Computational Insights into the Interaction between Neprilysin and α-Bisabolol: Proteolytic Activity against Beta-Amyloid Aggregates in Alzheimer’s Disease
Abstract
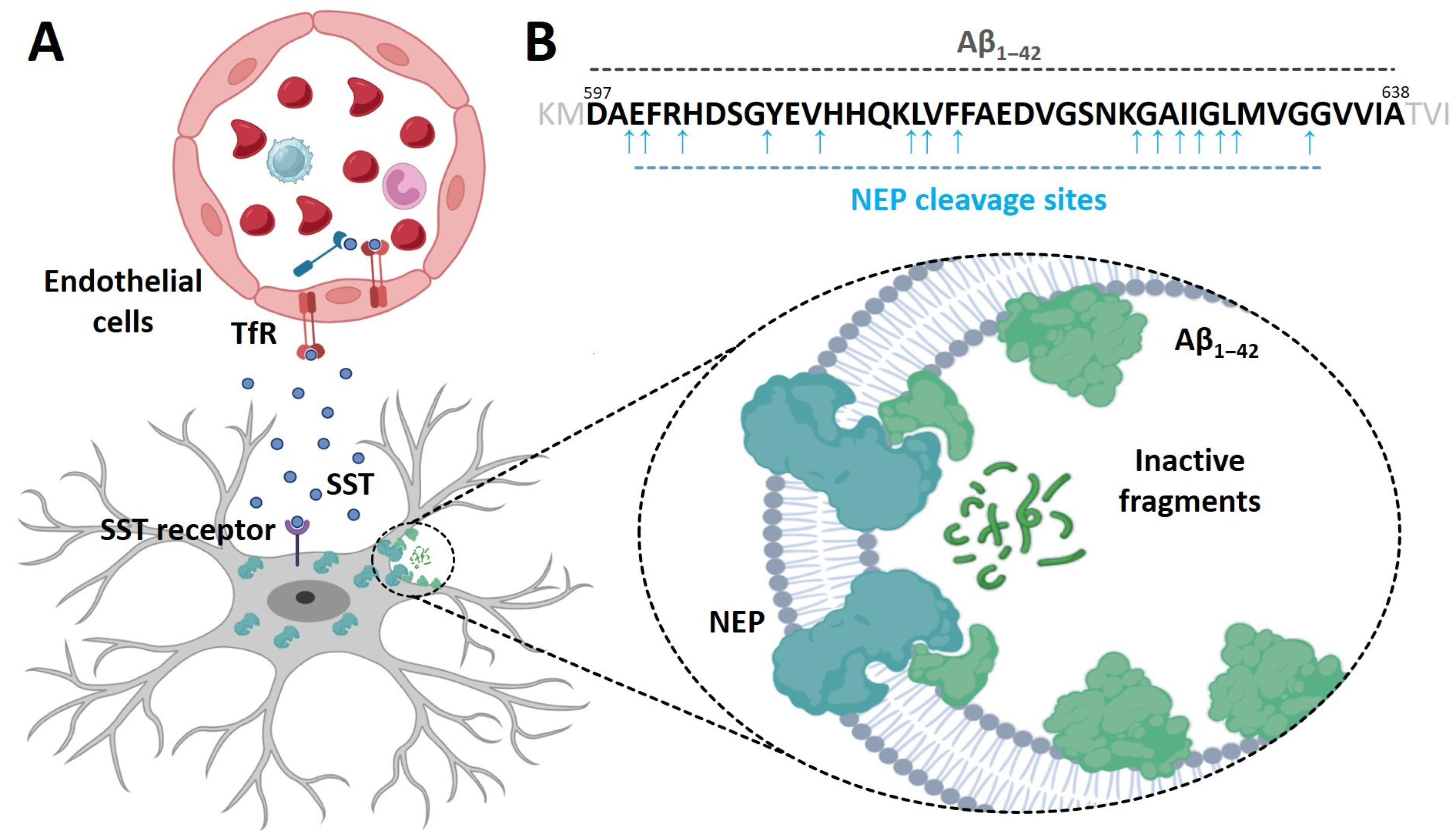
:1. Introduction
2. Materials and Methods
2.1. Domain and Motif Prediction
2.2. Structural Prediction of the Ectodomain of NEP2(m)
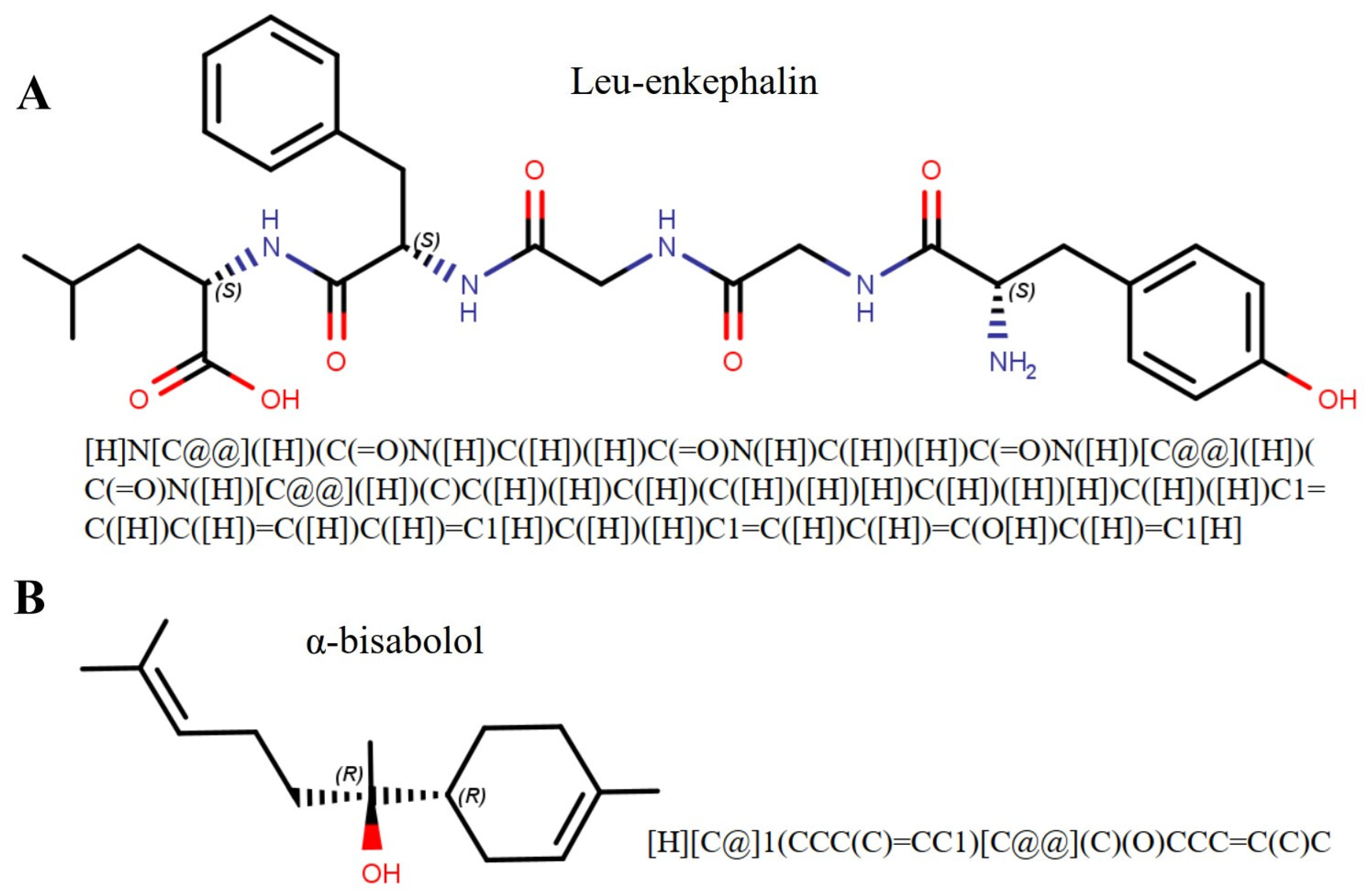
2.3. Bioactivity Prediction of the α-Bisabolol
2.4. Prediction of Pockets in NEP2(m) and Docking Calculations
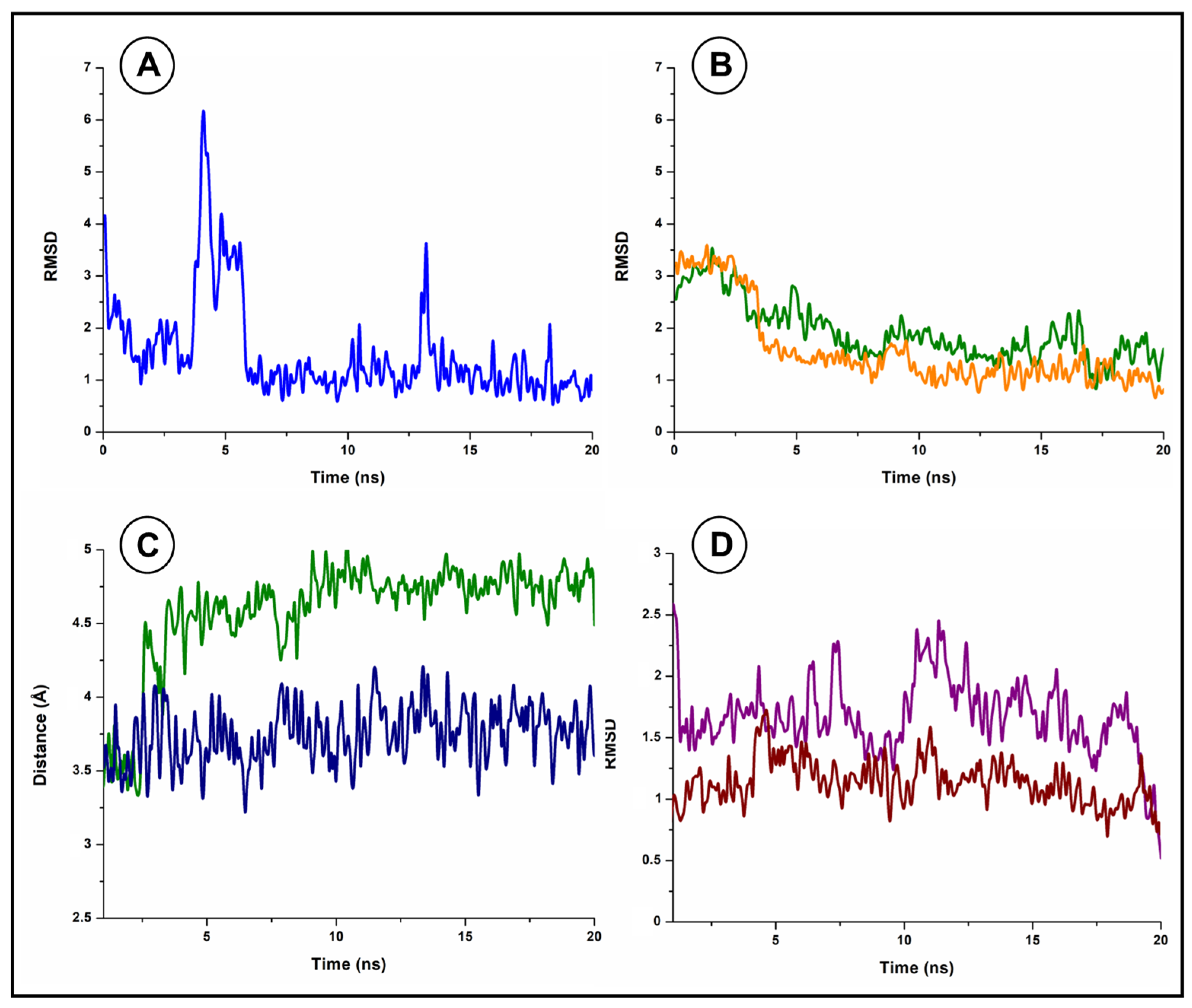
2.5. Molecular Dynamics Simulation and Schematic Representations
3. Results
3.1. Prediction of the Domains and Motifs of the Ectodomain of NEP2(m)
3.2. Structural Analysis of the Ectodomain of NEP2(m)
3.3. Ligand Bioactivity Prediction
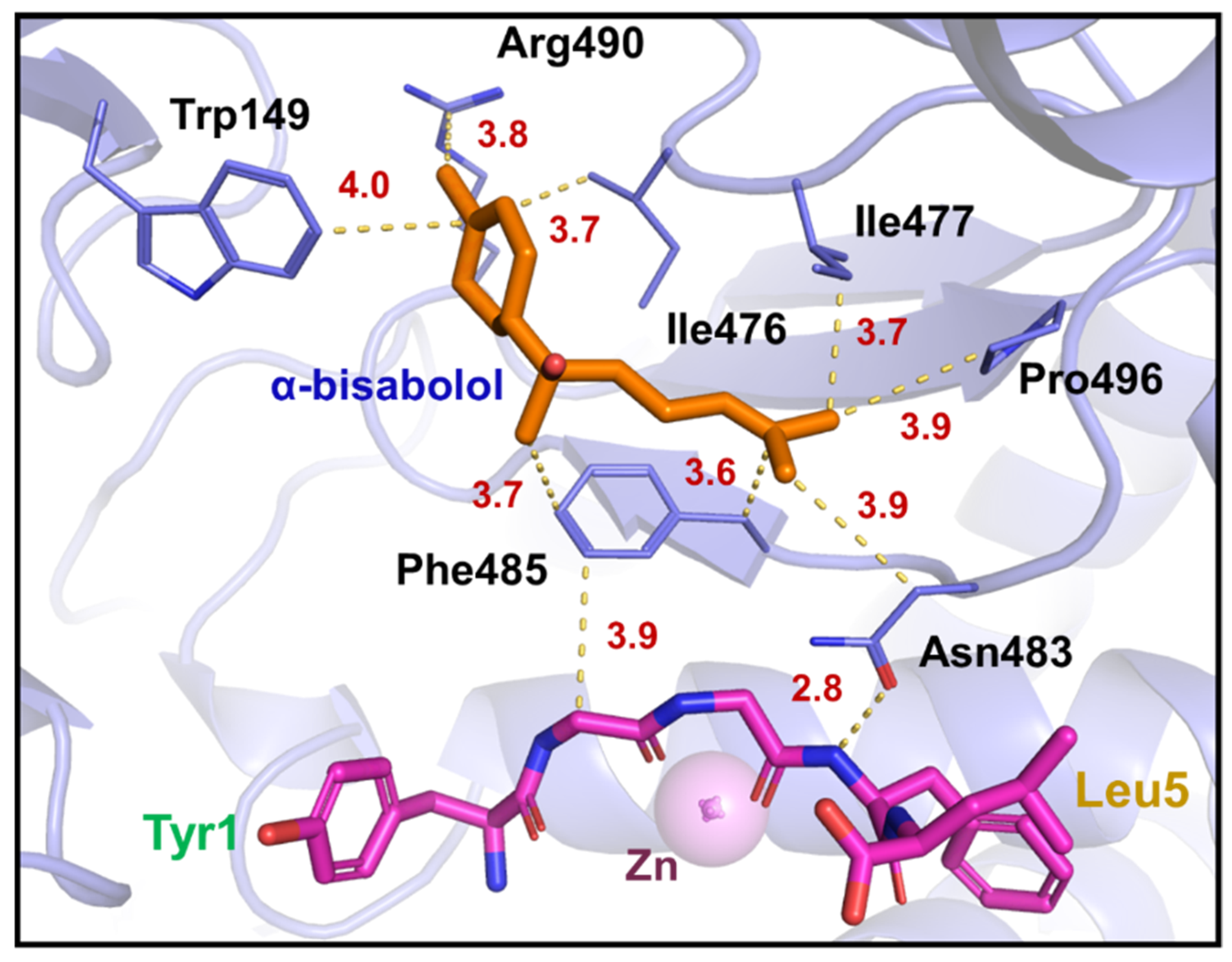
3.4. Molecular Docking Studies and Molecular Dynamics
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2019 Dementia Forecasting Collaborators. Estimation of the Global Prevalence of Dementia in 2019 and Forecasted Prevalence in 2050: An Analysis for the Global Burden of Disease Study 2019. Lancet Public Health 2022, 7, e105–e125. [Google Scholar] [CrossRef] [PubMed]
- Bovolenta, T.M.; Schumacher-Schuh, A.F.; Santos-Lobato, B.L.d.; Godeiro Júnior, C.d.O.; Silva, D.J.d.; Nicaretta, D.; Barbosa, E.R.; Cardoso, F.E.C.; Della Coletta, M.V.; Braga Neto, P.; et al. Average Annual Cost of Parkinson’s Disease in a Brazilian Multiethnic Population. Park. Relat. Disord. 2023, 117, 105897. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.J.; Xiong, C.; Benzinger, T.L.S.; Fagan, A.M.; Goate, A.; Fox, N.C.; Marcus, D.S.; Cairns, N.J.; Xie, X.; Blazey, T.M.; et al. Clinical and Biomarker Changes in Dominantly Inherited Alzheimer’s Disease. N. Engl. J. Med. 2012, 367, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Foster, E.M.; Fernandes, M.; Dangla-Valls, A.; Hublitz, P.; Pangalos, M.; Lovestone, S.; Ribe, E.M.; Buckley, N.J. Glycosylated Clusterin Species Facilitate Aβ Toxicity in Human Neurons. Sci. Rep. 2022, 12, 18639. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Ali, R.; Verma, S. Aβ-Oligomers: A Potential Therapeutic Target for Alzheimer’s Disease. Int. J. Biol. Macromol. 2023, 239, 124231. [Google Scholar] [CrossRef] [PubMed]
- Connor, J.P.; Quinn, S.D.; Schaefer, C. Sticker-and-Spacer Model for Amyloid Beta Condensation and Fibrillation. Front. Mol. Neurosci. 2022, 15, 962526. [Google Scholar] [CrossRef] [PubMed]
- Schraen-Maschke, S.; Duhamel, A.; Vidal, J.S.; Ramdane, N.; Vaudran, L.; Dussart, C.; Buée, L.; Sablonnière, B.; Delaby, C.; Allinquant, B.; et al. The Free Plasma Amyloid Aβ1–42/Aβ1–40 Ratio Predicts Conversion to Dementia for Subjects with Mild Cognitive Impairment with Performance Equivalent to That of the Total Plasma Aβ1–42/Aβ1–40 Ratio. The BALTAZAR Study. Neurobiol. Dis. 2024, 193, 106459. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.; Tam, K.Y. Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regen. Res. 2022, 17, 543. [Google Scholar] [CrossRef] [PubMed]
- Malfroy, B.; Kuang, W.-J.; Seeburg, P.H.; Mason, A.J.; Schofield, P.R. Molecular Cloning and Amino Acid Sequence of Human Enkephalinase (Neutral endopeptidase). FEBS Lett. 1988, 229, 206–210. [Google Scholar] [CrossRef]
- Malfroy, B.; Schofield, P.R.; Kuang, W.-J.; Seeburg, P.H.; Mason, A.J.; Henzel, W.J. Molecular Cloning and Amino Acid Sequence of Rat Enkephalinase. Biochem. Biophys. Res. Commun. 1987, 144, 59–66. [Google Scholar] [CrossRef]
