Abstract
Cholesterol regulation is an important aspect of human health. In this work we bring together and extend two recent mathematical models describing cholesterol biosynthesis and lipoprotein endocytosis to create an integrated model of lipoprotein metabolism in the context of a single hepatocyte. The integrated model includes a description of low density lipoprotein (LDL) receptor and cholesterol synthesis, delipidation of very low density lipoproteins (VLDLs) to LDLs and subsequent lipoprotein endocytosis. Model analysis shows that cholesterol biosynthesis produces the majority of intracellular cholesterol. The availability of free receptors does not greatly effect the concentration of intracellular cholesterol, but has a detrimental effect on extracellular VLDL and LDL levels. We test our model by considering its ability to reproduce the known biology of Familial Hypercholesterolaemia and statin therapy. In each case the model reproduces the known biological behaviour. Quantitative differences in response to statin therapy are discussed in the context of the need to extend the work to a more in vivo setting via the incorporation of more dietary lipoprotein related processes and the need for further testing and parameterisation of in silico models of lipoprotein metabolism.
1. Introduction
Cholesterol is an intrinsic part of living cells. Every cell in the human body requires cholesterol in order to produce and maintain a healthy cell membrane. The formation of hormones and of bile acids that assist in the digestion of food, depend on cholesterol. Myelin, which covers nerve axioms to assist the conduction of electrical impulses, facilitating movement, vision, taste and the processing of sensory input is 20%, by weight, cholesterol [1]. This makes cholesterol vital for our nervous system and for memory and learning to take place. All cells have the ability to produce and regulate cholesterol, but the liver is primarily responsible for the metabolism of dietary cholesterol and is the only organ that can remove it from the body via the formation of bile.
Despite being such an important part of cellular health, irregular control of cholesterol homeostasis in hepatocytes (liver cells) can cause the liver to poorly process dietary cholesterol. This in turn can lead to high levels of circulating plasma cholesterol, which is widely known to be a major risk factor for a large number of cardiac diseases, for example coronary artery disease (CAD), cardiovascular disease (CVD) and coronary heart disease (CHD). The chance of developing CAD, CVD or CHD is determined by risk factors, some of which are modifiable. Modifiable risk factors include body weight, blood pressure and blood lipid levels, which are all influenced by exercise levels, smoking and diet. Non-modifiable risk factors include genetic predisposition, age, gender and ethnicity [2]. Whilst we may not be able to control these factors, the effects are sometimes modifiable with pharmaceutical interventions such as statins.
Dietary or exogenous cholesterol however, accounts for merely 20% of the body’s cholesterol. The other 80%, endogenous cholesterol, is produced mainly by hepatocytes, but also by cells in the central nervous system and reproductive organs [3]. Each cell is subject to the cholesterol biosynthesis cascade initiated by 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase (HMGCR). This signalling pathway cues the change in rate of production or inhibition of cholesterol in response to declining or increasing cellular cholesterol levels. In this case the transcription factor, sterol regulatory element-binding protein 2 (SREBP-2), is blocked from upregulating mRNA transcription of the HMGCR gene when cellular levels of cholesterol are high, but is free to upregulate transcription when levels are low. This allows the cell to change the rate of cholesterol production according to its needs.
Fats and cholesterol from a normal diet enter the blood stream, through the stomach, having been packed into carrier molecules known as a lipoproteins. Lipids are insoluble and must thus be packed into particles in order to be transported around the body. Lipoproteins are surrounded by phospholipids and apolipoproteins. As well as surrounding lipoprotein molecules, apolipoproteins play a vital role in the binding of lipoproteins to receptors on cell surfaces for removal from circulation. They also act as activators for lipolytic enzymes involved in metabolism.
There are five main classes of lipoprotein: chylomicrons, very low density lipoproteins (VLDL), intermediate density lipoprotein (IDL), low density lipoprotein (LDL) and high density lipoprotein (HDL). They are classed due to their varying triglyceride, cholesterol and apolipoprotein contents. Lipoproteins continuously exchange lipids and proteins with cells and other lipoproteins leading to a reduction in lipid content as the particles vary from being chylomicrons, to chylomicron remnants, VLDL, IDL and subsequently LDL particles.
Carrying fats as an energy source through the blood stream to cells in need, lipoproteins eventually end up in the liver where they are removed from circulation by a process known as receptor mediated endocytosis (RME). The rate of lipoprotein uptake is regulated by the number of available free receptors on the cell surface. Receptors are synthesised by the cell. Newly synthesised receptors are placed on the surface of the cell where they collect in clathrin coated pits. Apolipoproteins attach the lipoprotein to the receptor, after which the clathrin pit encloses around the lipoprotein and pinches off forming endocytotic vesicles which are internalised. Empty pits may also undergo this process. Following internalisation, the clathrin coating is shed and vesicles merge together to form larger endosomes within which the lipoprotein dissociates from the receptor. Some receptors are removed at this point and recycled to the cell surface. The endosomes then combine with lysosomes within the cell and the contents are degraded by lysosomal enzyme hydrolysis releasing amino acids and cholesterol for use in cellular metabolism [4].
Receptors, once synthesised or recycled, insert randomly on the cell surface before diffusing into clathrin coated pits. The concentration of receptors in the pits determine how many lipoproteins can bind and be internalised at any one time. LDL receptor (LDLR) synthesis is governed by SREBP-2. When intracellular cholesterol concentrations are low transcription of LDLR is upregulated, increasing the uptake of lipoproteins. Similarly high levels of cholesterol lead to downregulation of LDLR synthesis, decreasing lipoprotein endocytosis. In high cholesterol concentrations, receptor and cholesterol synthesis is inhibited. RME is the target of drugs, used in cardiovascular therapy, known as statins which inhibit cholesterol biosynthesis and up-regulate receptor synthesis, thus increasing the amount of lipoproteins cleared from the circulation.
Dysregulated cholesterol biosynthesis and lipoprotein metabolism can lead to a number of health conditions. Dyslipidemia, raised levels of LDL in blood plasma and/or reduced levels of high density lipoprotein (HDL), is a major health issue throughout the world [5], which has been linked to increases in dietary fat and sugar intake and sedimentary lifestyles. Hypercholesterolemia, elevated levels of cholesterol in blood plasma, has been linked to cardiovascular and pulmonary inflammation [6] and the overloading of macrophages with cholesterol in vitro has been shown to initiate immune responses. In contrast, unduly lowering the biosynthetic production of cholesterol levels, such as more recently shown via pathogenic infections which may lead to sepsis, can have dramatic adverse results [7,8].
There exists a growing literature on the mathematical modelling of lipoprotein metabolism as recently reviewed in [9]. Such models have generally been formulated using the theory of linear and nonlinear ordinary differential equations and parameterised and tested, to varying degrees, against the experimental literature. The mathematical models reviewed in [9] were tested for their ability to correctly predict the response of each to statin therapy. They found that only a small proportion of models within the literature correctly predicted the well known effect of statins on increasing LDL uptake from the circulation.
