.W. Surface enhanced Raman scattering spectroscopy for detection and identification of microbial pathogens isolated from human serum. Sens. Bio Sens. Res. 2016, 8, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.K.; Sharma, J.; Pathak, A.K.; Ghany, C.T.; Gondal, M.A. Laser-induced breakdown spectroscopy (LIBS): A novel technology for identifying microbes causing infectious diseases. Biophys. Rev. 2018, 10, 1221–1239. [Google Scholar] [CrossRef]
- Saari, S.; Järvinen, S.; Reponen, T.; Mensah-Attipoe, J.; Pasanen, P.; Toivonen, J.; Keskinen, J. Identification of single microbial particles using electro-dynamic balance assisted laser-induced breakdown and fluorescence spectroscopy. Aerosol Sci. Technol. 2016, 50, 126–132. [Google Scholar] [CrossRef] [Green Version]
- Cheeseman, S.; Shaw, Z.L.; Vongsvivut, J.; Crawford, R.J.; Dupont, M.F.; Boyce, K.J.; Gangadoo, S.; Bryant, S.J.; Bryant, G.; Cozzolino, D.; et al. Analysis of pathogenic bacterial and yeast biofilms using the combination of synchrotron ATR-FTIR microspectroscopy and chemometric approaches. Molecules 2021, 26, 3890. [Google Scholar] [CrossRef]
- Bosch, A.; Serra, D.; Prieto, C.; Schmitt, J.; Naumann, D.; Yantorno, O. Characterization of Bordetella pertussis growing as biofilm by chemical analysis and FT-IR spectroscopy. Appl. Microbiol. Biotechnol. 2006, 71, 736–747. [Google Scholar] [CrossRef]
- Wang, H.; Ding, S.; Wang, G.; Xu, X.; Zhou, G. In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach. Int. J. Food Microbiol. 2013, 167, 293–302. [Google Scholar] [CrossRef]
- Chen, P.; Wang, J.J.; Hong, B.; Tan, L.; Yan, J.; Zhang, Z.; Liu, H.; Pan, Y.; Zhao, Y. Characterization of mixed-species biofilm formed by Vibrio parahaemolyticus and Listeria monocytogenes. Front. Microbiol. 2019, 10, 2543. [Google Scholar] [CrossRef] [Green Version]
- Van Duuren, J.B.; Müsken, M.; Karge, B.; Tomasch, J.; Wittmann, C.; Häussler, S.; Brönstrup, M. Use of single-frequency impedance spectroscopy to characterize the growth dynamics of biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 2017, 7, 5223. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Li, H.Z.; Zhu, X.; Su, J.Q.; Ren, B.; Zhu, Y.G.; Cui, L. Rapid antibiotic susceptibility testing of pathogenic bacteria using heavy-water-labeled single-cell Raman spectroscopy in clinical samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef]
- Zarnowiec, P.; Lechowicz, L.; Czerwonka, G.; Kaca, W. Fourier transform infrared spectroscopy (FTIR) as a tool for the identification and differentiation of pathogenic bacteria. Curr. Med. Chem. 2015, 22, 1710–1718. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Asai, T.; Nakaya, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman imaging of pathogenic Candida auris: Visualization of structural characteristics and machine-learning identification. Front. Microbiol. 2021, 12, 769597. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.; Jupe, J.; Mack, H.; Coleman, T.P.; Lawrence, S.M.; Fraley, S.I. Emerging Technologies for Molecular Diagnosis of Sepsis. Clin. Microbiol. Rev. 2018, 31, e00089-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roth, A.; Dornuf, F.; Klein, O.; Mäntele, W. IR spectroscopy goes to the hospital: Progress in reagent-free blood analysis and haemodialysis monitoring. FTIR Spectrosc. Microbiol. Med. Diagn. 2011, 20, 46. [Google Scholar]
- Lopez-Reyes, G.; Pérez, F.R. A method for the automated Raman spectra acquisition. J. Raman Spectrosc. 2017, 48, 1654–1664. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhao, W. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. SLAS Technol. 2017, 22, 585. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Chen, Y.; Tang, H.; Zong, N.; Jiang, X. Microfluidics for biomedical analysis. Small Methods 2020, 4, 1900451. [Google Scholar] [CrossRef]
- Cowcher, D.P.; Xu, Y.; Goodacre, R. Portable, quantitative detection of Bacillus bacterial spores using surface-enhanced Raman scattering. Anal. Chem. 2013, 85, 3297–3302. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [Green Version]