- Devault, A.; Lazure, C.; Nault, C.; Le Moual, H.; Seidah, N.G.; Chrétien, M.; Kahn, P.; Powell, J.; Mallet, J.; Beaumont, A. Amino Acid Sequence of Rabbit Kidney Neutral Endopeptidase 24.11 (Enkephalinase) Deduced from a Complementary DNA. EMBO J. 1987, 6, 1317–1322. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Saito, Y. Roles of Natriuretic Peptides and the Significance of Neprilysin in Cardiovascular Diseases. Biology 2022, 11, 1017. [Google Scholar] [CrossRef]
- Nalivaeva, N.; Zhuravin, I.; Turner, A. Neprilysin Expression and Functions in Development, Ageing, and Disease. Mech. Ageing Dev. 2020, 192, 111363. [Google Scholar] [CrossRef] [PubMed]
- Zhi, J.; Yin, L.; Zhang, Z.; Lv, Y.; Wu, F.; Yang, Y.; Zhang, E.; Li, H.; Lu, N.; Zhou, M.; et al. Network Pharmacology-Based Analysis of Jin-Si-Wei on the Treatment of Alzheimer’s Disease. J. Ethnopharmacol. 2024, 319, 117291. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Emoto, N.; Giaid, A.; Slaughter, C.; Kaw, S.; deWit, D.; Yanagisawa, M. ECE-1: A Membrane-Bound Metalloprotease That Catalyzes the Proteolytic Activation of Big Endothelin-1. Cell 1994, 78, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Emoto, N.; Yanagisawa, M. Endothelin-Converting Enzyme-2 Is a Membrane-Bound, Phosphoramidon-Sensitive Metalloprotease with Acidic pH Optimum. J. Biol. Chem. 1995, 270, 15262–15268. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Quinto, J.; Clausen, D.; Pérez-González, R.; Peng, H.; Meszaros, A.; Eckman, C.B.; Levy, E.; Eckman, E.A. Intracellular Metalloprotease Activity Controls Intraneuronal Aβ Aggregation and Limits Secretion of Aβ via Exosomes. FASEB J. 2019, 33, 3758–3771. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.; Lee, E.J. Advances in Amyloid-β Clearance in the Brain and Periphery: Implications for Neurodegenerative Diseases. Exp. Neurobiol. 2023, 32, 216–246. [Google Scholar] [CrossRef]
- Bland, N.D.; Pinney, J.W.; Thomas, J.E.; Turner, A.J.; Isaac, R.E. Bioinformatic Analysis of the Neprilysin (M13) Family of Peptidases Reveals Complex Evolutionary and Functional Relationships. BMC Evol. Biol. 2008, 8, 16. [Google Scholar] [CrossRef]
- Turner, A.J.; Tanzawa, K. Mammalian Membrane Metallopeptidases: NEP, ECE, KELL, and PEX. FASEB J. 1997, 11, 355–364. [Google Scholar] [CrossRef]
- Lee, S.; Zambas, E.D.; Marsh, W.L.; Redman, C.M. Molecular Cloning and Primary Structure of Kell Blood Group Protein. Proc. Natl. Acad. Sci. USA 1991, 88, 6353–6357. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Couture, C.; Hirata, I.Y.; Juliano, M.A.; Loisel, T.P.; Crine, P.; Juliano, L.; Boileau, G.; Carmona, A.K. Human Recombinant Endopeptidase PHEX Has a Strict S1’ Specificity for Acidic Residues and Cleaves Peptides Derived from Fibroblast Growth Factor-23 and Matrix Extracellular Phosphoglycoprotein. Biochem. J. 2003, 373, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Desbarats, M.; Viel, J.; Glorieux, F.H.; Cawthorn, C.; Ecarot, B. cDNA Cloning of the Murine Pex Gene Implicated in X-Linked Hypophosphatemia and Evidence for Expression in Bone. Genomics 1996, 36, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Ul-Haq, Z.; Iqbal, S.; Moin, S.T. Dynamic Changes in the Secondary Structure of ECE-1 and XCE Account for Their Different Substrate Specificities. BMC Bioinform. 2012, 13, 285. [Google Scholar] [CrossRef] [PubMed]