In contrast there are few mathematical models of cholesterol biosynthesis. Those that do exist vary in the size of the mathematical models formulated (number of variables and parameters) and complexity. In [10] the authors derived, parameterised and analysed a three variable nonlinear ordinary differential equation (ODE) model of cholesterol biosynthesis via the HMGCR pathway. They demonstrated that whilst the system only exhibited one steady-state, three types of behaviour were possible; monotic, damped and oscillatory. A more recent ODE model of the mevalonate pathway has been formulated and analysed in detail by [11]. This model describes cholesterol biosynthesis via the HMGCR and squalene synthase pathways, demonstrates the effect of the cholesterol-SREBP-2 feedback on the network’s temporal responses, whilst more localised positive feedbacks within the network ensure cholesterol levels remained tightly bound should any products within the pathway be adversely increased or decreased.
Limited work has focused on integrating molecular scale cholesterol synthesis with lipoprotein endocytosis and LDLR synthesis. One exception is that of the unpublished work of [12], which integrated a description of LDL endocytosis [13] with that of cholesterol biosynthesis [10]. This was favourably evaluated by [9] in assessing how well the model reproduced the known cellular response to statins, but no mathematical or computational analysis of the model was undertaken.
In this work we present a model of integrated cholesterol biosynthesis, which includes a description of SREBP-2 regulation by cholesterol (as detailed in [14]), a full description of VLDL and LDL uptake (as detailed in [15]) coupled with a description of receptor biosynthesis. The work provides a full account of model formulation, analysis and testing thereof. We are motivated by the following considerations. Firstly, we wish to develop a well-informed integrated mathematical model of the core exogenous and endogenous cholesterol pathways within a hepatocyte. We wish to evaluate whether a simplified model formulated in an in vitro context can capture the known in vivo biological response of the system in respect of Familial Hypercholesterolaemia and statin therapy, without having to complicate the model by accounting for other in vivo aspects (for instance HDL, chylomicron remnants, VLDL hepatocyte recycling). Secondly, we wish to consider how rates of VLDL to LDL delipidation coupled with competition between the two particles for cell membrane level receptors may affect intracellular cholesterol levels. High levels of circulating LDL is an indicator of risk in a number cardiovascular diseases and one of the main sources of circulating LDL is delipidation. Because of this we have included VLDL to LDL delipidation in this model in order to explore it’s effects without the complication of a full description of dietary lipoprotein metabolism. Finally, we wish to evaluate any differences that the assumption of a continuum of receptors on the surface of a cell has (as per [15]) versus that of discrete description of receptors bound by differing numbers of VLDL and LDL particles (as per [13]).
Our paper is organised as follows. In Section 1.1 we discuss the main biological features included in our integrated mathematical model before presenting an ODE model of the system in Section 2.1. Details of the model parameterisation are discussed in Section 2.4, which is followed by numerical simulations of the governing system of equations in Section 3.1. Results of computational and mathematical model analysis are presented in Section 3.2, before we investigate the effect of different classes of FH on extracellular levels of LDL and intracellular cholesterol levels. The effect of varying levels of statin therapy are investigated in Section 3.4 before we summarise and discuss our findings in Section 4.
Whilst our model has been formulated and parameterised in an in vitro context, the extrapolation to an in vivo setting is not considerably different given hepatocytes will be surrounded by lipoproteins within the liver. We thus wish to test how well this extrapolation works by testing the model against known in vivo outcomes in respect of Familial Hypercholesterolaemia and response to statin therapy.
1.1. Cholesterol Biosynthesis and Lipoprotein Metabolism
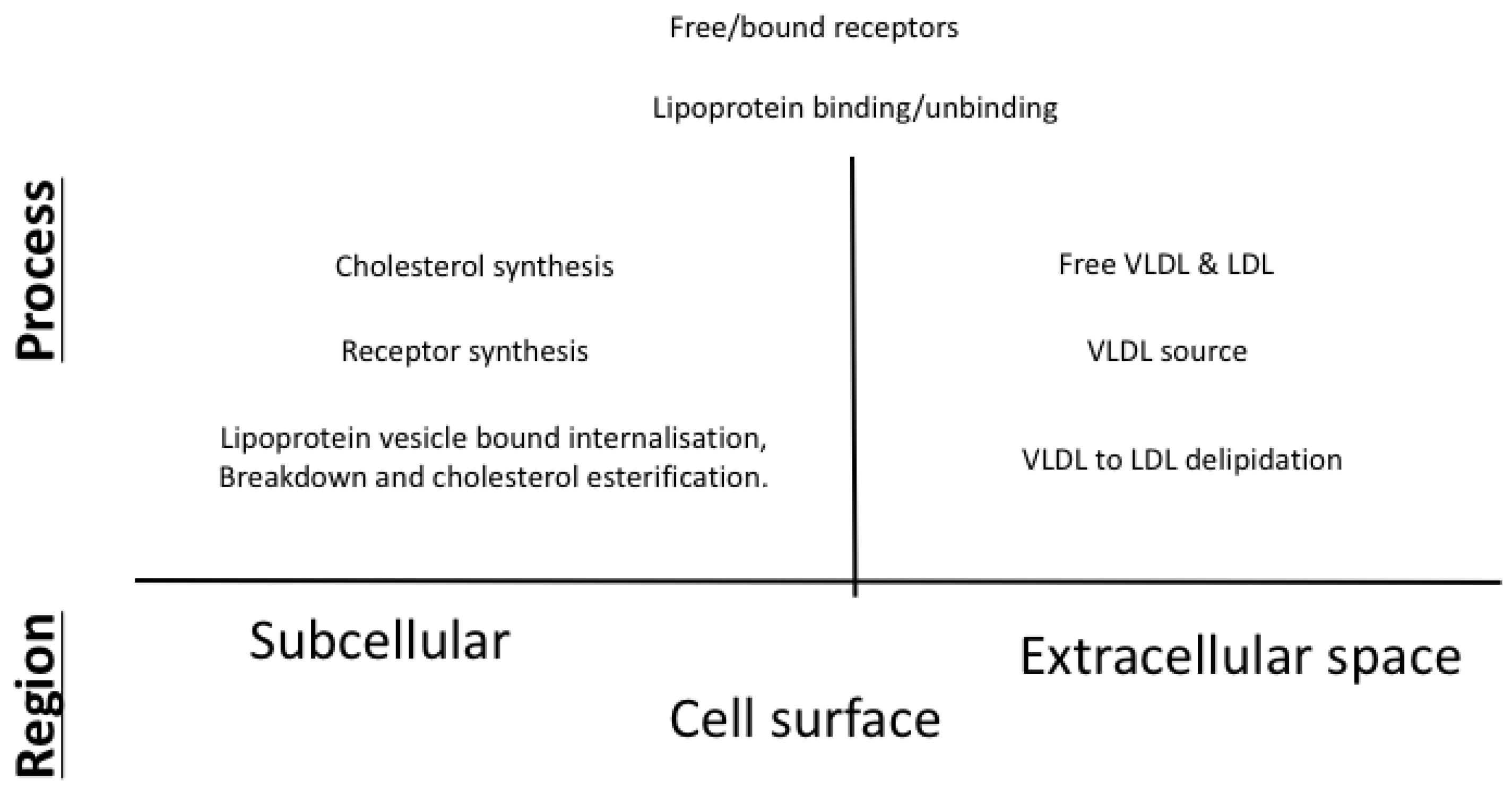
Our work here couples the endocytosis model of VLDL and LDL metabolism in an hepatocyte described in [15] with the description of cholesterol biosynthesis detailed in [10] and extends it with descriptions of LDLR synthesis, VLDL uptake and VLDL to LDL delipidation. An overview of the main processes included in our model is given in Figure 1 with further details on the exact mechanisms provided in Figure 2.
