Cloning, Expression and Enzymatic Characterization of Pectin Methyl Esterase from Populus trichocarpa and Its Application
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Reagents
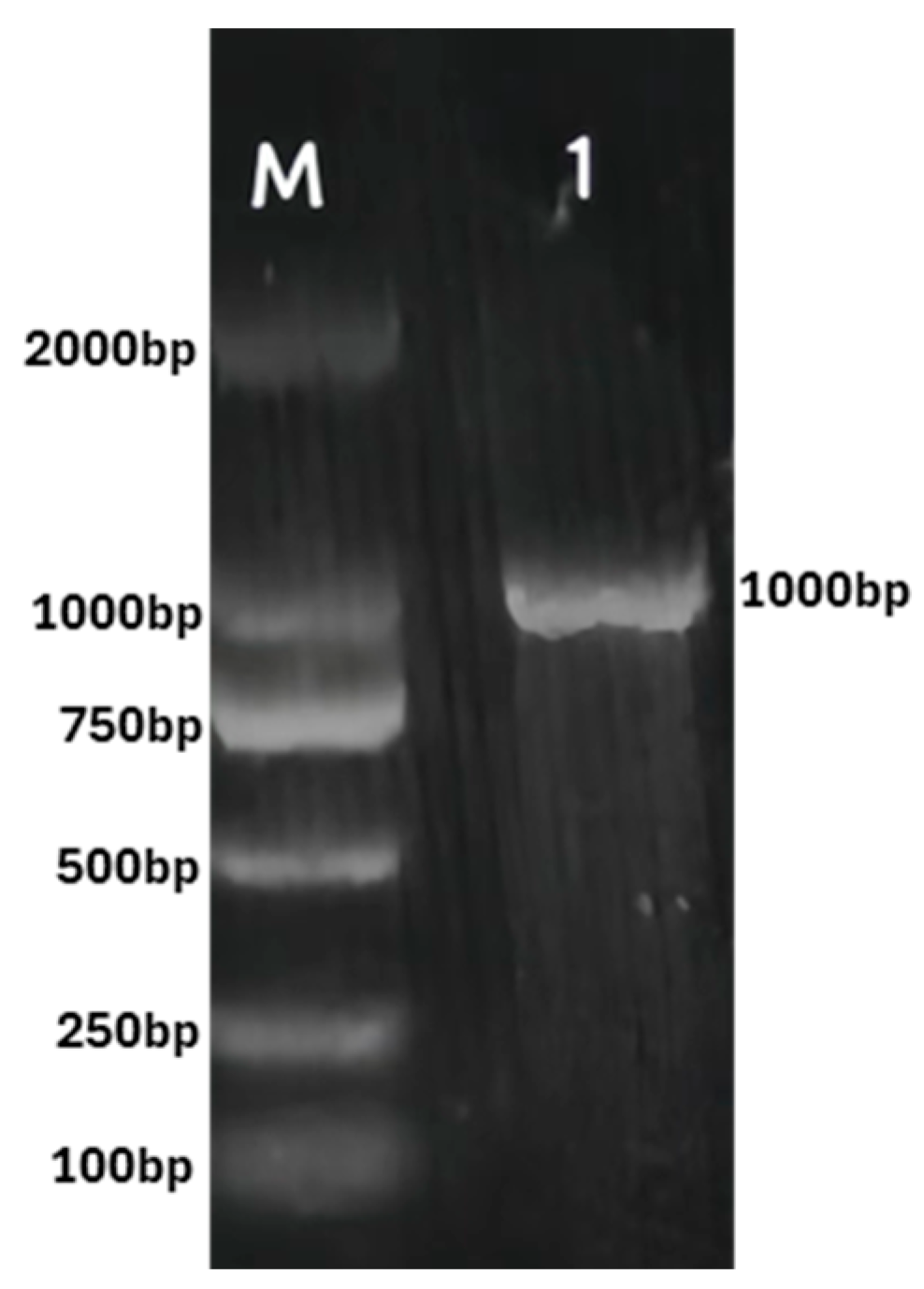
2.2. Molecular Cloning
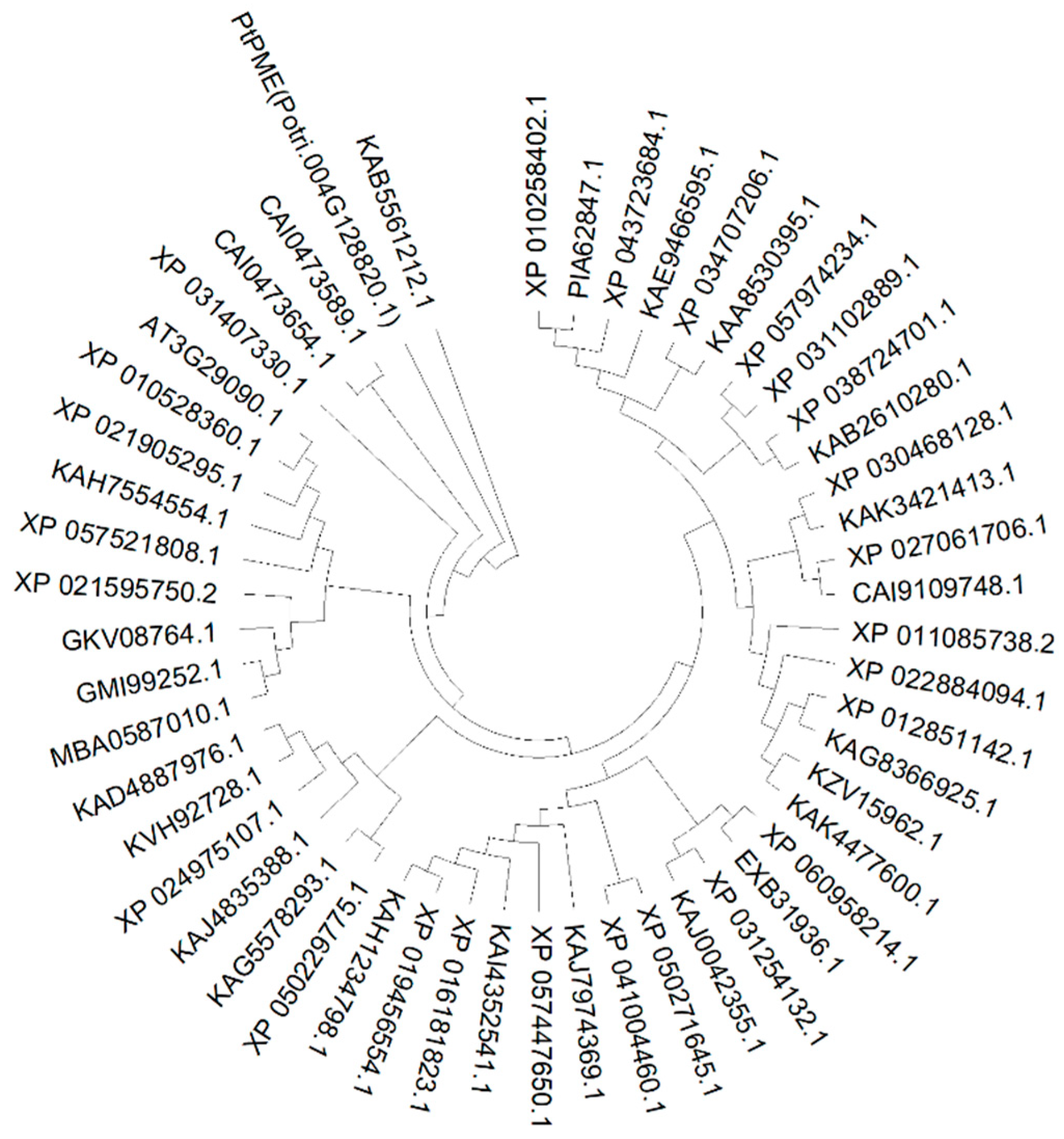
2.3. Sequence Analysis
2.4. Expression and Purification of Recombinant PtPME
2.5. Enzyme Activity Assay of Recombinant PtPME
2.6. Biochemical Characterization of the Purified Recombinant PtPME
2.7. Orthogonal Experiments
2.8. Determination of Esterification Degree
2.9. Homology Modelling
2.10. Molecular Docking
2.11. Statistical Analysis
3. Results and Discussion
3.1. Cloning and Sequence Analysis of PtPME Gene
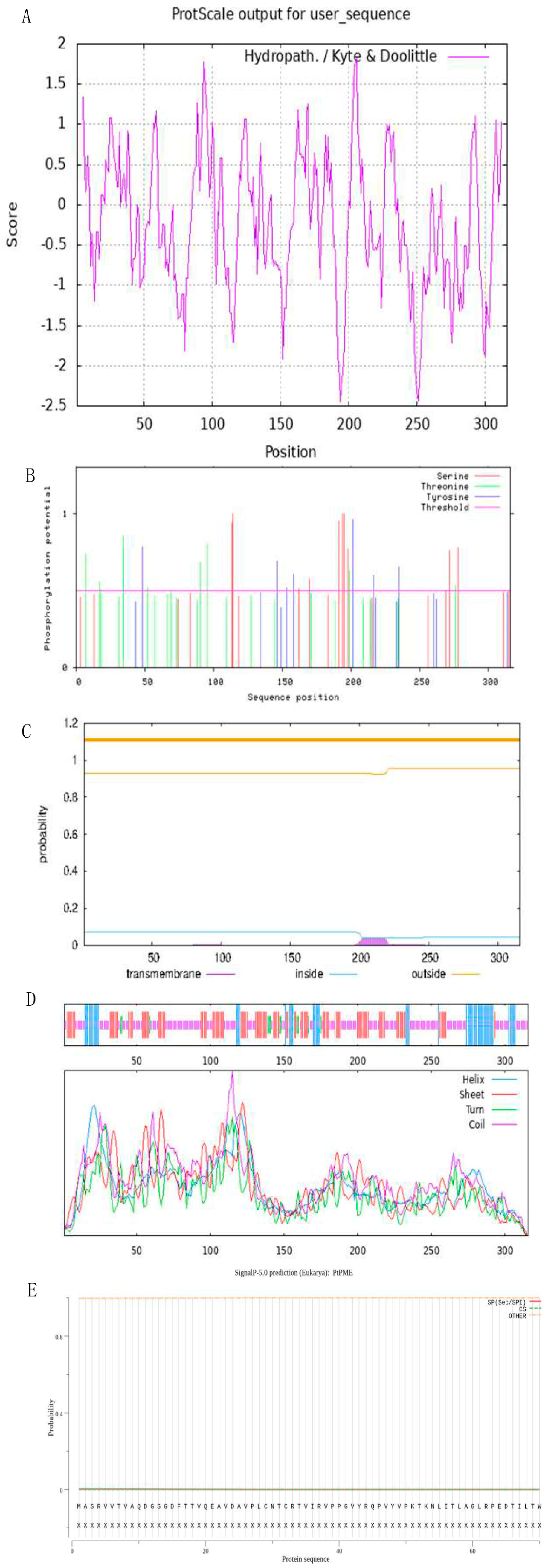
3.2. Physical and Chemical Properties
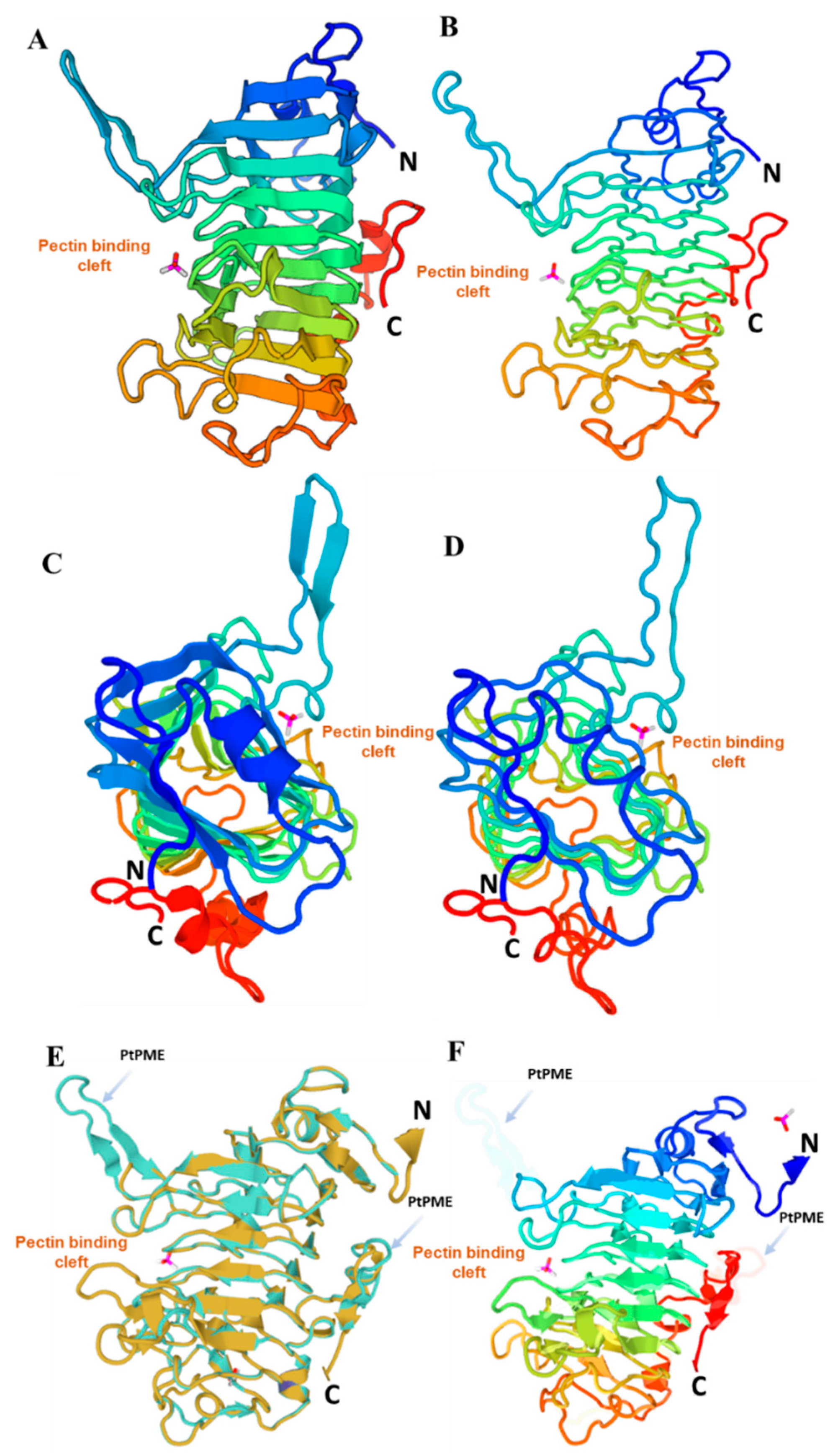
3.3. Molecular Modeling of PtPME
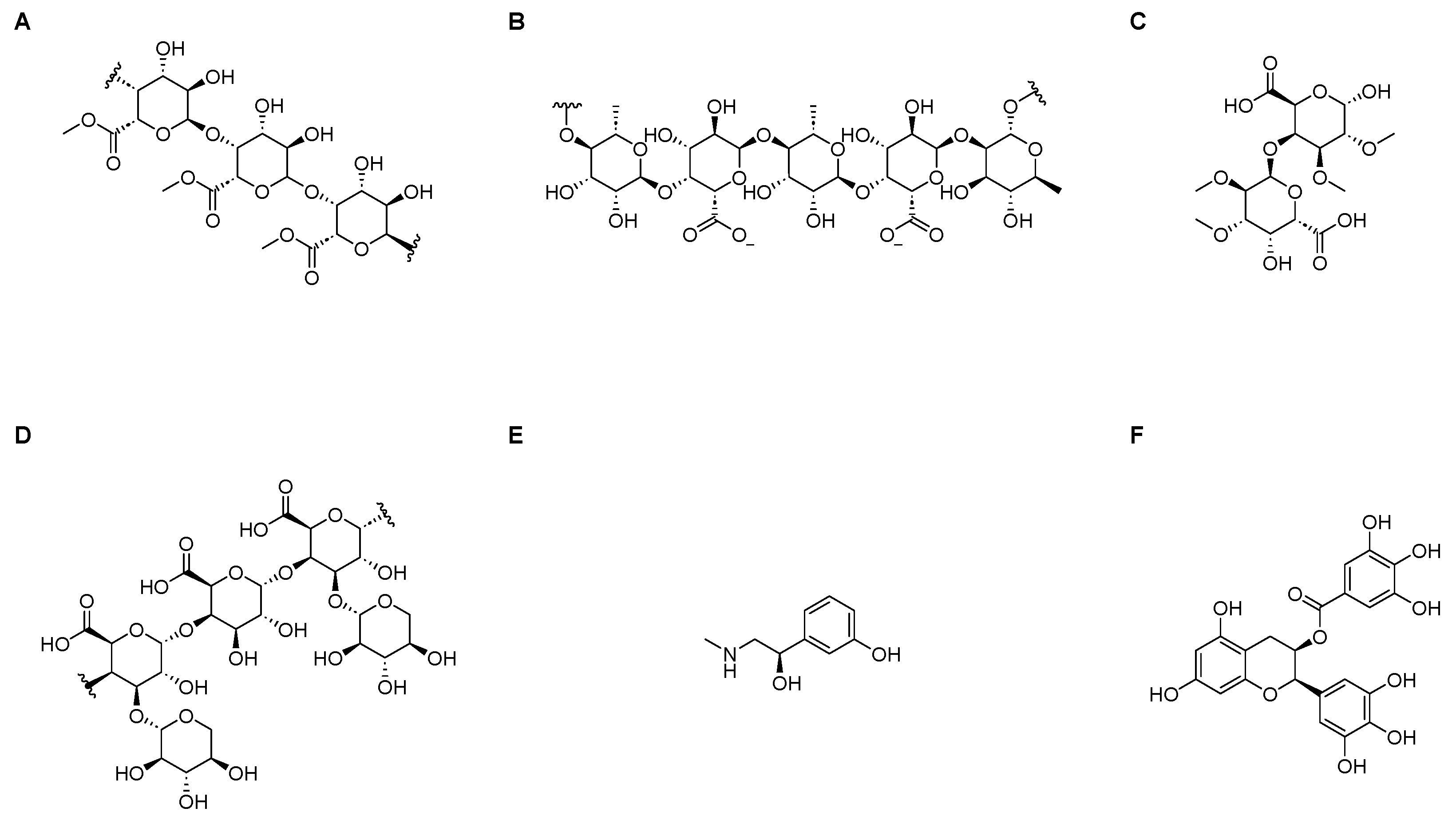
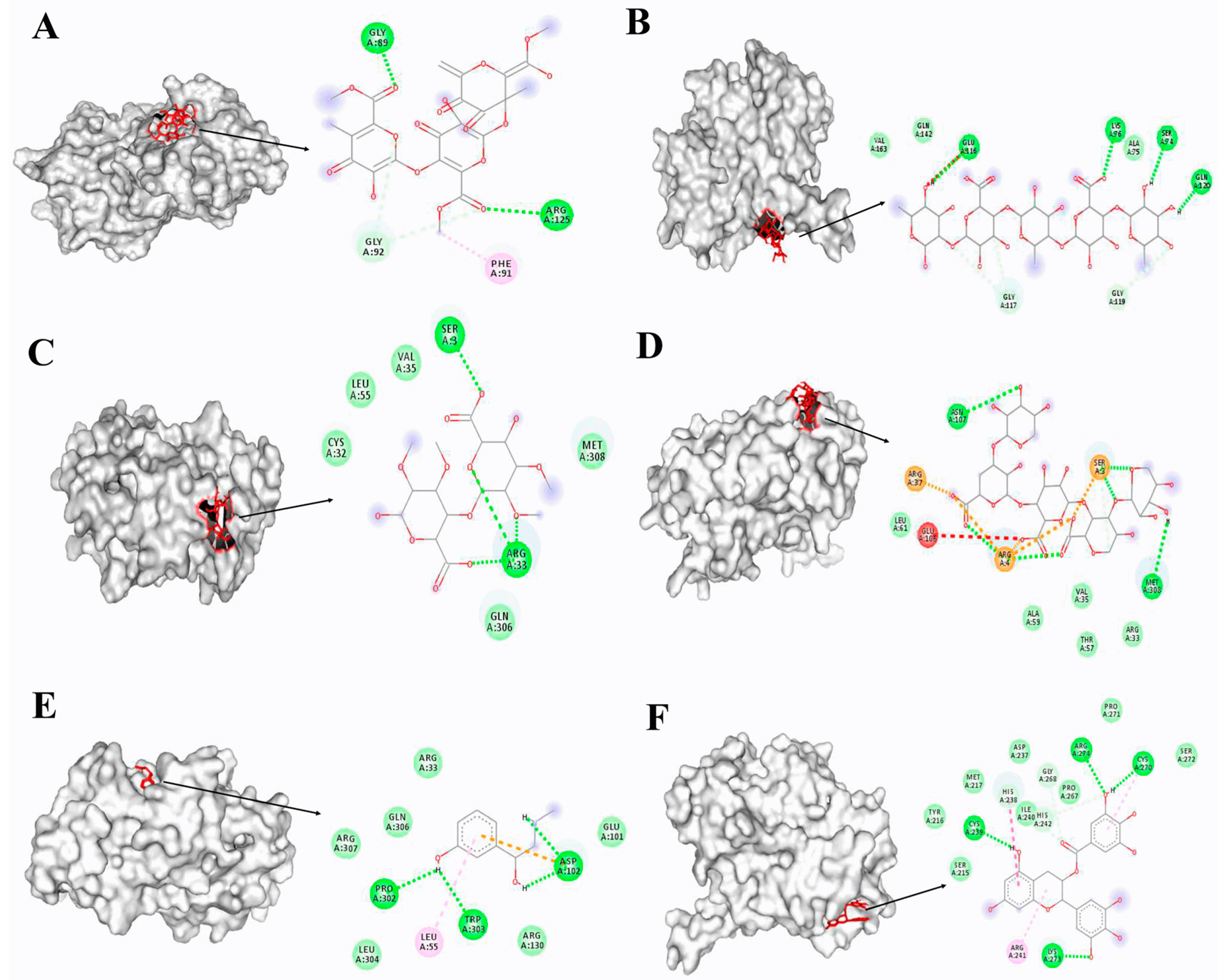
3.4. Docking Simulation of Different Substrates
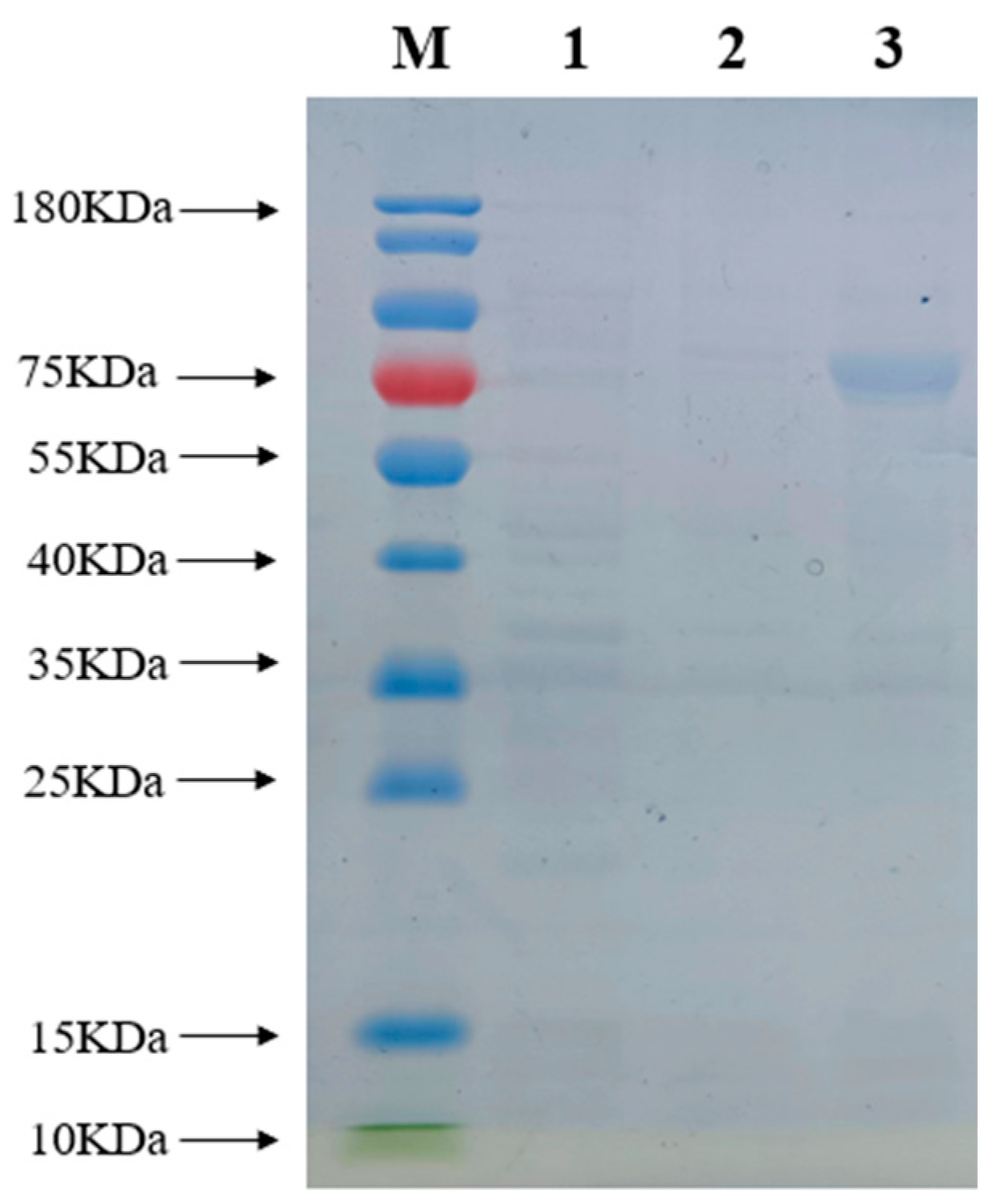
3.5. Prokaryotic Expression and Purification of PtPME Protein
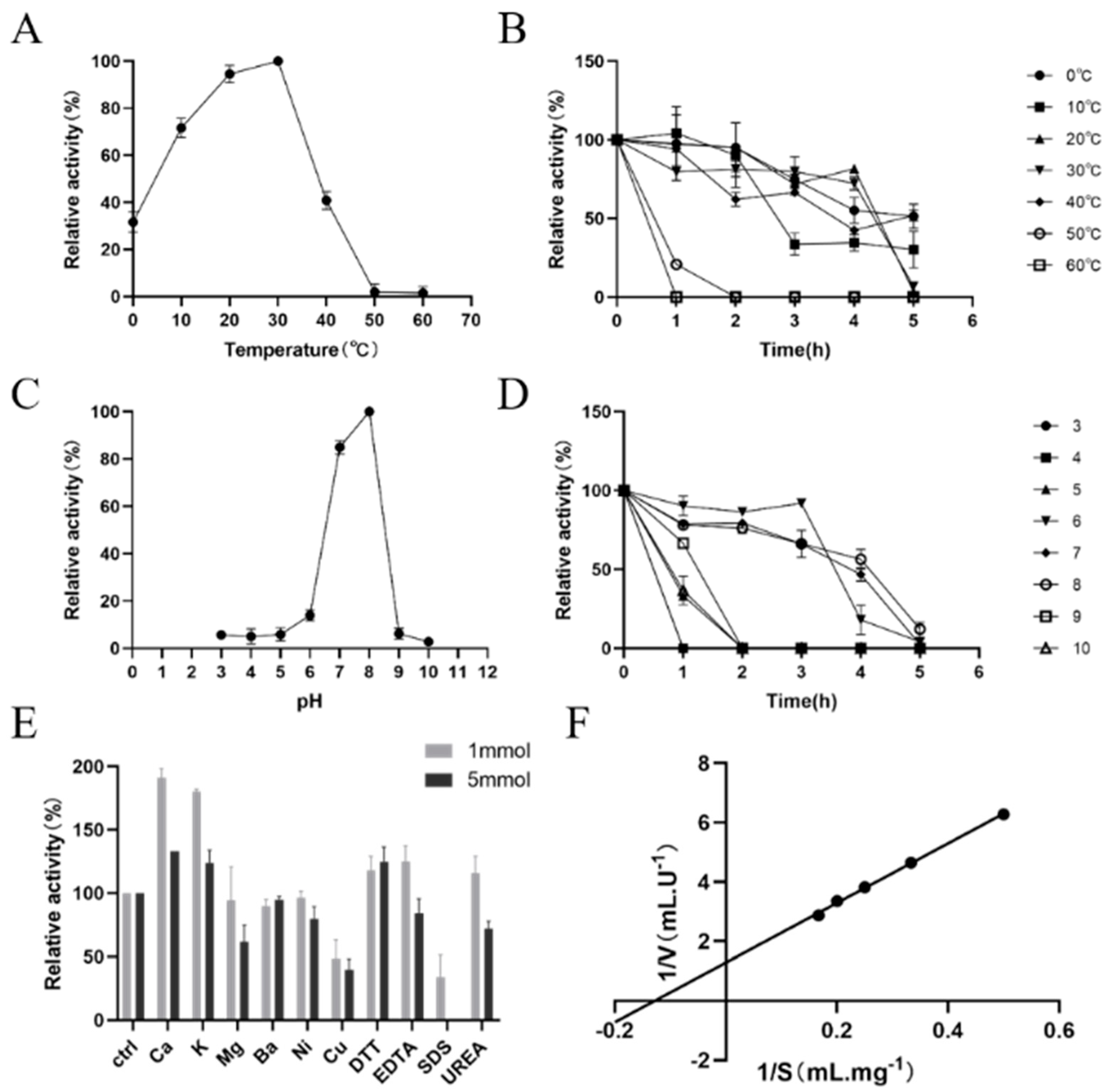
3.6. Analysis of the Enzymatic Properties of PtPME
3.7. Orthogonal Test of Enzyme Activity
3.8. Degree of Esterification
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lara-Espinoza, C.; Carvajal-Millán, E.; Balandrán-Quintana, R.; López-Franco, Y.; Rascón-Chu, A. Pectin and pectin-based composite materials: Beyond food texture. Molecules 2018, 23, 942. [Google Scholar] [CrossRef]