- Valdenaire, O.; Richards, J.G.; Faull, R.L.M.; Schweizer, A. XCE, a New Member of the Endothelin-Converting Enzyme and Neutral Endopeptidase Family, Is Preferentially Expressed in the CNS. Mol. Brain Res. 1999, 64, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.K.; Dankel, S.; Cai, W.; Sakaguchi, M.; Kasif, S.; Kahn, C.R. Membrane Metallo-Endopeptidase (Neprilysin) Regulates Inflammatory Response and Insulin Signaling in White Preadipocytes. Mol. Metab. 2019, 22, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Pope, D.; Madura, J.D.; Cascio, M. β-Amyloid and Neprilysin Computational Studies Identify Critical Residues Implicated in Binding Specificity. J. Chem. Inf. Model. 2014, 54, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Shanmuganathan, B.; Suryanarayanan, V.; Sathya, S.; Narenkumar, M.; Singh, S.K.; Ruckmani, K.; Pandima Devi, K. Anti-Amyloidogenic and Anti-Apoptotic Effect of α-Bisabolol against Aβ Induced Neurotoxicity in PC12 Cells. Eur. J. Med. Chem. 2018, 143, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.S.M.; Lilyanna, S.; Ng, J.Y.X.; Chong, J.P.C.; Lin, Q.; Yong, X.E.; Lim, T.K.; Lin, Q.; Richards, A.M.; Liew, O.W. Multiple Circulating Forms of Neprilysin Detected with Novel Epitope-Directed Monoclonal Antibodies. Cell. Mol. Life Sci. 2024, 81, 42. [Google Scholar] [CrossRef] [PubMed]
- Moss, S.; Subramanian, V.; Acharya, K.R. High-Resolution Crystal Structure of Substrate-Free Human Neprilysin. J. Struct. Biol. 2018, 204, 19–25. [Google Scholar] [CrossRef]
- Bayes-Genis, A.; Barallat, J.; Richards, A.M. A Test in Context: Neprilysin. J. Am. Coll. Cardiol. 2016, 68, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Oefner, C.; D’Arcy, A.; Hennig, M.; Winkler, F.K.; Dale, G.E. Structure of Human Neutral Endopeptidase (Neprilysin) Complexed with Phosphoramidon. J. Mol. Biol. 2000, 296, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.-J.; Martin, D.B.; Aebersold, R. Identification and Quantification of N-Linked Glycoproteins Using Hydrazide Chemistry, Stable Isotope Labeling and Mass Spectrometry. Nat. Biotechnol. 2003, 21, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Sato, B.; Katagiri, Y.U.; Iijima, K.; Yamada, H.; Ito, S.; Kawasaki, N.; Okita, H.; Fujimoto, J.; Kiyokawa, N. The Human CD10 Lacking an N-Glycan at Asn628 Is Deficient in Surface Expression and Neutral Endopeptidase Activity. Biochim. Biophys. Acta (BBA) Gen. Subj. 2012, 1820, 1715–1723. [Google Scholar] [CrossRef] [PubMed]
- Deeb, S.J.; Cox, J.; Schmidt-Supprian, M.; Mann, M. N-Linked Glycosylation Enrichment for in-Depth Cell Surface Proteomics of Diffuse Large b-Cell Lymphoma Subtypes. Mol. Cell. Proteom. 2014, 13, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Miners, J.S.; Van Helmond, Z.; Kehoe, P.G.; Love, S. Changes with Age in the Activities of β-Secretase and the Aβ-Degrading Enzymes Neprilysin, Insulin-Degrading Enzyme and Angiotensin-Converting Enzyme. Brain Pathol. 2010, 20, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Shirotani, K.; Tsubuki, S.; Iwata, N.; Takaki, Y.; Harigaya, W.; Maruyama, K.; Kiryu-Seo, S.; Kiyama, H.; Iwata, H.; Tomita, T.; et al. Neprilysin Degrades Both Amyloid β Peptides 1–40 and 1–42 Most Rapidly and Efficiently among Thiorphan- and Phosphoramidon-Sensitive Endopeptidases. J. Biol. Chem. 2001, 276, 21895–21901. [Google Scholar] [CrossRef] [PubMed]
- Iwata, N.; Mizukami, H.; Shirotani, K.; Takaki, Y.; Muramatsu, S.; Lu, B.; Gerard, N.P.; Gerard, C.; Ozawa, K.; Saido, T.C. Presynaptic Localization of Neprilysin Contributes to Efficient Clearance of Amyloid-β Peptide in Mouse Brain. J. Neurosci. 2004, 24, 991–998. [Google Scholar] [CrossRef]
- Iwata, N.; Tsubuki, S.; Hama, E.; Takaki, Y.; Shirotani, K.; Saido, T.C. Reply to: “Clearance of Amyloid β-Peptide from Brain: Transport or Metabolism?". Nat. Med. 2000, 6, 718–719. [Google Scholar] [CrossRef]
- Iwata, N.; Tsubuki, S.; Takaki, Y.; Shirotani, K.; Lu, B.; Gerard, N.P.; Gerard, C.; Hama, E.; Lee, H.-J.; Saido, T.C. Metabolic Regulation of Brain Aβ by Neprilysin. Science 2001, 292, 1550–1552. [Google Scholar] [CrossRef]