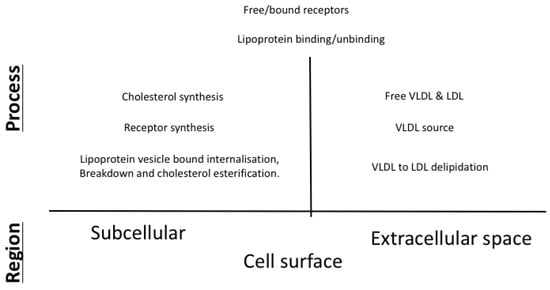
Figure 1.
An overview of the main features included in our integrated mathematical model of cholesterol and receptor biosynthesis coupled with lipoprotein (VLDL and LDL) endocytosis.
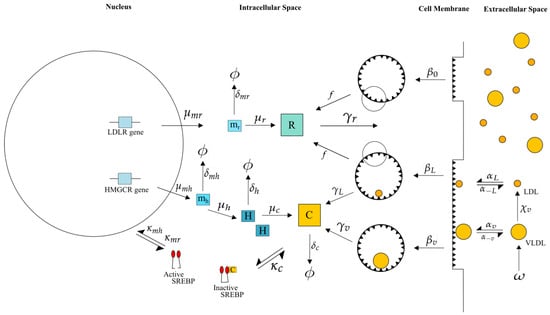
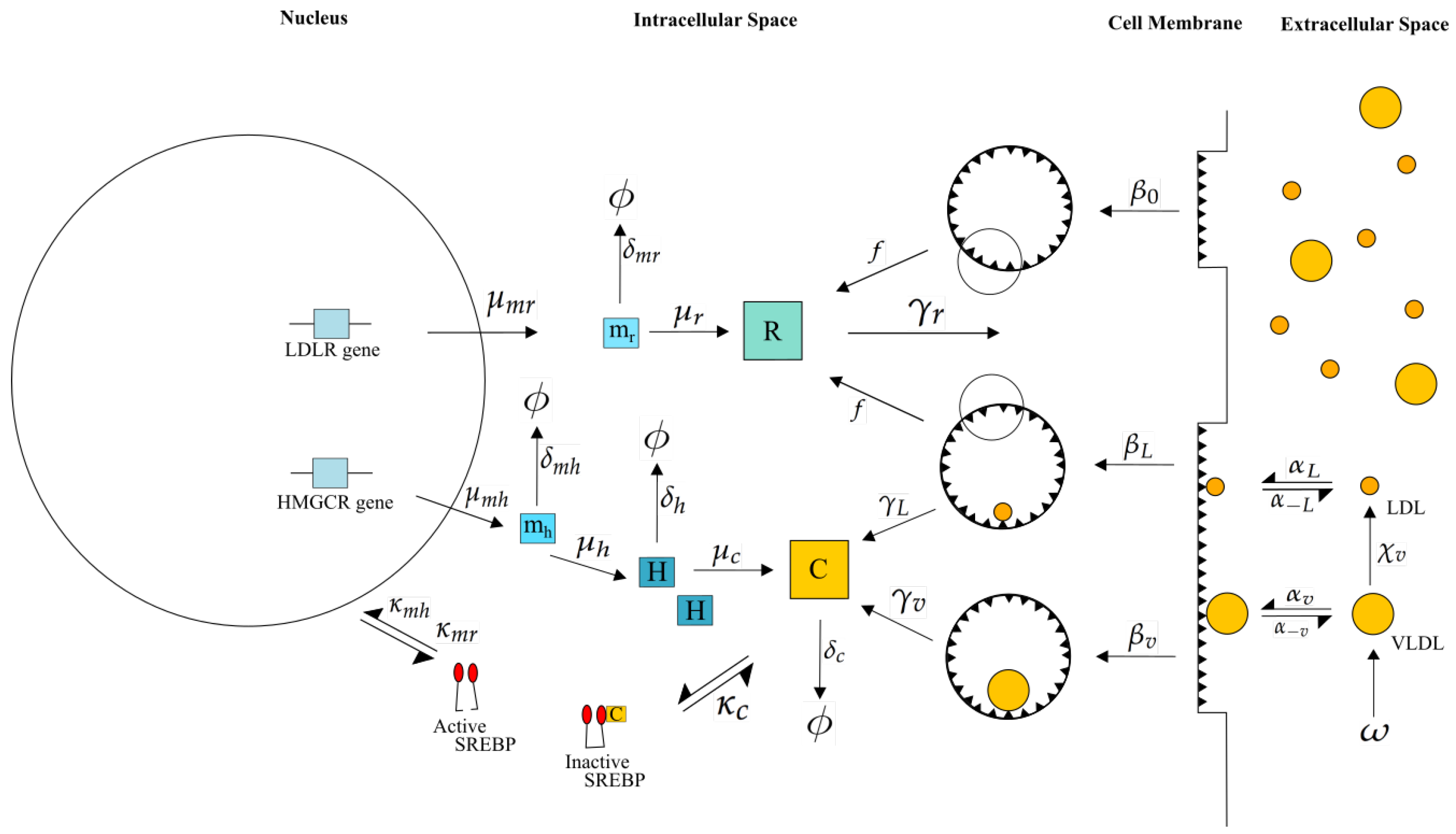
Figure 2.
Our integrated model of cholesterol biosynthesis and lipoprotein endocytosis in a hepatocyte. Transcription of HMGCR and LDLR mRNA (at rates and , respectively) by SREBP-2 (, ), leads to the synthesis of HMGCR (), LDLR () and cholesterol (), which negatively regulates SREBP-2 (). In the extracellular space, VLDL delipidates to LDL () and each bind/unbind to LDLR on the cell surface (, ). Finally VLDL and LDL occupied receptor pits are endocytosed (, ) as well as empty ones (), cholesterol extracted from internalised lipoproteins ( and ) and receptors recycled in the intracellular space (). Here represents HMGCR mRNA, LDLR mRNA, H HMGCR, receptors in the internal store, c intracellular cholesterol and degradation of the respective entity.
While the model described in this work extends previous descriptions of in vitro lipoprotein endocytosis, it has not been formulated with a specific in vitro cell experiment in mind, as was the case in [15]. Instead it seeks to describe relevant processes at the subcellular and extracellular scale, which can be found both in vitro and in vivo, thus providing a means of extrapolating between the two. We assume concentrations of VLDL and LDL are fed to hepatocytes in a controlled manner, thereby describing the basic mechanisms of cholesterol synthesis and LDL and VLDL metabolism without the added complexity of describing other dietary lipoprotein metabolism.