- Chandel, V.; Biswas, D.; Roy, S.; Vaidya, D.; Verma, A.; Gupta, A.J.F. Current advancements in pectin: Extraction, properties and multifunctional applications. Foods 2022, 11, 2683. [Google Scholar] [CrossRef]
- Zdunek, A.; Pieczywek, P.M.; Cybulska, J.J.C.R.i.F.S.; Safety, F. The primary, secondary, and structures of higher levels of pectin polysaccharides. Compr. Rev. Food Sci. Food Saf. 2021, 20, 1101–1117. [Google Scholar] [CrossRef]
- Saffer, A.M. Expanding roles for pectins in plant development. J. Integr. Plant Biol. 2018, 60, 910–923. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Zhang, D.; Wang, J.; Qin, X.; Liu, B.; Xu, X.; Zhang, W.; Zhang, Y. Expression and characterization of a pectin methylesterase from aspergillus niger zj5 and its application in fruit processing. J. Biosci. Bioeng. 2018, 126, 690–696. [Google Scholar] [CrossRef]
- Coculo, D.; Lionetti, V. The plant invertase/pectin methylesterase inhibitor superfamily. Front. Plant Sci. 2022, 13, 863892. [Google Scholar] [CrossRef]
- Shrestha, S.; Rahman, M.S.; Qin, W. New insights in pectinase production development and industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 9069–9087. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, J.; Mahajan, R. Microbial pectinases: An ecofriendly tool of nature for industries. 3 Biotech 2016, 6, 47. [Google Scholar] [CrossRef]
- Li, Y.; He, H.; He, L.-F. Genome-wide analysis of the pectin methylesterase gene family in potato. Potato Res. 2020, 64, 1–19. [Google Scholar] [CrossRef]