- El-Amouri, S.S.; Zhu, H.; Yu, J.; Marr, R.; Verma, I.M.; Kindy, M.S. Neprilysin: An Enzyme Candidate to Slow the Progression of Alzheimer’s Disease. Am. J. Pathol. 2008, 172, 1342–1354. [Google Scholar] [CrossRef]
- Tampellini, D.; Rahman, N.; Lin, M.T.; Capetillo-Zarate, E.; Gouras, G.K. Impaired β-Amyloid Secretion in Alzheimer’s Disease Pathogenesis. J. Neurosci. 2011, 31, 15384–15390. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, J.; Liu, J.; Liu, F. A Novel System for in Vivo Neprilysin Gene Delivery Using a Syringe Electrode. J. Neurosci. Methods 2010, 193, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Marr, R.A.; Guan, H.; Rockenstein, E.; Kindy, M.; Gage, F.H.; Verma, I.; Masliah, E.; Hersh, L.B. Neprilysin Regulates Amyloid β Peptide Levels. J. Mol. Neurosci. 2004, 22, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Hemming, M.L.; Patterson, M.; Reske-Nielsen, C.; Lin, L.; Isacson, O.; Selkoe, D.J. Reducing Amyloid Plaque Burden via Ex Vivo Gene Delivery of an Aβ-Degrading Protease: A Novel Therapeutic Approach to Alzheimer Disease. PLoS Med. 2007, 4, e262. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Liu, Y.; Daily, A.; Police, S.; Kim, M.-H.; Oddo, S.; LaFerla, F.M.; Pauly, J.R.; Murphy, M.P.; Hersh, L.B. Peripherally Expressed Neprilysin Reduces Brain Amyloid Burden: A Novel Approach for Treating Alzheimer’s Disease. J. Neurosci. Res. 2009, 87, 1462–1473. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Studzinski, C.; Beckett, T.; Guan, H.; Hersh, M.A.; Murphy, M.P.; Klein, R.; Hersh, L.B. Expression of Neprilysin in Skeletal Muscle Reduces Amyloid Burden in a Transgenic Mouse Model of Alzheimer Disease. Mol. Ther. 2009, 17, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Studzinski, C.; Beckett, T.; Murphy, M.P.; Klein, R.L.; Hersh, L.B. Circulating Neprilysin Clears Brain Amyloid. Mol. Cell. Neurosci. 2010, 45, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Spencer, B.; Marr, R.A.; Gindi, R.; Potkar, R.; Michael, S.; Adame, A.; Rockenstein, E.; Verma, I.M.; Masliah, E. Peripheral Delivery of a CNS Targeted, Metalo-Protease Reduces Aβ Toxicity in a Mouse Model of Alzheimer’s Disease. PLoS ONE 2011, 6, e16575. [Google Scholar] [CrossRef]
- Roques, B.P.; Noble, F.; Daugé, V.; Fournié-Zaluski, M.C.; Beaumont, A. Neutral Endopeptidase 24.11: Structure, Inhibition, and Experimental and Clinical Pharmacology. Pharmacol. Rev. 1993, 45, 87–146. [Google Scholar]
- Iijima-Ando, K.; Hearn, S.A.; Granger, L.; Shenton, C.; Gatt, A.; Chiang, H.-C.; Hakker, I.; Zhong, Y.; Iijima, K. Overexpression of Neprilysin Reduces Alzheimer Amyloid-Β42 (Aβ42)-Induced Neuron Loss and Intraneuronal Aβ42 Deposits but Causes a Reduction in Camp-Responsive Element-Binding Protein-Mediated Transcription, Age-Dependent Axon Pathology, and Premature Death in Drosophila. J. Biol. Chem. 2008, 283, 19066–19076. [Google Scholar] [CrossRef] [PubMed]
- Nagore, P.; Lokhande, P.; Mujawar, H. In-Vitro Bioevaluation, Pharmacokinetics and Molecular Docking Study of Unexplored Bisabolol-Rich Curcuma Inodora Blatt. Essential Oil from Konkan Region: A Biodiversity Hotspot. Plant Sci. Today 2023, 10, 223–231. [Google Scholar] [CrossRef]
- Nazarinia, D.; Moslehi, A.; Hashemi, P. (-)-α-Bisabolol Exerts Neuroprotective Effects against Pentylenetetrazole-Induced Seizures in Rats by Targeting Inflammation and Oxidative Stress. Physiol. Behav. 2023, 272, 114351. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yu, S.; Kim, J.; Khaliq, N.U.; Choi, W.I.; Kim, H.; Sung, D. Facile Fabrication of α-Bisabolol Nanoparticles with Improved Antioxidant and Antibacterial Effects. Antioxidants 2023, 12, 207. [Google Scholar] [CrossRef] [PubMed]