These two models have been individually parameterised, analysed and their behaviour tested against published in vitro experimental data as detailed in each publication [10,15]. The integrated model consists of three main compartments: (i) the cell nucleus in which genetic regulation of HMGCR and LDLR occurs; (ii) the cell cytoplasm surrounded by the cell membrane in which all processes related to VLDL and LDL binding and breakdown, cell receptor and cholesterol regulation take place; and (iii) the extracellular space around the cell containing sources of VLDL and LDL, as shown in Figure 2. Whilst full details on each of the mathematical models can be found in each of the respective references, we provide here a summary of the main processes incorporated into the model for completeness, along with descriptions of the additional processes required for an integrated model. To ensure these mechanisms are clear in the context of the mathematical formulation presented in Section 2.1, we define the respective model variables as each mechanism is discussed. Parameter values associated with each process are detailed in Figure 2 and Table 1.

Table 1.
Dimensional model parameters. Molec. denotes molecules, r receptors and conc. concentration.
At the genetic level SREBP-2 transcribes the HMGCR gene to produce HMGCR mRNA, HMGCR and cholesterol (at rates , and respectively) as described in [10]. Our integrated model also requires a description of LDLR synthesis as a result of SREBP-2 transcription which is described by the following biochemical equation
where unbound free LDLR gene is represented by , free SREBP-2 is represented by S and represents SREBP-2 bound to the LDLR gene. Once SREBP-2 and the LDLR gene become bound transcription of LDLR mRNA, is upregulated and accordingly, translation of the mRNA to create receptors, occurs. These are placed in the internal receptor store of the cell, assumed to be in the cell cytoplasm.
The reaction rate constants of represent SREBP-2 binding to the LDLR free gene, whilst represents the reaction rate of SREBP-2 unbinding from the LDLR gene. Here is the number of binding sites on the LDLR gene SREBP-2 binds to. The rate of transcription of mRNA responsible for receptor synthesis is and is the rate of translation of LDLR from LDLR mRNA.
The incorporated mechanisms of VLDL and LDL endocytosis and their respective dynamics at the cellular level are as follows. LDL particles in the medium surrounding the cell (denoted ) bind to free receptors () in clathrin pits on the cell surface at rate and unbind at rate . In binding, LDL particles effectively bind to one receptor, but can occlude up to surrounding receptors. The bound receptor and LDL () and the free and occluded receptors in the pit are then internalised (at rate ) to become internalised vesicles ().
Internalised vesicles and their contents subsequently break down whereby the cholesterol contained within the LDL particles is added to the internal cholesterol pool of the cell (), where it is esterified for other cellular processes at a rate proportional to it’s concentration (). A proportion f of the free receptors in the vesicle are placed in the cells internal cell store, ready for recycling back to the cell surface (). The rest are degraded.
VLDL particle binding, internalisation and subsequent breakdown are governed by a similar series of processes represented by variables denoted with instead of , for example for the concentration of extracellular VLDL and for binding of VLDL to receptors. Pits consisting of P empty receptors (no LDL or VLDL bound) may also be internalised .
Further to these processes, VLDL supply to the serum and VLDL to LDL delipidation is described by
where is a constant supply of VLDL to the serum and is the respective reaction rate constant of delipidation.
2. Materials and Methods
2.1. Mathematical Formulation
Applying the Law of Mass Action to those mechanisms described in Section 1.1 coupled with the models of [10,15] leads to the system of Equations (3)–(20), that integrate cholesterol and receptor biosynthesis with lipoprotein metabolism metabolism.
Biosynthesis of cholesterol and LDLR via SREBP-2 are described by
where the time dependent variables represent concentrations of each of the respective entities detailed in Figure 2, such that is the concentration of free HMGCR gene, that of LDLR gene, is free unbound SREBP-2, is SREBP-2 bound to the HMGCR gene, is SREBP-2 bound to the LDLR gene, is the concentration of HMGCR mRNA, is that of LDLR mRNA and is HMGCR. We note that is the number of binding sites for cholesterol to bind to SREBP-2, is the number of binding sites on the LDLR gene for SREBP to bind to and is the number of binding sites on the HMGCR gene to which SREBP binds.
The mechanisms of LDL and VLDL endocytosis and the subsequent processing of cholesterol and receptors, is given by the following
where the concentration of free LDL surrounding the cell is , is receptor bound LDL, is internalised LDL, is free VLDL surrounding the cell, is receptor bound VLDL, is internalised VLDL, are free unbound receptors, are internalised receptors, is cholesterol and is the SREBP-2 cholesterol bound complex.
Equations (3)–(9) and (20) describe the synthesis of HMGCR mRNA and LDLR mRNA via SREBP-2 as regulated by cholesterol (Equation (19)). Equation (10) describes the production of HMGCR from HMGCR mRNA while production of LDLR, which are assumed to automatically join the internal receptor store, is accounted for in Equation (18). Equations (11)–(20) are essentially those as detailed in [15] with the addition of the respective genetic synthesis terms describing cholesterol and LDLR production. Equations (11) and (14) detail the association of extracellular LDL and VLDL, respectively, to receptors and VLDL to LDL delipidation with a constant source of VLDL. Equations (12) and (15) describe bound LDL and VLDL formation, unbinding and internalisation, respectively, whilst Equations (13) and (16) detail the internalisation of LDL and VLDL bound pits and their respective breakdown. Equations (17) and (18) describe free and internalised receptor dynamics, whilst Equations (19) and (20) detail endogenous and exogenous cholesterol regulation, the latter the effect of cholesterol binding/unbinding to free SREBP-2.
The scaling parameters J and W are ratios representing the difference in volume between the compartments in which the reactions take place. The nucleus of the cell constitutes approximately 10% [16] of the volume of the total cell and thus we set . We assume the volume surrounding the cell is considerably greater than that of the cell, which from Jackson et al. [17] gives . This accounts for the proportional concentrations between the three compartments: serum, cell and nucleus.
Many of the initial conditions are set equal to zero in order to help understand if the overall system response is in agreement with the known biological behaviour and are given by
2.2. Model Reduction
Our model consists of 18 coupled linear and non-linear ODEs. We now seek to reduce the number of equations by application of the quasi-steady state approximation and conservation laws as detailed in Appendix A. This reduces the number of equations to 12 such that
with the initial conditions
2.3. Non-Dimensionalisation
The model was non-dimensionalised according to the following rescalings
where molecules/mL and molecules/mL are the total concentrations of cholesterol and SREBP-2 in a hepatocyte [12,18], respectively. Substituting these rescalings into Equations (22)–(34) leads to
with the initial conditions
All non-dimensional parameters in terms of dimensional ones and their values are shown in Table 2.

Table 2.
Table of non-dimensional parameters including their definition and value.
2.4. Parameter Estimation
We have utilised parameterisations detailed in [10,15] to inform each of the models, respectively. Table 1 details each dimensional parameter, their value and source. Non-dimensional parameter values and their definitions (in terms of dimensional ones) are given in Table 2.