- Ishii, S.; Kiho, K.; Sugiyama, S.; Sugimoto, H. Low-methoxyl pectin prepared by pectinesterase from aspergillus japonicus. J. Food Sci. 1979, 44, 611–614. [Google Scholar] [CrossRef]
- Gupta, A.; Gangotia, D.; Mani, I. Bioinformatics tools and software. In Advances in Bioinformatics; Singh, V., Kumar, A., Eds.; Springer: Singapore, 2021; pp. 15–35. [Google Scholar]
- He, G.; Guan, C.-N.; Chen, Q.-X.; Gou, X.-J.; Liu, W.; Zeng, Q.-Y.; Lan, T. Genome-wide analysis of the glutathione s-transferase gene family in capsella rubella: Identification, expression, and biochemical functions. Front. Plant Sci. 2016, 7, 1325. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. Mega11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Salas-Tovar, J.A.; Flores-Gallegos, A.C.; Contreras-Esquivel, J.C.; Escobedo-García, S.; Morlett-Chávez, J.A.; Rodríguez-Herrera, R. Analytical methods for pectin methylesterase activity determination: A review. Food Anal. Methods 2017, 10, 3634–3646. [Google Scholar] [CrossRef]
- Deytieux-Belleau, C.; Vallet, A.; Doneche, B.; Geny, L. Pectin methylesterase and polygalacturonase in the developing grape skin. Plant Physiol. Biochem. 2008, 46, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, P.; Chakraborty, S. Influence of pulsed light, ultrasound, and series treatments on quality attributes, pectin methyl esterase, and native flora inactivation in sweet orange juice (citrus sinensis l. Osbeck). Food Bioprocess Technol. 2023, 16, 2095–2112. [Google Scholar] [CrossRef]