- Kamatou, G.P.P.; Viljoen, A.M. A Review of the Application and Pharmacological Properties of α-Bisabolol and α-Bisabolol-Rich Oils. J. Am. Oil Chem. Soc. 2010, 87, 1–7. [Google Scholar] [CrossRef]
- Eddin, L.B.; Jha, N.K.; Goyal, S.N.; Agrawal, Y.O.; Subramanya, S.B.; Bastaki, S.M.A.; Ojha, S. Health Benefits, Pharmacological Effects, Molecular Mechanisms, and Therapeutic Potential of α-Bisabolol. Nutrients 2022, 14, 1370. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.Y.D.; Carmo, M.R.S.d.; Fonteles, A.A.; Neves, J.C.d.S.; Silva, A.T.A.d.; Pereira, J.F.; Ferreira, E.d.O.; Lima, N.M.R.d.; Neves, K.R.T.; Andrade, G.M.d. (-)-α-Bisabolol Prevents Neuronal Damage and Memory Deficits through Reduction of Proinflammatory Markers Induced by Permanent Focal Cerebral Ischemia in Mice. Eur. J. Pharmacol. 2019, 842, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Braga, P.C.; Dal Sasso, M.; Fonti, E.; Culici, M. Antioxidant Activity of Bisabolol: Inhibitory Effects on Chemiluminescence of Human Neutrophil Bursts and Cell-Free Systems. Pharmacology 2009, 83, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Zargaran, A.; Borhani Haghighi, A.; Faridi, P.; Daneshamouz, S.; Kordafshari, G.; Mohagheghzadeh, A. Potential Effect and Mechanism of Action of Topical Chamomile (Matricaria chamomila L.) Oil on Migraine Headache: A Medical Hypothesis. Med. Hypotheses 2014, 83, 566–569. [Google Scholar] [CrossRef]
- Nascimento, T.V.C. Evidence for the Involvement of TNF-α and IL-1β in the Antinociceptive and Anti-Inflammatory Activity of Stachys lavandulifolia Vahl. (Lamiaceae) Essential Oil and (-)-α-Bisabolol, Its Main Compound, in Mice. J. Ethnopharmacol. 2016, 191, 9–18. [Google Scholar]
- Sathya, S.; Shanmuganathan, B.; Devi, K.P. Deciphering the Anti-Apoptotic Potential of α-Bisabolol Loaded Solid Lipid Nanoparticles against Aβ Induced Neurotoxicity in Neuro-2a Cells. Colloids Surf. B Biointerfaces 2020, 190, 110948. [Google Scholar] [CrossRef] [PubMed]
- Jeyakumar, M.; Sathya, S.; Gandhi, S.; Tharra, P.; Suryanarayanan, V.; Singh, S.K.; Baire, B.; Pandima Devi, K. α-Bisabolol β-D-Fucopyranoside as a Potential Modulator of β-Amyloid Peptide Induced Neurotoxicity: An in vitro & in silico study. Bioorganic Chem. 2019, 88, 102935. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Boratyn, G.M.; Joukov, V.; Vera Alvarez, R.; Madden, T.L. ElasticBLAST: Accelerating Sequence Search via Cloud Computing. BMC Bioinform. 2023, 24, 117. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Research and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Moss, S.; Subramanian, V.; Acharya, K.R. Crystal Structure of Peptide-Bound Neprilysin Reveals Key Binding Interactions. FEBS Lett. 2020, 594, 327–336. [Google Scholar] [CrossRef]
- Shen, M.; Sali, A. Statistical Potential for Assessment and Prediction of Protein Structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B.; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-Atom Contacts and Structure Validation for Proteins and Nucleic Acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef]
- Prisant, M.G.; Williams, C.J.; Chen, V.B.; Richardson, J.S.; Richardson, D.C. New Tools in MolProbity Validation: CaBLAM for CryoEM Backbone, UnDowser to Rethink “Waters,” and NGL Viewer to Recapture Online 3D Graphics. Protein Sci. 2020, 29, 315–329. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. DoGSiteScorer: A Web Server for Automatic Binding Site Prediction, Analysis and Druggability Assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed Atlas of Surface Topography of Proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef] [PubMed]