In the case of the additional reactions involving LDLR synthesis (LDLR transcription, translation, mRNA degradation and the dissociation of SREBP-2 for the LDLR gene) these were determined in a similar manner to that detailed in [10] with further details provided in Appendix B. The rates of VLDL and LDL delipidation and the source of extracellular VLDL were calculated as also shown in Appendix B.
Initially the integrated model was informed with the parameter values detailed in [10,15] and derived in Appendix B. A local sensitivity analysis was then used to determine which parameters required variation such that the integrated model reproduced known intracellular cholesterol concentrations [12]. Where adjustments to parameter values determined in [10,15] where made, this is detailed in Table 1 (denoted “This study" along with either the citing of [10,15]) and Appendix B.
3. Results
3.1. Numerical Simulations
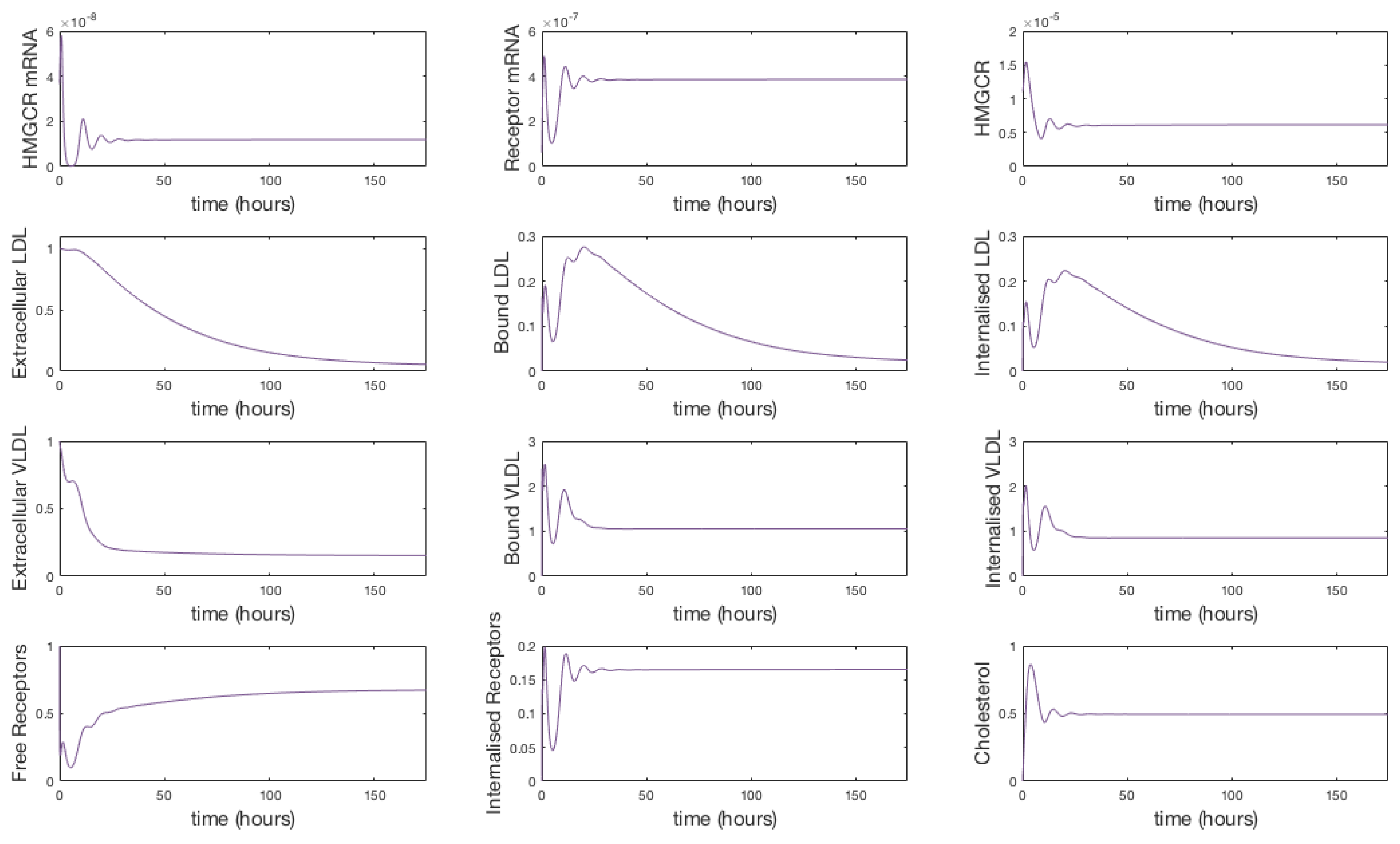
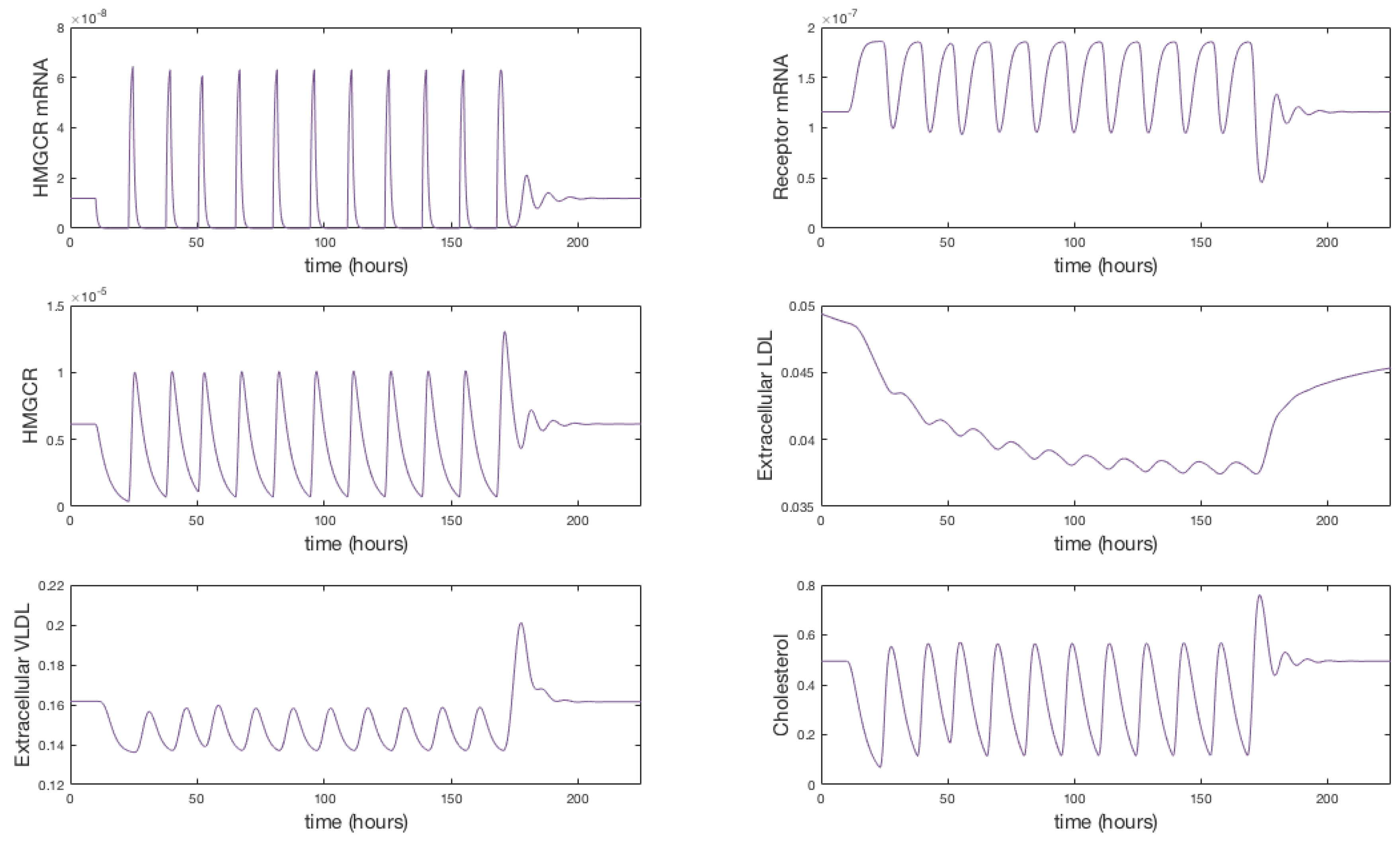
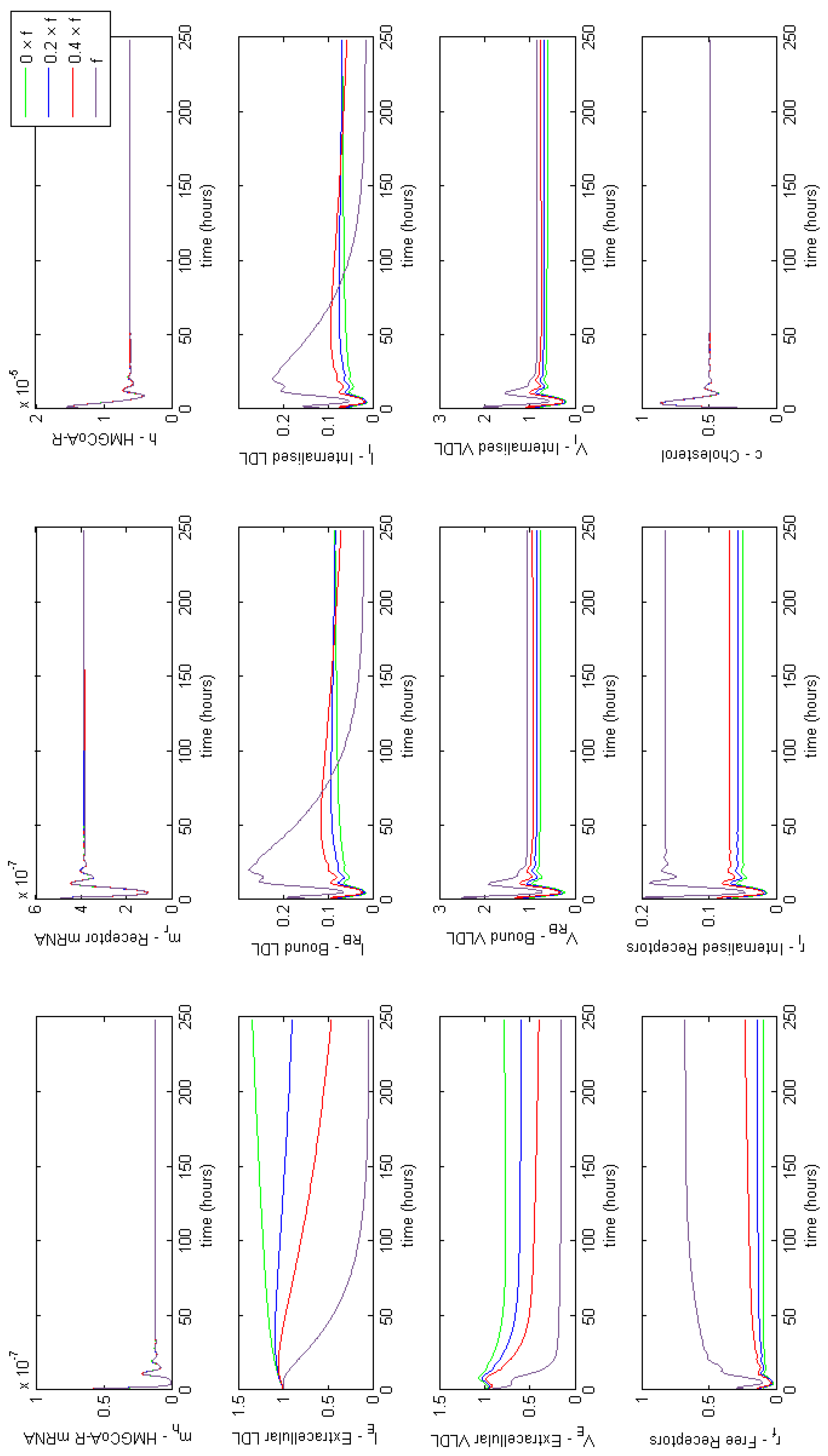
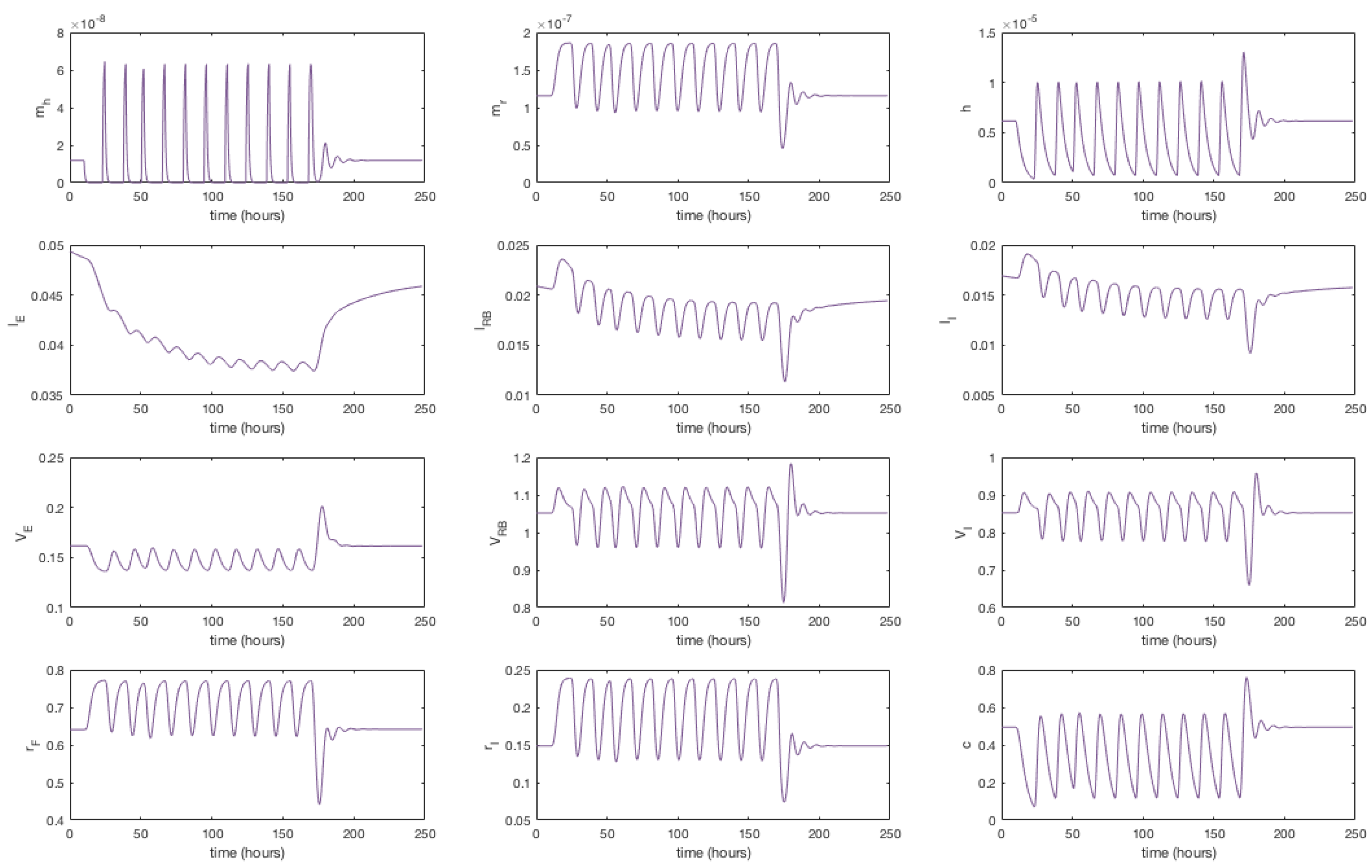
The system of Equations (36)–(48), parameterised with Table 1, was solved using the Matlab stiff differential equation solver ODE15s, given the stiffness coefficient of the system was determined to be . A plot of the simulation is shown in Figure 3. We have re-dimensionalised time on the horizontal axis and run the system for approximately 175 h (until steady state) to capture the whole range of behaviours exhibited.
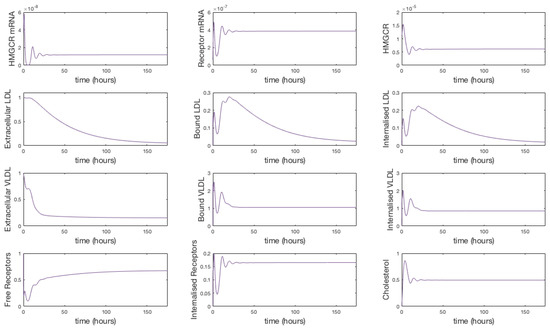
Figure 3.
Numerical simulation of Equations (36)–(48). Initially mRNA levels increase in response to zero cholesterol in the system, which leads to an increase in HMGCR and internal receptor levels. VLDL and LDL bind to receptors and are internalised where cholesterol is extracted. The increase in HMGCR and extraction of cholesterol from internalised lipoproteins cause intracellular cholesterol concentrations to increase. Damped oscillations can