- Chaiwarit, T.; Rachtanapun, P.; Kantrong, N.; Jantrawut, P. Preparation of clindamycin hydrochloride loaded de-esterified low-methoxyl mango peel pectin film used as a topical drug delivery system. Polymers 2020, 12, 1006. [Google Scholar] [CrossRef] [PubMed]
- Sabzevari, A.G.; Sabahi, H.; Nikbakht, M.; McInnes, S.J.P. Development and characteristics of layered egcg/montmorillonite hybrid: An oral controlled-release formulation of egcg. J. Drug Deliv. Sci. Technol. 2022, 76, 103750. [Google Scholar] [CrossRef]
- Cheong, M.S.; Lee, D.Y.; Seo, K.H.; Choi, G.H.; Song, Y.H.; Park, K.H.; Kim, J.H. Phenylephrine, a small molecule, inhibits pectin methylesterases. Biochem. Biophys. Res. Commun. 2019, 508, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.J.; Johnson, D.L.; Bohlmann, J.; van Vuuren, H.J.J.; Jones, S.J.M.; Pretorius, I.S.; Schmidt, S.A.; Borneman, A.R. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar chardonnay. PLoS Genet. 2018, 14, e1007807. [Google Scholar] [CrossRef]
- Kim, S.; Park, J.; Yeom, S.I.; Kim, Y.M.; Seo, E.; Kim, K.T.; Kim, M.S.; Lee, J.M.; Cheong, K.; Shin, H.S.; et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 2017, 18, 210. [Google Scholar] [CrossRef]
- Zhong, Y.; Chen, Y.; Zheng, D.; Pang, J.; Liu, Y.; Luo, S.; Meng, S.; Qian, L.; Wei, D.; Dai, S.; et al. Chromosomal-level genome assembly of the orchid tree bauhinia variegata (leguminosae; cercidoideae) supports the allotetraploid origin hypothesis of bauhinia. DNA Res. 2022, 29, dsac012. [Google Scholar] [CrossRef]