- da Cruz Freire, J.E.; Júnior, J.E.M.; Pinheiro, D.P.; da Cruz Paiva Lima, G.E.; do Amaral, C.L.; Veras, V.R.; Madeira, M.P.; Freire, E.B.L.; Ozório, R.G.; Fernandes, V.O.; et al. Evaluation of the Anti-Diabetic Drug Sitagliptin as a Novel Attenuate to SARS-CoV-2 Evidence-Based in Silico: Molecular Docking and Molecular Dynamics. 3 Biotech 2022, 12, 344. [Google Scholar] [CrossRef]
- Gonzatti, M.B.; Júnior, J.E.M.; Rocha, A.J.; de Oliveira, J.S.; Evangelista, A.J.d.J.; Fonseca, F.M.P.; Ceccatto, V.M.; de Oliveira, A.C.; da Cruz Freire, J.E. Mechanism of Molecular Interaction of Sitagliptin with Human DPP4 Enzyme—New Insights. Adv. Med. Sci. 2023, 68, 402–408. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A Smooth Particle Mesh Ewald Method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular Dynamics with Coupling to an External Bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A Parallel Linear Constraint Solver for Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Schiering, N.; D’Arcy, A.; Villard, F.; Ramage, P.; Logel, C.; Cumin, F.; Ksander, G.M.; Wiesmann, C.; Karki, R.G.; Mogi, M. Structure of Neprilysin in Complex with the Active Metabolite of Sacubitril. Sci. Rep. 2016, 6, 27909. [Google Scholar] [CrossRef]
- Pankow, K.; Schwiebs, A.; Becker, M.; Siems, W.-E.; Krause, G.; Walther, T. Structural Substrate Conditions Required for Neutral Endopeptidase-Mediated Natriuretic Peptide Degradation. J. Mol. Biol. 2009, 393, 496–503. [Google Scholar] [CrossRef]
- Salazar, J.; Rojas-Quintero, J.; Cano, C.; Pérez, J.L.; Ramírez, P.; Carrasquero, R.; Torres, W.; Espinoza, C.; Chacín-González, M.; Bermúdez, V. Neprilysin: A Potential Therapeutic Target of Arterial Hypertension? Curr. Cardiol Rev. 2020, 16, 25–35. [Google Scholar] [CrossRef]
- Campbell, D.J.; Anastasopoulos, F.; Duncan, A.M.; James, G.M.; Kladis, A.; Briscoe, T.A. Effects of Neutral Endopeptidase Inhibition and Combined Angiotensin Converting Enzyme and Neutral Endopeptidase Inhibition on Angiotensin and Bradykinin Peptides in Rats. J. Pharmacol. Exp. Ther. 1998, 287, 567–577. [Google Scholar]
- Stephenson, S.L.; Kenny, A.J. The Hydrolysis of Alpha-Human Atrial Natriuretic Peptide by Pig Kidney Microvillar Membranes Is Initiated by Endopeptidase-24.11. Biochem J. 1987, 243, 183–187. [Google Scholar] [CrossRef]
- Jiang, W.; Jiang, H.-F.; Pan, C.-S.; Cai, D.-Y.; Qi, Y.-F.; Pang, Y.-Z.; Tang, C.-S. Relationship between the Contents of Adrenomedullin and Distributions of Neutral Endopeptidase in Blood and Tissues of Spontaneously Hypertensive Rats. Hypertens. Res. 2004, 27, 109–117. [Google Scholar] [CrossRef]
- Kokkonen, J.O.; Kuoppala, A.; Saarinen, J.; Lindstedt, K.A.; Kovanen, P.T. Kallidin- and Bradykinin-Degrading Pathways in Human Heart: Degradation of Kallidin by Aminopeptidase M-like Activity and Bradykinin by Neutral Endopeptidase. Circulation 1999, 99, 1984–1990. [Google Scholar] [CrossRef] [PubMed]
- Mangiafico, S.; Costello-Boerrigter, L.C.; Andersen, I.A.; Cataliotti, A.; Burnett, J.C. Neutral Endopeptidase Inhibition and the Natriuretic Peptide System: An Evolving Strategy in Cardiovascular Therapeutics. Eur. Heart J. 2013, 34, 886–893c. [Google Scholar] [CrossRef] [PubMed]
- Abassi, Z.; Golomb, E.; Keiser, H.R. Neutral Endopeptidase Inhibition Increases the Urinary Excretion and Plasma Levels of Endothelin. Metabolism 1992, 41, 683–685. [Google Scholar] [CrossRef]
- Jaffe, A.S.; Apple, F.S.; Mebazaa, A.; Vodovar, N. Unraveling N-Terminal pro-B-Type Natriuretic Peptide: Another Piece to a Very Complex Puzzle in Heart Failure Patients. Clin. Chem. 2015, 61, 1016–1018. [Google Scholar] [CrossRef] [PubMed]