be seen as HMGCR mRNA responds to changing cholesterol concentrations and the receptor mechanism responds accordingly. Following this initial transient behaviour, the molecular and cholesterol parts of the system settle to a relatively stable steady-state whilst VLDL and LDL continues to be extracted from the extracellular environment until a final steady-state is reached.
The solutions in Figure 3 show initially (up to 10 h) HMGCR and receptor mRNA increase in response to the initially low cholesterol levels which leads to an increase in HMGCR, internal receptors and cholesterol. VLDL and LDL bind rapidly to free receptors, however VLDL molecules bind more rapidly than LDL due to their greater binding affinity. The rapid binding of lipoproteins leads to an increase in bound LDL and VLDL and hence internalised LDL and VLDL increase. Intracellular cholesterol concentrations increase as cholesterol is extracted from the internalised lipoproteins, and receptors are stored internally and recycled to the cell surface. As intracellular cholesterol concentrations increase, the negative feedbacks from SREBP-2 inhibit HMGCR and LDLR mRNA transcription and hence less HMGCR and LDLR are synthesised. This decrease activates the feedforward/feedback mechanisms and the cell exhibits transient oscillatory type behaviour as a result of the system dynamics.
After this initial period, the molecular components of the system settle to a stable steady-state whilst the longer timescale events of VLDL and LDL endocytosis continue to occur. Eventually each component of this part of the system settles down to a non-zero stable steady-state, a result of the constant influx of VLDL to the system; extracellular VLDL settle before that of extracellular LDL given delipidation and an increased receptor-molecule binding affinity for VLDL than LDL. Internalised receptors tend to a non-zero steady state as the cell is constantly producing receptors to keep in the internal store ready for insertion onto the cell surface.