- Zapata, L.; Ding, J.; Willing, E.M.; Hartwig, B.; Bezdan, D.; Jiao, W.B.; Patel, V.; Velikkakam James, G.; Koornneef, M.; Ossowski, S.; et al. Chromosome-level assembly of arabidopsis thaliana ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA 2016, 113, E4052–E4060. [Google Scholar] [CrossRef]
- Li, J.; Bi, Z.; Ma, S.; Chen, B.; Cai, C.; He, J.; Schwarz, S.; Sun, C.; Zhou, Y.; Yin, J.; et al. Inter-host transmission of carbapenemase-producing escherichia coli among humans and backyard animals. Environ. Health Perspect. 2019, 127, 107009. [Google Scholar] [CrossRef]
- Teller, D.C.; Behnke, C.A.; Pappan, K.; Shen, Z.; Reese, J.C.; Reeck, G.R.; Stenkamp, R.E. The structure of rice weevil pectin methylesterase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1480–1484. [Google Scholar] [CrossRef]
- Singh, P.; Yadav, S.; Shah, S.; Shanker, K.; Sundaresan, V.; Shukla, A.K. Characterization of a crpme indicates its possible role in determining vindoline accumulation in catharanthus roseus leaves. Physiol. Plant. 2024, 176, e14276. [Google Scholar] [CrossRef]
- Hua, T.; Li, Y.; Wang, K.; Tu, T.; Huang, H.; Luo, H.; Yao, B. High-level expression and characterization of pectin methylesterase pmet from talaromyces leycettanus jcm12802 in pichia pastoris. Acta Microbiol. Sin. 2018, 58, 122–130. [Google Scholar]
- Guo, F.; Li, X.; Zhao, J.; Li, G.; Gao, P.; Han, X. Optimizing culture conditions by statistical approach to enhance production of pectinase from bacillus sp. Y1. BioMed Res. Int. 2019, 2019, 8146948. [Google Scholar] [CrossRef]