- Carbonnelle, E.; Raskine, L. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Bio Trib. Mag. 2011, 39, 35–42. [Google Scholar] [CrossRef]
- Madonna, A.J.; van Cuyk, S.; Voorhees, K.J. Detection of Escherichia coli using immunomagnetic separation and bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 257–263. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.; Kaushik, K.S.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in chemical and biological methods to identify microorganisms—From past to present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [Green Version]
- Braga, P.A.; Tata, A.; dos Santos, V.G.; Barreiro, J.R.; Schwab, N.V.; dos Santos, M.V.; Eberlin, M.N.; Ferreira, C.R. Bacterial identification: From the agar plate to the mass spectrometer. RSC Adv. 2013, 3, 994–1008. [Google Scholar] [CrossRef]
- Trivedi, N.; Dubey, A. Degradation studies of pendimethalin by indigenous soil bacterium Pseudomonas strain PD1 using spectrophotometric scanning and FTIR. Arch. Microbiol. 2021, 203, 4499–4507. [Google Scholar] [CrossRef]
- Myers, J.A.; Curtis, B.S.; Curtis, W.R. Improving accuracy of cell and chromophore concentration measurements using optical density. BMC Biophys. 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- McBirney, S.E.; Trinh, K.; Wong-Beringer, A.; Armani, A.M. Wavelength-normalized spectroscopic analysis of Staphylococcus aureus and Pseudomonas aeruginosa growth rates. Biomed. Opt. Express 2016, 7, 4034–4042. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hong, H.; Myklejord, D.V.; Cai, W. Molecular imaging with SERS-active nanoparticles. Small. 2011, 7, 3261–3269. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Zhang, S.; Yue, S. Raman Spectroscopy and Imaging for Cancer Diagnosis. J. Healthc. Eng. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227. [Google Scholar] [CrossRef] [Green Version]
- Neng, J.; Harpster, M.H.; Wilson, W.C.; Johnson, P.A. Surface-enhanced Raman scattering (SERS) detection of multiple viral antigens using magnetic capture of SERS-active nanoparticles. Biosens. Bioelectron. 2013, 41, 316–321. [Google Scholar] [CrossRef]
- Ho, C.S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.; Ermon, S.; Dionne, J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef] [Green Version]
- Efrima, S.; Zeiri, L. Understanding SERS of bacteria. J. Raman Spectr. 2009, 40, 277–288. [Google Scholar] [CrossRef]
- Kloß, S.; Rösch, P.; Pfister, W.; Kiehntopf, M.; Popp, J. Toward culture-free Raman spectroscopic identification of pathogens in ascitic fluid. Anal. Chem. 2015, 87, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; Ngo-Thi, N.A.; van Vreeswijk, T.; Stämmler, M.; Endtz, H.P.; Bruining, H.A.; Naumann, D.; Puppels, G.J. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 2003, 41, 324–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Hu, Q.; Li, C.; Zhang, F.; Gu, H.; Wang, X.; Li, S.; Xue, L.; Madl, T.; Zhang, Y.; et al. Wide-Range, Rapid, and Specific Identification of Pathogenic Bacteria by Surface-Enhanced Raman Spectroscopy. ACS Sens. 2021, 6, 2911–2919. [Google Scholar] [CrossRef]
- Singh, K.S.; Majik, M.S.; Tilvi, S. Vibrational spectroscopy for structural characterization of bioactive compounds. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2014; Volume 65, pp. 115–148. [Google Scholar]
- Gieroba, B.; Krysa, M.; Wojtowicz, K.; Wiater, A.; Pleszczyńska, M.; Tomczyk, M.; Sroka-Bartnicka, A. The FT-IR and Raman Spectroscopies as Tools for Biofilm Characterization Created by Cariogenic Streptococci. Int. J. Mol. Sci. 2020, 21, 3811. [Google Scholar] [CrossRef]
- Ozaki, Y. Infrared spectroscopy—Mid-infrared, near-infrared, and far-infrared/terahertz spectroscopy. Anal. Sci. 2021, 37, 1193–1212. [Google Scholar] [CrossRef]
- How an FTIR Spectrometer Operates—Chemistry LibreTexts. 2022. Available online: https://chem.libretexts.org/Book-shelves/Physical_and_Theoretical_Chemistry_Textbook_Maps/Supplemental_Modules_(Physical_and_Theoretical_Chemis-try)/Spectroscopy/Vibrational_Spectroscopy/Infrared_Spectroscopy/How_an_FTIR_Spectrometer_Operates (accessed on 23 February 2022).