- Riddell, E.; Vader, J.M. Potential Expanded Indications for Neprilysin Inhibitors. Curr. Heart Fail. Rep. 2017, 14, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Laws III, J.S.; Shrestha, S.; Smid, S.D. Cannabis Terpenes Display Variable Protective and Anti-Aggregatory Actions against Neurotoxic β Amyloid in Vitro: Highlighting the Protective Bioactivity of α-Bisabolol in Motorneuronal-like NSC-34 Cells. Neuro Toxicol. 2022, 90, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Sathya, S.; Shanmuganathan, B.; Manirathinam, G.; Ruckmani, K.; Devi, K.P. α-Bisabolol Loaded Solid Lipid Nanoparticles Attenuates Aβ Aggregation and Protects Neuro-2a Cells from Aβ Induced Neurotoxicity. J. Mol. Liq. 2018, 264, 431–441. [Google Scholar] [CrossRef]
- Khan, M.F.; Kundu, D.; Hazra, C.; Patra, S. A Strategic Approach of Enzyme Engineering by Attribute Ranking and Enzyme Immobilization on Zinc Oxide Nanoparticles to Attain Thermostability in Mesophilic Bacillus Subtilis Lipase for Detergent Formulation. Int. J. Biol. Macromol. 2019, 136, 66–82. [Google Scholar] [CrossRef]
- Sharma, U.; Cozier, G.E.; Sturrock, E.D.; Acharya, K.R. Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme. J. Med. Chem. 2020, 63, 5488–5500. [Google Scholar] [CrossRef]
- Verma, R.; Mitchell-Koch, K. In Silico Studies of Small Molecule Interactions with Enzymes Reveal Aspects of Catalytic Function. Catalysts 2017, 7, 212. [Google Scholar] [CrossRef]
- Agarwal, P.K.; Doucet, N.; Chennubhotla, C.; Ramanathan, A.; Narayanan, C. Conformational Sub-States and Populations in Enzyme Catalysis. Methods Enzym. 2016, 578, 273–297. [Google Scholar] [CrossRef]
- Bruice, T.C. Computational Approaches: Reaction Trajectories, Structures, and Atomic Motions. Enzyme Reactions and Proficiency. Chem. Rev. 2006, 106, 3119–3139. [Google Scholar] [CrossRef]
- Ramanathan, A.; Savol, A.; Burger, V.; Chennubhotla, C.S.; Agarwal, P.K. Protein Conformational Populations and Functionally Relevant Substates. Acc Chem. Res. 2014, 47, 149–156. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Jeyakumar, M.; Sathya, S.; Gandhi, S.; Tharra, P.; Aarthy, M.; Balan, D.J.; Kiruthiga, C.; Baire, B.; Singh, S.K.; Devi, K.P. α-Bisabolol β-D-Fucopyranoside Inhibits β-Amyloid (Aβ)25–35 Induced Oxidative Stress in Neuro-2a Cells via Antioxidant Approaches. Process Biochem. 2022, 121, 493–503. [Google Scholar] [CrossRef]

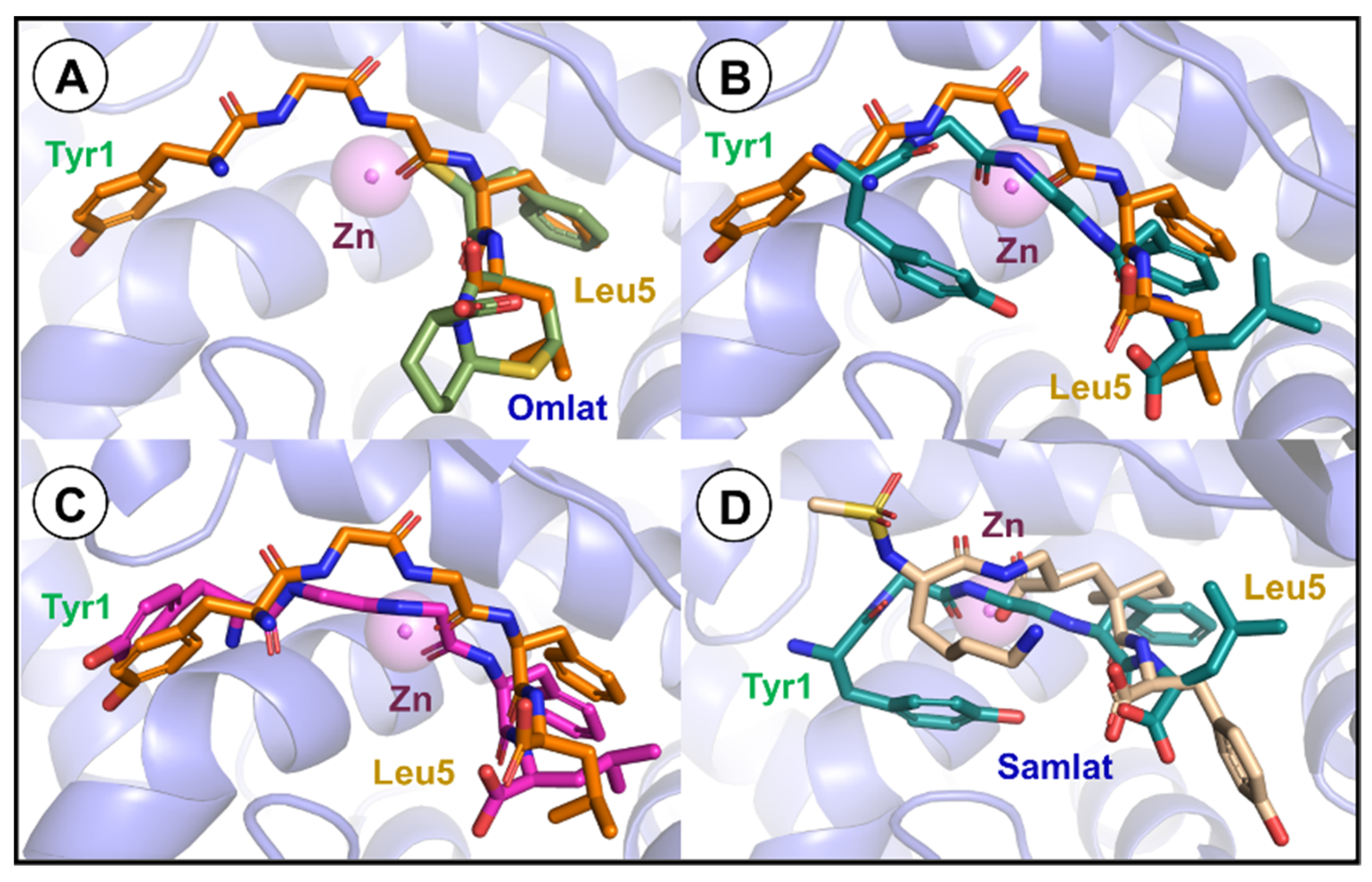

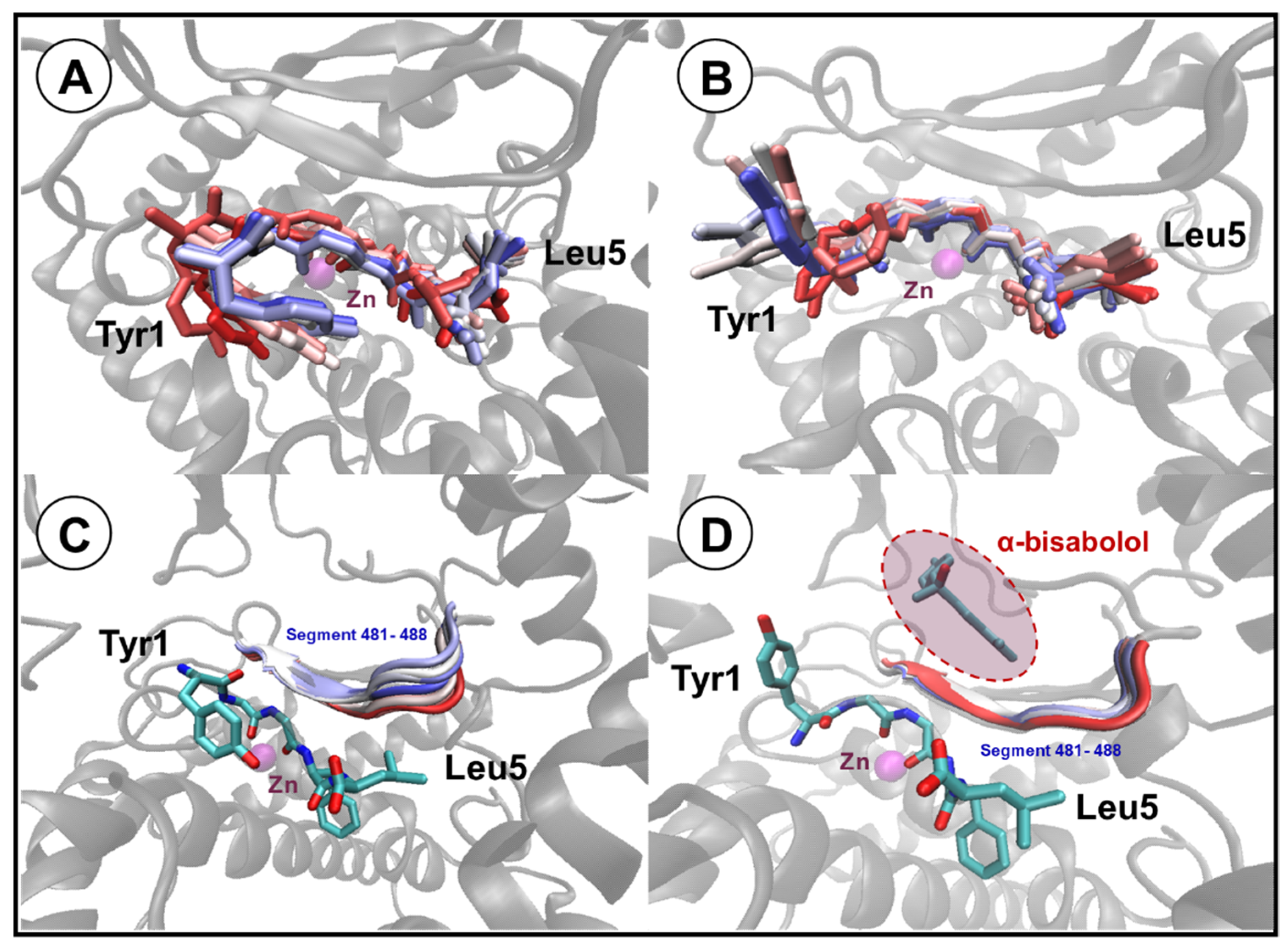

| Neprilysin | Leu-Enkephalin | Bond Properties | |||
|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | Type of Interaction | Distance (Å) |
| His237 | NE2 | Tyr1 | OH | H-bond | 3.3 |
| His258 | ND1 | Tyr1 | O | Van der Waals | 3.6 |
| Ser538 | O | Tyr1 | OH | H-bond | 3.0 |
| Val651 | CG2 | Tyr1 | CD1 | Hydrophobic | 3.6 |
| Glu525 | OE1 | Gly2 | O | H-bond | 3.2 |
| Phe485 | CD2 | Gly3 | CA | Hydrophobic | 3.8 |
| Glu587 | OE2 | Gly3 | O | H-bond | 3.5 |
| ZN | ZN | Gly3 | O | Metal coordination | 2.0 |
| Phe50 | CZ | Phe4 | CE1 | Hydrophobic | 3.3 |
| Asn483 | OD1 | Phe4 | N | H-bond | 3.3 |
| Ala484 | CB | Phe4 | CB | Hydrophobic | 3.9 |
| Phe504 | CE1 | Phe4 | CE1 | Hydrophobic | 3.5 |
| Val521 | CG2 | Phe4 | CE2 | Hydrophobic | 3.7 |
| Glu587 | OE2 | Phe4 | N | Van der Waals | 3.6 |
| Trp634 | CZ2 | Phe4 | CZ | Hydrophobic | 3.6 |
| His652 | NE2 | Phe4 | O | H-bond | 3.2 |
| Arg658 | NH1 | Phe4 | O | H-bond | 3.0 |
| Phe50 | CG | Leu5 | CG | Hydrophobic | 3.6 |
| Asn483 | OD1 | Leu5 | N | H-bond | 3.0 |
| Asn483 | ND2 | Leu5 | OXT | Hydrophobic | 2.9 |
| His652 | ND1 | Leu5 | O | Van der Waals | 3.6 |
| Neprilysin | Leu-Enkephalin | Bond Properties | |||
|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | Type of Interaction | Distance (Å) |
| Glu587 | CG | Tyr1 | CE1 | Hydrophobic | 3.6 |
| Glu587 | OE2 | Tyr1 | OH | H-bond | 2.8 |