The concentration of intracellular cholesterol increases initially as a result of the biosynthesis cascade with cholesterol extracted from internalised VLDL and LDL having a significantly less impact on cholesterol levels after the first 10 h.
3.2. Model Analysis
3.2.1. Steady-State Analysis
Given the occurrence of negative and positive feedbacks (genetic and whole cell scale) within the system, we undertook a steady-state analysis of Equations (36)–(48) to understand how many biologically feasible steady-states it may exhibit; more than one real, positive steady-state may indicate more complex underlying system dynamics which have not been previously elucidated experimentally. This analysis was conducted in the absence () and presence () of a source of VLDL particles. In the case of we obtained the expected result that all of the extracellular lipoproteins are internalised and esterified, leading to an abundance of free receptors on the cell surface and LDLR and cholesterol being produced via their respective biosynthetic pathways
where the notation indicates steady-state. This result was also verified numerically.
In the case of the system of equations could only be reduced to the four state system of
where
and
3.2.2. Sensitivity analysis
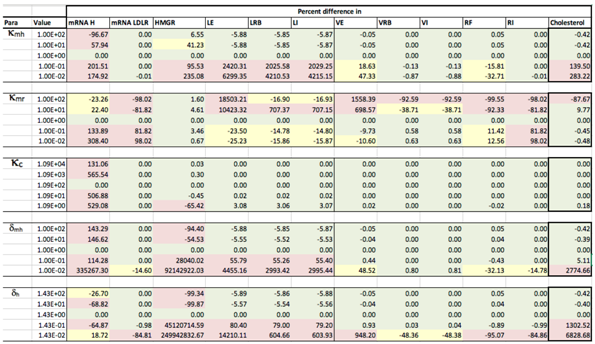
Sensitivity analysis was conducted to determine how the output of our model can be apportioned to the varying sources of input. We conducted local sensitivity analysis, varying each parameter 100-fold above and below its initial value, whilst keeping all the other variables constant as shown in Table 2. We quantitatively measured, primarily, the effect of mechanisms on the steady state intracellular cholesterol concentrations whilst also looking for significant variations in key elements of the system, for example extracellular LDL levels which are an indicator of risk in CVD. What follows is a summary of our findings with more details provided in Appendix C.