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Hong, S.; Jang, J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017, 7, 42771. [Google Scholar] [CrossRef] [Green Version]
- Al-Qadiri, H.M.; Lin, M.; Cavinato, A.G.; Rasco, B.A. Fourier transform infrared spectroscopy, detection and identification of Escherichia coli O157:H7 and Alicyclobacillus strains in apple juice. Int. J. Food Microbiol. 2006, 111, 73–80. [Google Scholar] [CrossRef]
- Donlan, R.M.; Piede, J.A.; Heyes, C.D.; Sanii, L.; Murga, R.; Edmonds, P.; El-Sayed, I.; El-Sayed, M.A. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl. Environ. Microbiol. 2004, 70, 4980–4988. [Google Scholar] [CrossRef] [Green Version]
- Stewart, G.N. The charges produced by the growth of bacteria in the molecular concentration and electrical conductivity of culture media. J. Exp. Med. 1899, 4, 235. [Google Scholar] [CrossRef] [Green Version]
- Brosel-Oliu, S.; Uria, N.; Abramova, N.; Bratov, A. Impedimetric sensors for bacteria detection. Biosens. Micro Nanoscale Appl. 2015, 24, 257–288. [Google Scholar]
- Lagier, J.C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. 2015, 28, 208. [Google Scholar] [CrossRef] [Green Version]
- Dean, D.A.; Ramanathan, T.; Machado, D.; Sundararajan, R. Electrical impedance spectroscopy study of biological tissues. J. Electrost. 2008, 66, 165–177. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Bashir, R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol. Adv. 2008, 26, 135–150. [Google Scholar] [CrossRef]
- Jahnke, H.G.; Heimann, A.; Azendorf, R.; Mpoukouvalas, K.; Kempski, O.; Robitzki, A.A.; Charalampaki, P. Impedance spectroscopy—An outstanding method for label-free and real-time discrimination between brain and tumor tissue in vivo. Biosens. Bioelectron. 2013, 46, 8–14. [Google Scholar] [CrossRef]
- Permeh, S.; Lau, K.; Duncan, M. Characterization of biofilm formation and coating degradation by electrochemical impedance spectroscopy. Coatings 2019, 9, 518. [Google Scholar] [CrossRef] [Green Version]
- Gogichaeva, N.V.; Williams, T.; Alterman, M.A. MALDI TOF/TOF tandem mass spectrometry as a new tool for amino acid analysis. J. Am. Soc. Mass Spectrom. 2007, 18, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Goloborodko, A.A.; Gorshkov, M.V.; Good, D.M.; Zubarev, R.A. Sequence scrambling in shotgun proteomics is negligible. J. Am. Soc. Mass Spectrom. 2011, 22, 1121–1124. [Google Scholar] [CrossRef] [Green Version]
- Juiz, P.M.; Almela, M.; Melción, C.; Campo, I.; Esteban, C.; Pitart, C.; Marco, F.; Vila, J. A comparative study of two different methods of sample preparation for positive blood cultures for the rapid identification of bacteria using MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1353–1358. [Google Scholar] [CrossRef]
- Hou, T.Y.; Chiang-Ni, C.; Teng, S.H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Haiko, J.; Savolainen, L.E.; Hilla, R.; Pätäri-Sampo, A. Identification of urinary tract pathogens after 3-hours urine culture by MALDI-TOF mass spectrometry. J. Microbiol. Methods 2016, 129, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rychert, J. Benefits and limitations of MALDI-TOF mass spectrometry for the identification of microorganisms. J. Infect. Epidemiol. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Lasch, P.; Nattermann, H.; Erhard, M.; Stämmler, M.; Grunow, R.; Bannert, N.; Appel, B.; Naumann, D. MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal. Chem. 2008, 80, 2026–2034. [Google Scholar] [CrossRef]
- Jeong, Y.S.; Choi, S.; Chong, E.; Kim, J.H.; Kim, S.J. Rapid detection of B acillus spore aerosol particles by direct in situ analysis using MALDI-TOF mass spectrometry. Lett. Appl. Microbiol. 2014, 59, 177–183. [Google Scholar] [CrossRef]
- Johansson, Å.; Nagy, E.; Sóki, J. Detection of carbapenemase activities of Bacteroides fragilis strains with matrix-assisted laser desorption ionization—time of flight mass spectrometry (MALDI-TOF MS). Anaerobe 2014, 26, 49–52. [Google Scholar] [CrossRef]
- Hoyos-Mallecot, Y.; Cabrera-Alvargonzalez, J.; Miranda-Casas, C.; Rojo-Martín, M.D.; Liebana-Martos, C.; Navarro-Marí, J. MALDI-TOF MS, a useful instrument for differentiating metallo-β-lactamases in Enterobacteriaceae and Pseudomonas spp. Lett. Appl. Microbiol. 2014, 58, 325–329. [Google Scholar] [CrossRef]