- Jeong, H.Y.; Nguyen, H.P.; Eom, S.H.; Lee, C. Integrative analysis of pectin methylesterase (pme) and pme inhibitors in tomato (Solanum lycopersicum): Identification, tissue-specific expression, and biochemical characterization. Plant Physiol. Biochem. 2018, 132, 557–565. [Google Scholar] [CrossRef]
- Kotnala, B.; N, S.M.; Vasu, P. Purification and characterization of a salt-dependent pectin methylesterase from carica papaya fruit mesocarp-exocarp tissue. J. Food Sci. 2018, 83, 2062–2070. [Google Scholar] [CrossRef]
- L’Enfant, M.; Domon, J.M.; Rayon, C.; Desnos, T.; Ralet, M.C.; Bonnin, E.; Pelloux, J.; Pau-Roblot, C. Substrate specificity of plant and fungi pectin methylesterases: Identification of novel inhibitors of pmes. Int. J. Biol. Macromol. 2015, 81, 681–691. [Google Scholar] [CrossRef]
- Yan, R.; Han, C.; Fu, M.; Jiao, W.; Wang, W. Inhibitory effects of cacl2 and pectin methylesterase on fruit softening of raspberry during cold storage. Horticulturae 2022, 8, 1. [Google Scholar] [CrossRef]
- Li, J.; Yang, Z.-L.; Ding, T.; Song, Y.-J.; Li, H.-C.; Li, D.-Q.; Chen, S.; Xu, F. The role of surface functional groups of pectin and pectin-based materials on the adsorption of heavy metal ions and dyes. Carbohydr. Polym. 2022, 276, 118789. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Wang, H.; Zhu, Y.; Pan, S.; Cai, R.; Liu, F.; Pan, S. Comparative study on gelling properties of low methoxyl pectin prepared by high hydrostatic pressure-assisted enzymatic, atmospheric enzymatic, and alkaline de-esterification. Carbohydr. Polym. 2019, 226, 115285. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Wang, K.; Yang, R.; Kang, J.; Zhang, J. Rheological properties of natural low-methoxyl pectin extracted from sunflower head. Food Hydrocoll. 2015, 44, 122–128. [Google Scholar] [CrossRef]