| His652 | CB | Tyr1 | CE1 | Hydrophobic | 3.8 |
| ZN | ZN | Gly2 | O | Metal coordination | 2.0 |
| ZN | ZN | Gly3 | O | Metal coordination | 2.1 |
| Ala484 | CB | Phe4 | CB | Hydrophobic | 3.8 |
| Val521 | CG2 | Phe4 | CE1 | Hydrophobic | 3.7 |
| His524 | CD2 | Phe4 | CD2 | Hydrophobic | 3.6 |
| Glu587 | OE2 | Phe4 | O | H-bond | 3.4 |
| Arg658 | NH1 | Phe4 | O | H-bond | 3.0 |
| Phe50 | CE2 | Leu5 | CA | Hydrophobic | 3.6 |
| Leu53 | CD2 | Leu5 | CD2 | Hydrophobic | 3.8 |
| Val482 | CG1 | Leu5 | CD1 | Hydrophobic | 3.5 |
| Ile499 | CD | Leu5 | CD1 | Hydrophobic | 3.8 |
| His652 | NE2 | Leu5 | OT2 | H-bond | 2.7 |
| Neprilysin | α-Bisabolol | Bond Properties | ||
|---|---|---|---|---|
| Residue | Atom | Atom | Type of Interaction | Distance (Å) |
| Trp149 | CH2 | C13 | Hydrophobic | 4.1 |
| Ile476 | CG2 | C13 | Hydrophobic | 3.7 |
| Ile476 | CB | C13 | Hydrophobic | 3.9 |
| Ile476 | O | H14 | Van der Waals | 2.6 |
| Ile477 | CB | C12 | Hydrophobic | 4.0 |
| Asn483 | ND2 | H11 | Van der Waals | 3.0 |
| Phe485 | Centroid | C7 | π-alkyl | 4.1 |
| Arg490 | NH2 | H6 | Van der Waals | 3.0 |
| Pro496 | CG | C4 | Hydrophobic | 4.1 |
| Neprilysin | Leu-Enkephalin | Bond Properties | |||
|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | Type of Interaction | Distance (Å) |
| His237 | ND1 | Tyr1 | OH | H-bond | 3.0 |
| Glu587 | OE2 | Tyr1 | N | H-bond | 2.7 |
| Glu587 | OE2 | Tyr1 | O | H-bond | 3.1 |
| Phe485 | CE1 | Gly2 | CA | Hydrophobic | 3.9 |
| ZN | ZN | Gly2 | O | Metal coordination | 2.1 |
| ZN | ZN | Gly3 | O | Metal coordination | 2.1 |
| Asn483 | OD1 | Phe4 | N | H-bond | 2.8 |
| Glu525 | OE2 | Phe4 | N | Van der Waals | 4.0 |
| His524 | CE1 | Phe4 | CD2 | Hydrophobic | 3.9 |
| Val521 | CG2 | Phe4 | CZ | Hydrophobic | 3.9 |
| Trp634 | CE2 | Phe4 | CZ | Hydrophobic | 4.0 |
| Val633 | CG2 | Phe4 | CZ | Hydrophobic | 4.0 |
| Phe50 | CZ | Phe4 | CE1 | Hydrophobic | 3.9 |
| Arg654 | NH1 | Phe4 | O | H-bond | 2.8 |
| Asn483 | OD1 | Leu5 | N | H-bond | 2.9 |
| Asn483 | ND2 | Leu5 | OT2 | Van der Waals | 4.0 |
| His652 | NE2 | Leu5 | OT1 | H-bond | 2.7 |
| Val482 | CG1 | Leu5 | CD1 | Hydrophobic | 3.8 |
| Phe50 | CG | Leu5 | CD2 | Hydrophobic | 3.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, J.E.R.; da Cruz Freire, J.E.; Vasconcelos-Filho, F.S.L.; da Silva de Almeida, D.; Ceccatto, V.M.; de Sousa, B.L. Computational Insights into the Interaction between Neprilysin and α-Bisabolol: Proteolytic Activity against Beta-Amyloid Aggregates in Alzheimer’s Disease. Processes 2024, 12, 885. https://doi.org/10.3390/pr12050885
Martins JER, da Cruz Freire JE, Vasconcelos-Filho FSL, da Silva de Almeida D, Ceccatto VM, de Sousa BL. Computational Insights into the Interaction between Neprilysin and α-Bisabolol: Proteolytic Activity against Beta-Amyloid Aggregates in Alzheimer’s Disease. Processes. 2024; 12(5):885. https://doi.org/10.3390/pr12050885
Chicago/Turabian StyleMartins, Jonathan Elias Rodrigues, José Ednésio da Cruz Freire, Francisco Sérgio Lopes Vasconcelos-Filho, Diego da Silva de Almeida, Vânia Marilande Ceccatto, and Bruno Lopes de Sousa. 2024. "Computational Insights into the Interaction between Neprilysin and α-Bisabolol: Proteolytic Activity against Beta-Amyloid Aggregates in Alzheimer’s Disease" Processes 12, no. 5: 885. https://doi.org/10.3390/pr12050885
APA StyleMartins, J. E. R., da Cruz Freire, J. E., Vasconcelos-Filho, F. S. L., da Silva de Almeida, D., Ceccatto, V. M., & de Sousa, B. L. (2024). Computational Insights into the Interaction between Neprilysin and α-Bisabolol: Proteolytic Activity against Beta-Amyloid Aggregates in Alzheimer’s Disease. Processes, 12(5), 885. https://doi.org/10.3390/pr12050885