- Hart, P.J.; Wey, E.; McHugh, T.D.; Balakrishnan, I.; Belgacem, O. A method for the detection of antibiotic resistance markers in clinical strains of Escherichia coli using MALDI mass spectrometry. J. Microbiol. Methods 2015, 111, 1–8. [Google Scholar] [CrossRef]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell AG, M. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1–24. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Popp, J. Vibrational spectroscopy—A powerful tool for the rapid identification of microbial cells at the single-cell level. Cytom. Part A 2009, 75, 104–113. [Google Scholar] [CrossRef]
- Roggo, Y.; Chalus, P.; Maurer, L.; Lema-Martinez, C.; Edmond, A.; Jent, N. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J. Pharm. Biomed. Anal. 2007, 44, 683–700. [Google Scholar] [CrossRef]
- Hamprecht, J.; Corsten, D.; Noll, M.; Meier, E. Controlling the sustainability of food supply chains. Supply Chain. Manag. Int. J. 2005, 10, 7–10. [Google Scholar] [CrossRef]
- Curto, B.; Moreno, V.; García-Esteban, J.A.; Blanco, F.J.; González, I.; Vivar, A.; Revilla, I. Accurate prediction of sensory attributes of cheese using near-infrared spectroscopy based on artificial neural network. Sensors 2020, 20, 3566. [Google Scholar] [CrossRef]
- Li, B.; Lin, Y.; Yu, W.; Wilson, D.I.; Young, B.R. Application of mechanistic modelling and machine learning for cream cheese fermentation pH prediction. J. Chem. Technol. Biotechnol. 2020, 96, 125–133. [Google Scholar] [CrossRef]
- Sipos, A. A knowledge-based system as a sustainable software application for the supervision and intelligent control of an alcoholic fermentation process. Sustainability 2020, 12, 10205. [Google Scholar] [CrossRef]
- Viejo, C.G.; Torrico, D.D.; Dunshea, F.R.; Fuentes, S. Development of artificial neural network models to assess beer acceptability based on sensory properties using a robotic pourer: A comparative model approach to achieve an artificial intelligence system. Beverages 2019, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Cao, S.; Horn, A.L. Emerging Applications of Machine Learning in Food Safety. Annu. Rev. Food Sci. Technol. 2021, 12, 513–538. [Google Scholar] [CrossRef]
- Vajdi, M.; Varidi, M.J.; Varidi, M.; Mohebbi, M. Using electronic nose to recognize fish spoilage with an optimum classifier. J. Food Meas. Charact. 2019, 13, 1205–1217. [Google Scholar] [CrossRef]
- Gutiérrez, P.; Godoy, S.E.; Torres, S.; Oyarzún, P.; Sanhueza, I.; Díaz-García, V.; Contreras-Trigo, B.; Coelho, P. Improved anti-biotic detection in raw milk using machine learning tools over the absorption spectra of a problem-specific nanobiosensor. Sensors 2020, 20, 4552. [Google Scholar] [CrossRef]
- Karami, H.; Rasekh, M.; Mirzaee-Ghaleh, E. Application of the E-nose machine system to detect adulterations in mixed edible oils using chemometrics methods. J. Food Process Preserv. 2020, 44, 1–12. [Google Scholar] [CrossRef]
- Ayari, F.; Mirzaee- Ghaleh, E.; Rabbani, H.; Heidarbeigi, K. Using an E-nose machine for detecting the adulteration of margarine in cow ghee. J. Food Process. Eng. 2018, 41, e12806. [Google Scholar] [CrossRef]
- Amigo, J.M.; Martí, I.; Gowen, A. Hyperspectral imaging and chemometrics: A perfect combination for the analysis of food structure, composition and quality. Data Handl. Sci. Technol. 2013, 28, 343–370. [Google Scholar]
- Spyrelli, E.D.; Papachristou, C.K.; Nychas, G.J.E.; Panagou, E.Z. Microbiological Quality Assessment of Chicken Thigh Fillets Using Spectroscopic Sensors and Multivariate Data Analysis. Foods 2021, 10, 2723. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, E.; Núñez, F.; Rodríguez, M.; Grassi, S.; González-Mohino, A. Potential of Near Infrared Spectroscopy as a Rapid Method to Discriminate OTA and Non-OTA-Producing Mould Species in a Dry-Cured Ham Model System. Toxins 2021, 13, 620. [Google Scholar] [CrossRef] [PubMed]
- Alexandrakis, D.; Downey, G.; Scannell, A.G.M. Rapid non-destructive detection of spoilage of intact chicken breast muscle using near-infrared and Fourier transform mid-infrared spectroscopy and multivariate statistics. Food Bioprocess Technol. 2012, 5, 338–347. [Google Scholar] [CrossRef]
- Azadshahraki, F.; Sharifi, K.; Jamshidi, B.; Karimzadeh, R.; Naderi, H. Diagnosis of Early Blight Disease in Tomato Plant based on Visible/Near-Infrared Spectroscopy and Principal Components Analysis-Artificial Neural Network Prior to Visual Disease Symptoms. J. Agric. Mach. 2022, 12, 81–94. [Google Scholar]
- Rahi, S.; Mobli, H.; Jamshidi, B.; Azizi, A.; Sharifi, M. Achieving a robust Vis/NIR model for microbial contamination detection of Persian leek by spectral analysis based on genetic, iPLS algorithms and VIP scores. Postharvest Biol. Technol. 2021, 175, 111413. [Google Scholar] [CrossRef]