- Yuliarti, O.; Mardyiah Binte Othman, R. Temperature dependence of acid and calcium-induced low-methoxyl pectin gel extracted from cyclea barbata miers. Food Hydrocoll. 2018, 81, 300–311. [Google Scholar] [CrossRef]
- Limberg, G.; Körner, R.; Buchholt, H.C.; Christensen, T.M.I.E.; Roepstorff, P.; Mikkelsen, J.D. Analysis of different de-esterification mechanisms for pectin by enzymatic fingerprinting using endopectin lyase and endopolygalacturonase II from A. Niger. Carbohydr. Res. 2000, 327, 293–307. [Google Scholar] [CrossRef]

| Level | A (Ph) | B (T/°C) | C (S/mg·mL−1) |
|---|---|---|---|
| 1 | 7 | 20 | 6 |
| 2 | 8 | 30 | 7 |
| 3 | 9 | 40 | 8 |
| Substrate | Bing Energy (KJ/mol) | Binding Pocket | RMSD |
|---|---|---|---|
| HGA-unit | −3.29 | Gly89, Arg125 | 2.059 |
| RGI-unit | 1.62 | Ser74, Lys76, Gln120, Glu116 | 4.438 |
| RGII-unit | −2.83 | Ser3, Arg33 | 1.340 |
| XGA-unit | −1.16 | Ser3, Arg4 | 2.903 |
| PE | −4.85 | Asp102, Pro302, Trp303 | 1.687 |
| EGCG | −3.57 | Cys270, Lys273, Arg274, Cys239 | 3.073 |
| Exp. Number | Ph (A) | T(B) (°C) | Pectin Concentration (C) (mg/mL) | Absorbance | Enzymatic Activity |
|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 0.369 | 0.219 |
| 2 | 1 | 2 | 3 | 0.347 | 0.188 |
| 3 | 1 | 3 | 2 | 0.423 | 0.275 |
| 4 | 2 | 1 | 3 | 0.289 | 0.173 |
| 5 | 2 | 2 | 2 | 0.345 | 0.234 |
| 6 | 2 | 3 | 1 | 0.291 | 0.151 |