- Sirisomboon, C.D.; Wongthip, P.; Sirisomboon, P. Potential of near infrared spectroscopy as a rapid method to detect aflatoxins in brown rice. J. Near Infrared Spectrosc. 2019, 27, 232–240. [Google Scholar] [CrossRef]
- Tao, F.; Yao, H.; Hruska, Z.; Liu, Y.; Rajasekaran, K.; Bhatnagar, D. Detection of aflatoxin B1 on corn kernel surfaces using visible-near infrared spectroscopy. J. Near Infrared Spectrosc. 2020, 28, 59–69. [Google Scholar] [CrossRef]
- Pan, W.; Zhao, J.; Chen, Q. Classification of foodborne pathogens using near infrared (NIR) laser scatter imaging system with multivariate calibration. Sci. Rep. 2015, 5, 9524. [Google Scholar] [CrossRef] [Green Version]
- Duan, C.; Chen, C.; Khan, M.N.; Liu, Y.; Zhang, R.; Lin, H.; Cao, L. Non-destructive determination of the total bacteria in flounder fillet by portable near infrared spectrometer. Food Control 2014, 42, 18–22. [Google Scholar] [CrossRef]
- Tito, N.B.; Rodemann, T.; Powell, S.M. Use of near infrared spectroscopy to predict microbial numbers on Atlantic salmon. Food Microbiol. 2012, 32, 431–436. [Google Scholar] [CrossRef]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-care diagnostics for infectious diseases: From methods to devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef]
- El-Bouri, K.; Johnston, S.; Rees, E.; Thomas, I.; Bome-Mannathoko, N.; Jones, C.; Reid, M.; Ben-Ismaeil, B.; Davies, A.P.; Harris, L.G.; et al. Comparison of bacterial identification by MALDI-TOF mass spectrometry and conventional diagnostic microbiology methods: Agreement, speed and cost implications. Br. J. Biomed. Sci. 2012, 69, 47–55. [Google Scholar] [CrossRef]
- Yeh, Y.T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V.; et al. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Yeh, Y.T.; Xue, Y.; Wang, Z.; Zhang, N.; Liu, H.; Zhang, K.; Yu, Z.; Roder, A.; Lopez, N.P.; et al. Accurate Virus Identification with Interpretable Raman Signatures by Machine Learning. bioRxiv 2021. [Google Scholar] [CrossRef]
- Rebrošová, K.; Bernatová, S.; Šiler, M.; Uhlirova, M.; Samek, O.; Ježek, J.; Holá, V.; Růžička, F.; Zemanek, P. Raman spectroscopy-a tool for rapid differentiation among microbes causing urinary tract infections. Anal. Chim. Acta 2022, 1191, 339292. [Google Scholar] [CrossRef]
- Zhao, Y.; Tian, S.; Yu, L.; Zhang, Z.; Zhang, W. Analysis and Classification of Hepatitis Infections Using Raman Spectroscopy and Multiscale Convolutional Neural Networks. J. Appl. Spectrosc. 2021, 88, 441–451. [Google Scholar] [CrossRef]
- Tiwari, D.; Jakhmola, S.; Pathak, D.K.; Kumar, R.; Jha, H.C. Temporal In Vitro Raman Spectroscopy for Monitoring Replication Kinetics of Epstein-Barr Virus Infection in Glial Cells. ACS Omega 2020, 5, 29547–29560. [Google Scholar] [CrossRef]
- Zyubin, A.; Lavrova, A.; Manicheva, O.; Dogonadze, M.; Belik, V.; Samusev, I. Raman spectroscopy for glutathione measurements in Mycobacterium tuberculosis strains with different antibiotic resistance. J. Raman Spectrosc. 2021, 52, 1661–1666. [Google Scholar] [CrossRef]
- Barker, K.R.; Santino, M.; LiPuma, J.J.; Tullis, E.; Muller, M.P.; Matukas, L.M.; Tadros, M. Fourier Transform Infrared Spectroscopy for Typing Burkholderia cenocepacia ET12 Isolates. Microbiol. Spectr. 2021, 9, e0183121. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Liu, Z.; Peng, Y.; Hou, G.; Chen, W.; Xiao, R. Automatic and sensitive detection of West Nile virus non-structural protein 1 with a portable SERS-LFIA detector. Mikrochim. Acta 2021, 188, 206. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Purrà, M.; Carré-Camps, M.; de Puig, H.; Bosch, I.; Gehrke, L.; Hamad-Schifferli, K. Surface-Enhanced Raman Spectroscopy-Based Sandwich Immunoassays for Multiplexed Detection of Zika and Dengue Viral Biomarkers. ACS Infect. Dis. 2017, 3, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Girard, A.; Cooper, A.; Mabbott, S.; Bradley, B.; Asiala, S.; Jamieson, L.; Clucas, C.; Capewell, P.; Marchesi, F.; Gibbins, M.P.; et al. Raman spectroscopic analysis of skin as a diagnostic tool for Human African Trypanosomiasis. PLoS Pathog. 2021, 17, e1010060. [Google Scholar] [CrossRef]
- Goh, B.; Ching, K.; Magalhães, R.J.S.; Ciocchetta, S.; Edstein, M.D.; Maciel-de-Freitas, R.; Sikulu-Lord, M.T. The application of spectroscopy techniques for diagnosis of malaria parasites and arboviruses and surveillance of mosquito vectors: A systematic review and critical appraisal of evidence. PLoS Negl. Trop. Dis. 2021, 15, e0009218. [Google Scholar] [CrossRef]
- Sanchez, L.; Ermolenkov, A.; Tang, X.T.; Tamborindeguy, C.; Kurouski, D. Non-invasive diagnostics of Liberibacter disease on tomatoes using a hand-held Raman spectrometer. Planta 2020, 251, 64. [Google Scholar] [CrossRef]
- Vallejo-Pérez, M.R.; Sosa-Herrera, J.A.; Navarro-Contreras, H.R.; Álvarez-Preciado, L.G.; Rodríguez-Vázquez, Á.G.; Lara-Ávila, J.P. Raman Spectroscopy and Machine-Learning for Early Detection of Bacterial Canker of Tomato: The Asymptomatic Disease Condition. Plants 2021, 10, 1542. [Google Scholar] [CrossRef]
- Strycker, B.D.; Han, Z.; Duan, Z.; Commer, B.; Wang, K.; Shaw, B.D.; Sokolov, A.V.; Scully, M.O. Identification of toxic mold species through Raman spectroscopy of fungal conidia. PLoS ONE 2020, 15, e0242361. [Google Scholar] [CrossRef]
- Saif, F.A.; Yaseen, S.A.; Alameen, A.S.; Mane, S.B.; Undre, P.B. Identification and characterization of Aspergillus species of fruit rot fungi using microscopy, FT-IR, Raman and UV-Vis spectroscopy. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2021, 246, 119010. [Google Scholar] [CrossRef]
- Mandrile, L.; Rotunno, S.; Miozzi, L.; Vaira, A.M.; Giovannozzi, A.M.; Rossi, A.M.; Noris, E. Nondestructive Raman Spectroscopy as a Tool for Early Detection and Discrimination of the Infection of Tomato Plants by Two Economically Important Viruses. Anal. Chem. 2019, 91, 9025–9031. [Google Scholar] [CrossRef]
- Huang, C.C.; Hsu, Z.H.; Lai, Y.S. Raman spectroscopy for virus detection and the implementation of unorthodox food safety. Trends Food Sci. Technol. 2021, 116, 525–532. [Google Scholar] [CrossRef]
- Petersen, M.; Yu, Z.; Lu, X. Application of Raman Spectroscopic Methods in Food Safety: A Review. Biosensors 2021, 11, 187. [Google Scholar] [CrossRef]
- Deidda, F.; Cionci, N.B.; Cordovana, M.; Campedelli, I.; Fracchetti, F.; Di Gioia, D.; Ambretti, S.; Pane, M. Bifidobacteria Strain Typing by Fourier Transform Infrared Spectroscopy. Front. Microbiol. 2021, 12, 692975. [Google Scholar] [CrossRef]
- Yakes, B.J.; Ellsworth, Z.; Karunathilaka, S.R.; Crump, E. Evaluation of Portable Sensor and Spectroscopic Devices for Seafood Decomposition Determination. Food Anal. Methods 2021, 14, 2346–2356. [Google Scholar] [CrossRef]
- Wang, K.; Chen, L.; Ma, X.; Ma, L.; Chou, K.C.; Cao, Y.; Khan, I.; Gölz, G.; Lu, X. Arcobacter Identification and Species Determination Using Raman Spectroscopy Combined with Neural Networks. Appl. Environ. Microbiol. 2020, 86, e00924-20. [Google Scholar] [CrossRef]
- Du, Y.; Han, D.; Liu, S.; Sun, X.; Ning, B.; Han, T.; Wang, J.; Gao, Z. Raman spectroscopy-based adversarial network combined with SVM for detection of foodborne pathogenic bacteria. Talanta 2022, 237, 122901. [Google Scholar] [CrossRef]
- Yan, S.; Wang, S.; Qiu, J.; Li, M.; Li, D.; Xu, D.; Li, D.; Liu, Q. Raman spectroscopy combined with machine learning for rapid detection of food-borne pathogens at the single-cell level. Talanta 2021, 226, 122195. [Google Scholar] [CrossRef]
- Xu, J.L.; Herrero-Langreo, A.; Lamba, S.; Ferone, M.; Scannell, A.; Caponigro, V.; Gowen, A.A. Characterisation and Classification of Foodborne Bacteria Using Reflectance FTIR Microscopic Imaging. Molecules 2021, 26, 6318. [Google Scholar] [CrossRef]
- Lu, J.; Chen, J.; Liu, C.; Zeng, Y.; Sun, Q.; Li, J.; Shen, Z.; Chen, S.; Zhang, R. Identification of antibiotic resistance and virulence-encoding factors in Klebsiella pneumoniae by Raman spectroscopy and deep learning. Microb. Biotechnol. 2022, 15, 1270–1280. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-resistant Staphylococcus aureus bacteria detection by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef]
- Chen, X.; Tang, M.; Liu, Y.; Huang, J.; Liu, Z.; Tian, H.; Zheng, Y.; de la Chapelle, M.L.; Zhang, Y.; Fu, W. Surface-enhanced Raman scattering method for the identification of methicillin-resistant Staphylococcus aureus using positively charged silver nanoparticles. Mikrochim. Acta 2019, 186, 102. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, L.; Chou, K.C.; Lu, X. Campylobacter jejuni Antimicrobial Resistance Profiles and Mechanisms Determined Using a Raman Spectroscopy-Based Metabolomic Approach. Appl. Environ. Microbiol. 2021, 87, e0038821. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Song, Y.; Xu, X.; Peng, D.; Wang, J.; Qie, X.; Lin, K.; Yu, M.; Ge, M.; Wang, Y.; et al. Development of a Fast Raman-Assisted Antibiotic Susceptibility Test (FRAST) for the Antibiotic Resistance Analysis of Clinical Urine and Blood Samples. Anal. Chem. 2021, 93, 5098–5106. [Google Scholar] [CrossRef] [PubMed]