| 7 | 3 | 1 | 2 | 0.277 | 0.087 |
| 8 | 3 | 2 | 1 | 0.242 | 0.042 |
| 9 | 3 | 3 | 3 | 0.281 | 0.044 |
| K1 | 0.682 | 0.479 | 0.412 | ||
| K2 | 0.558 | 0.464 | 0.596 | ||
| K3 | 0.173 | 0.47 | 0.405 | ||
| K1 | 0.227 | 0.159 | 0.137 | ||
| K2 | 0.186 | 0.154 | 0.198 | ||
| K3 | 0.057 | 0.156 | 0.135 | ||
| R | 0.169 | 0.005 | 0.063 | ||
| Best level | A1B1C2 | ||||
| Pectin | Degree of Esterification (%) |
|---|---|
| High-ester pectin | 68.8 ± 1.06 |
| De-esterified pectin | 21.83 ± 1.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Y.; Huang, L.; Zeng, Y.; Zhang, Y.; Liu, W.; He, G. Cloning, Expression and Enzymatic Characterization of Pectin Methyl Esterase from Populus trichocarpa and Its Application. Processes 2024, 12, 1511. https://doi.org/10.3390/pr12071511
Feng Y, Huang L, Zeng Y, Zhang Y, Liu W, He G. Cloning, Expression and Enzymatic Characterization of Pectin Methyl Esterase from Populus trichocarpa and Its Application. Processes. 2024; 12(7):1511. https://doi.org/10.3390/pr12071511
Chicago/Turabian StyleFeng, Yanjiao, Lifen Huang, Yue Zeng, Yiyuan Zhang, Wei Liu, and Gang He. 2024. "Cloning, Expression and Enzymatic Characterization of Pectin Methyl Esterase from Populus trichocarpa and Its Application" Processes 12, no. 7: 1511. https://doi.org/10.3390/pr12071511
APA StyleFeng, Y., Huang, L., Zeng, Y., Zhang, Y., Liu, W., & He, G. (2024). Cloning, Expression and Enzymatic Characterization of Pectin Methyl Esterase from Populus trichocarpa and Its Application. Processes, 12(7), 1511. https://doi.org/10.3390/pr12071511