- Gannesen, A.V.; Zdorovenko, E.L.; Botchkova, E.A.; Hardouin, J.; Massier, S.; Kopitsyn, D.S.; Gorbachevskii, M.V.; Kadykova, A.A.; Shashkov, A.S.; Zhurina, M.V.; et al. Composition of the Biofilm Matrix of Cutibacterium acnes Acneic Strain RT5. Front. Microbiol. 2019, 10, 1284. [Google Scholar] [CrossRef]
- Do, H.; Kwon, S.R.; Fu, K.; Morales-Soto, N.; Shrout, J.D.; Bohn, P.W. Electrochemical Surface-Enhanced Raman Spectroscopy of Pyocyanin Secreted by Pseudomonas aeruginosa Communities. Langmuir ACS J. Surf. Colloids 2019, 35, 7043–7049. [Google Scholar] [CrossRef]
- Horiue, H.; Sasaki, M.; Yoshikawa, Y.; Toyofuku, M.; Shigeto, S. Raman spectroscopic signatures of carotenoids and polyenes enable label-free visualization of microbial distributions within pink biofilms. Sci. Rep. 2020, 10, 7704. [Google Scholar] [CrossRef]
- Kriem, L.S.; Wright, K.; Ccahuana-Vasquez, R.A.; Rupp, S. Mapping of a Subgingival Dual-Species Biofilm Model Using Confocal Raman Microscopy. Front. Microbiol. 2021, 12, 729720. [Google Scholar] [CrossRef]
- World Health Organization. Clinical Management of COVID-19: Living Guideline, 13 January 2023; World Health Organization: Geneva, Switzerland, 2023. [Google Scholar]
- Wood, B.R.; Kochan, K.; Bedolla, D.E.; Salazar-Quiroz, N.; Grimley, S.L.; Perez-Guaita, D.; Baker, M.J.; Vongsvivut, J.; Tobin, M.J.; Bambery, K.R.; et al. Infrared Based Saliva Screening Test for COVID-19. Angew. Chem. 2021, 60, 17102–17107. [Google Scholar] [CrossRef]
- Huang, J.; Wen, J.; Zhou, M.; Ni, S.; Le, W.; Chen, G.; Wei, L.; Zeng, Y.; Qi, D.; Pan, M.; et al. On-Site Detection of SARS-CoV-2 Antigen by Deep Learning-Based Surface-Enhanced Raman Spectroscopy and Its Biochemical Foundations. Anal. Chem. 2021, 93, 9174–9182. [Google Scholar] [CrossRef]
- Yadav, S.; Sadique, M.A.; Ranjan, P.; Kumar, N.; Singhal, A.; Srivastava, A.K.; Khan, R. SERS Based Lateral Flow Immunoassay for Point-of-Care Detection of SARS-CoV-2 in Clinical Samples. ACS Appl. Bio Mater. 2021, 4, 2974–2995. [Google Scholar] [CrossRef]
- Zavyalova, E.; Ambartsumyan, O.; Zhdanov, G.; Gribanyov, D.; Gushchin, V.; Tkachuk, A.; Rudakova, E.; Nikiforova, M.; Kuznetsova, N.; Popova, L.; et al. SERS-Based Aptasensor for Rapid Quantitative Detection of SARS-CoV-2. Nanomaterials 2021, 11, 1394. [Google Scholar] [CrossRef]
- Desai, S.; Mishra, S.V.; Joshi, A.; Sarkar, D.; Hole, A.; Mishra, R.; Dutt, S.; Chilakapati, M.K.; Gupta, S.; Dutt, A. Raman spectroscopy-based detection of RNA viruses in saliva: A preliminary report. J. Biophotonics 2020, 13, e202000189. [Google Scholar] [CrossRef]
- Guleken, Z.; Jakubczyk, P.; Wiesław, P.; Krzysztof, P.; Bulut, H.; Öten, E.; Depciuch, J.; Tarhan, N. Characterization of Covid-19 infected pregnant women sera using laboratory indexes, vibrational spectroscopy, and machine learning classifications. Talanta 2022, 237, 122916. [Google Scholar] [CrossRef]
- Guleken, Z.; Tok, Y.T.; Jakubczyk, P.; Paja, W.; Pancerz, K.; Shpotyuk, Y.; Cebulski, J.; Depciuch, J. Development of novel spectroscopic and machine learning methods for the measurement of periodic changes in COVID-19 antibody level. Measurement 2022, 196, 111258. [Google Scholar] [CrossRef]
- Wu, X.; Zhao, Y.; Zughaier, S.M. Highly Sensitive Detection and Differentiation of Endotoxins Derived from Bacterial Pathogens by Surface-Enhanced Raman Scattering. Biosensors 2021, 11, 234. [Google Scholar] [CrossRef]
- Ge, X.; Pereira, F.C.; Mitteregger, M.; Berry, D.; Zhang, M.; Wagner, M.; Cheng, J.X. SRS-FISH: High-Throughput Platform Linking Microbiome Function to Identity at the Single Cell Level. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Cui, D.; Kong, L.; Chen, S.; Xie, W.; Zhang, C. Classification and Identification of Archaea Using Single-Cell Raman Ejection and Artificial Intelligence: Implications for Investigating Uncultivated Microorganisms. Anal. Chem. 2021, 93, 17012–17019. [Google Scholar] [CrossRef]
- Ren, Y.; Ji, Y.; Teng, L.; Zhang, H. Using Raman spectroscopy and chemometrics to identify the growth phase of Lactobacillus casei Zhang during batch culture at the single-cell level. Microb. Cell Factories 2017, 16, 233. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.; Qiu, J.; Guo, L.; Li, D.; Xu, D.; Liu, Q. Development overview of Raman-activated cell sorting devoted to bacterial detection at single-cell level. Appl. Microbiol. Biotechnol. 2021, 105, 1315–1331. [Google Scholar] [CrossRef]
- Song, Y.; Kaster, A.K.; Vollmers, J.; Song, Y.; Davison, P.A.; Frentrup, M.; Preston, G.M.; Thompson, I.P.; Murrell, J.C.; Yin, H.; et al. Single-cell genomics based on Raman sorting reveals novel carotenoid-containing bacteria in the Red Sea. Microb. Biotechnol. 2017, 10, 125–137. [Google Scholar] [CrossRef] [Green Version]
- García-Timermans, C.; Props, R.; Zacchetti, B.; Sakarika, M.; Delvigne, F.; Boon, N. Raman Spectroscopy-Based Measurements of Single-Cell Phenotypic Diversity in Microbial Populations. mSphere 2020, 5, e00806-20. [Google Scholar] [CrossRef] [PubMed]
- Skolrood, L.; Wang, Y.; Zhang, S.; Wei, Q. Single-molecule and particle detection on true portable microscopy platforms. Sens. Actuators Rep. 2022, 4, 100063. [Google Scholar] [CrossRef]

| Application | Chemometric Method | Main Finding | Reference |
|---|---|---|---|
| Early detection of blight disease in tomato with Vis-NIR spectroscopy | PCA-ANN | Early detection of blight disease and the associated pathogen type was achieved with about 93–100% accuracy | [88] |
| Detection of E. coli contamination in Persian leek with Vis/NIR spectroscopy | PLSDA with Genetic Algorithm (GA), interval PLS, variable influence on projection scores | GA exhibited high sensitivity (100%) and specificity (98%) and low classification error (0.8) in E. coli detection | [89] |
| Estimation of total viable counts and Pseudomonas spp. in chicken thigh fillets with FTIR and MSI | PLSR, LDA, QDA, SVM, and QSVM | SVM coupled with multispectral imaging showed the highest performance with about 94.4% overall accuracy | [85] |
| Detection of ochratoxin A-producing fungi from non-ochratoxin-producing fungi in dried meat with NIRS | PCA with SVM-DA | The SVM-DA model could differentiate between ochratoxin and non-ochratoxin-producing fungi with 86% specificity and 85% accuracy | [86] |
| Detection of aflatoxin B1 in corn kernel using Vis-NIRS | PCA-LDA and PLS-DA | Both discriminant and classification models exhibited over 90% accurate performance | [91] |
| Detection of aflatoxin contamination in brown rice with NIRS | PLSR | The model showed good predictive performance with a prediction coefficient of 0.95% | [90] |
| Classification of foodborne pathogens (E. coli, S. aureus, S. typhimurium, and mixed bacteria) using NIR-LSIS | Linear (PLSDA, KNN, and LDA) and nonlinear (BPANN, OSELM, and SVM) | Nonlinear methods performed better than linear methods, with OSELM exhibiting a performance accuracy of 95% | [92] |
| Quantification of total bacteria in fish fillets with a portable NIR spectrometer | PLS, GA combined with BPANN | GA combined with BPANN exhibited a better efficiency of prediction (about 96% accuracy) than PLS | [93] |
| Non-invasive and non-destructive detection of spoilage in chicken breast muscles via NIRS and FTIR | PCA, PLS-DA, and outer product analysis (OPA) | OPA performed better compared to PCA and PLS-DA in discriminating between bacterial loads | [87] |
| Detection and prediction of microbial spoilage in salmon with NIRS | PCA and PLS | The validation curve exhibited a large error of R2 = 0.64, although the calibration equation presented a good R2 of 0.95 | [94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandian, S.; Lakshmi, S.A.; Priya, A.; Balasubramaniam, B.; Zaukuu, J.-L.Z.; Durgadevi, R.; Abe-Inge, V.; Sohn, S.-I. Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview. Processes 2023, 11, 1191. https://doi.org/10.3390/pr11041191
Pandian S, Lakshmi SA, Priya A, Balasubramaniam B, Zaukuu J-LZ, Durgadevi R, Abe-Inge V, Sohn S-I. Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview. Processes. 2023; 11(4):1191. https://doi.org/10.3390/pr11041191
Chicago/Turabian StylePandian, Subramani, Selvaraj Alagu Lakshmi, Arumugam Priya, Boopathi Balasubramaniam, John-Lewis Zinia Zaukuu, Ravindran Durgadevi, Vincent Abe-Inge, and Soo-In Sohn. 2023. "Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview" Processes 11, no. 4: 1191. https://doi.org/10.3390/pr11041191
APA StylePandian, S., Lakshmi, S. A., Priya, A., Balasubramaniam, B., Zaukuu, J.-L. Z., Durgadevi, R., Abe-Inge, V., & Sohn, S.-I. (2023). Spectroscopic Methods for the Detection of Microbial Pathogens and Diagnostics of Infectious Diseases—An Updated Overview. Processes, 11(4), 1191. https://doi.org/10.3390/pr11041191