Sensitivity analysis of our fully integrated model demonstrated that the respective components (cholesterol biosynthesis and endocytosis) reproduced similar effects on intracellular cholesterol levels as they did when considered in isolation. For instance the integrated model shows the same competition effect between LDL and VLDL for LDLR as detailed in [13,15]; a result of VLDL having a greater receptor binding affinity than LDL. Likewise, this behaviour can be exacerbated by the number of receptors occluded by a bound lipoprotein and by the rate of receptor synthesis. However, where there are limited binding sites on the cell surface, smaller LDL particles are able to bind in spaces that larger VLDL are unable to. We found that variation in parameters affecting LDLR synthesis does not greatly affect cholesterol levels.
We found that biosynthesis of cholesterol has a greater effect on cellular cholesterol levels than the uptake of extracellular lipoproteins, which agrees with the known biology [3]. The concentration of free LDLR does not greatly affect intracellular cholesterol levels, but does have a much greater effect on the concentration of extracellular LDL and VLDL, both of which are major risk factors for health conditions such as CVD. Therefore we know the receptor mechanism plays an important role in lipoprotein clearance which agrees with experimental evidence [39].
The amount of cholesterol within an LDL or VLDL molecule (, ) was found to have very little effect on intracellular cholesterol levels. However, when and were increased, the concentration of HMGCR mRNA and LDLR mRNA were reduced. For 10-fold increased cholesterol in VLDL, HMGCR mRNA was reduced by nearly 20% and receptor mRNA reduced by around 5%. To induce similar levels of mRNA reduction, the cholesterol content of LDL needs to be increased 100-fold. This is due to the increased amount of cholesterol being brought in to the cell via receptor mediated endocytosis and shows very tight genetic regulatory control of intracellular cholesterol concentrations.
We have also found for certain values that parameters linked with cholesterol and receptor biosynthesis ( and ) can produce damped periodic behaviour; the damping a result of the difference in the volumes of the cell nucleus and cytoplasm.
Sensitivity analysis of the VLDL-LDL delipidation parameter, , provided some interesting results. Increasing the rate of delipidation results in a 40% decrease in extracellular VLDL concentrations, but a 450% increase in extracellular LDL concentrations. Conversely, however, a 10-fold decrease in the delipidation rate produces around an 8% increase in extracellular VLDL concentrations but, significantly, nearly a 90% decrease in extracellular LDL concentrations. Furthermore, there is no other significant changes in the rest of the system due to the perturbation of . Our model would suggest, then, that delipidation of VLDL to LDL would be a good candidate as a target for LDL reduction therapies.
3.3. Investigating Familial Hypercholesterolaemia (FH)
In this section we investigate whether our model is able to reproduce the known effects of the disease Familial Hypercholesterolaemia (FH), the aetiology of which is well known. By altering specific model parameters we can quantitatively represent the effect of genetic mutations on extracellular VLDL and LDL levels.
FH is a genetic disorder, primarily of the LDL receptor gene and is characterised by high levels of circulating LDL cholesterol. In certain populations (including French and Canadians) 1 in 67 people suffer FH, with an increased risk of heart disease 20 times greater than non-sufferers [40]. Furthermore, almost all people with FH will require plasma cholesterol-lowering drugs.
The gene pertaining to the LDL receptor is located on chromosome 19 and a number of mutations have been identified in the DNA of individuals affected by this disorder [41]. Hobbs et al. [42] identified five categories of LDL receptor defects, which are listed in Table 3 with a description of the biological traits and parameters in our model that are affected. We will model each case by multiplying each relevant parameter by 0, 0.2, 0.4 and 1. We are unable to model type II since our model does not, with the exception of the nucleus and cytoplasm, sub-compartmentalise the cell.

Table 3.
Familial hypercholesterolaemia class types and their relation to parameters in our model.
3.3.1. Class I FH
Let us first consider Class I where LDL receptors are not synthesised. In this case the associated parameter is , which we vary in order to investigate the model response. The results illustrated in Figure A1 show how the inhibition of receptor synthesis prolongs uptake of extracellular LDL, which would equate to higher circulating plasma LDL levels.
As expected, with , receptor numbers deplete and lipoproteins are unable to be internalised. Levels of extracellular VLDL increase because of continuous influx, as do LDL as VLDL are broken down into LDL. This increase in extracellular LDL concentration biologically would increase the risk of health problems. When varying successively we find that even a small increase in the number of receptors synthesised decreases the levels of extracellular LDL and VLDL. For instance increasing the value from 0 to 20% of normal function halves the concentration of circulating VLDL and LDL. Increasing the value from 0 to 40% of normal function decreases the concentration of circulating VLDL and LDL by 80–90%, respectively.
3.3.2. Class III FH
We now consider Class III where binding of LDL and receptors is ineffective. In this case the associated parameter is (LDL receptor binding), which we vary in order to investigate the model response. The results are illustrated in Figure A2 and show that the amount of extracellular LDL is affected significantly by the inability of LDL to bind to LDLR on the cell surface. We can see that increasing LDL-LDLR binding from 0 to just 20% of normal function decreases extracellular LDL concentrations by 67%. Subsequently restoring normal function reduces extracellular LDL concentrations by nearly 97%.
3.3.3. Class IV FH
Here LDL bound to receptors on the cell surface are not internalised properly. In this case we vary the associated parameter, and investigate the model response. The results illustrated in Figure A3 demonstrate the concentration of extracellular LDL is significantly affected if , but is only marginally altered if is increased. There is also a significant difference between the amount of bound LDL when and when is increased, however this does not appear to significantly affect extracellular VLDL concentrations. Aside from when , the system is fairly robust to changes in the internalisation rate of bound LDL particles.
3.3.4. Class V FH
Finally we consider Class V where LDL receptors are not recycled effectively. To investigate this case we vary the associated parameter f. The results in Figure A4 show the number of free and internalised receptors declines significantly for reduced receptor recycling which causes an increase in extracellular concentrations of LDL and VLDL. However the number of bound and internalised VLDL is not affected as significantly as the number of bound and internalised LDL. This is because VLDL have a greater binding affinity and so are more successful in binding competition. We also see a reduction by more than half in both free and internalised receptor concentrations between the usual value and altered values .
3.3.5. Individual Class FH Summary
Having explored the effects of different classes of FH, we have found that Class I has the greatest effect on extracellular LDL and Class IV the least.
The lack of variation in intracellular cholesterol, HMGCR mRNA and HMGCR levels suggest that despite the effects of FH, the cell is able to maintain intracellular cholesterol levels genetically. This makes sense as without this control the concentration of cholesterol may decline to cytotoxic levels and the cell perish. However we know that humans are able to survive with FH indicating their cells do not perish.
We now wish to simulate the effects of being afflicted with a combination of all four cases.
3.3.6. Combined FH
We used Latin Hypercube Sampling [43], to consider the possible outcomes for a range of combined effects of FH Class types I, III, IV and V. Latin Hypercube Sampling generates a sample of plausible collections of parameter values from a multidimensional distribution. The method takes the midpoint of each quartile for parameters selected and randomly samples the combination of the effects of the four FH class types. In this case this leads to the four hypothetical combined FH cases detailed below.
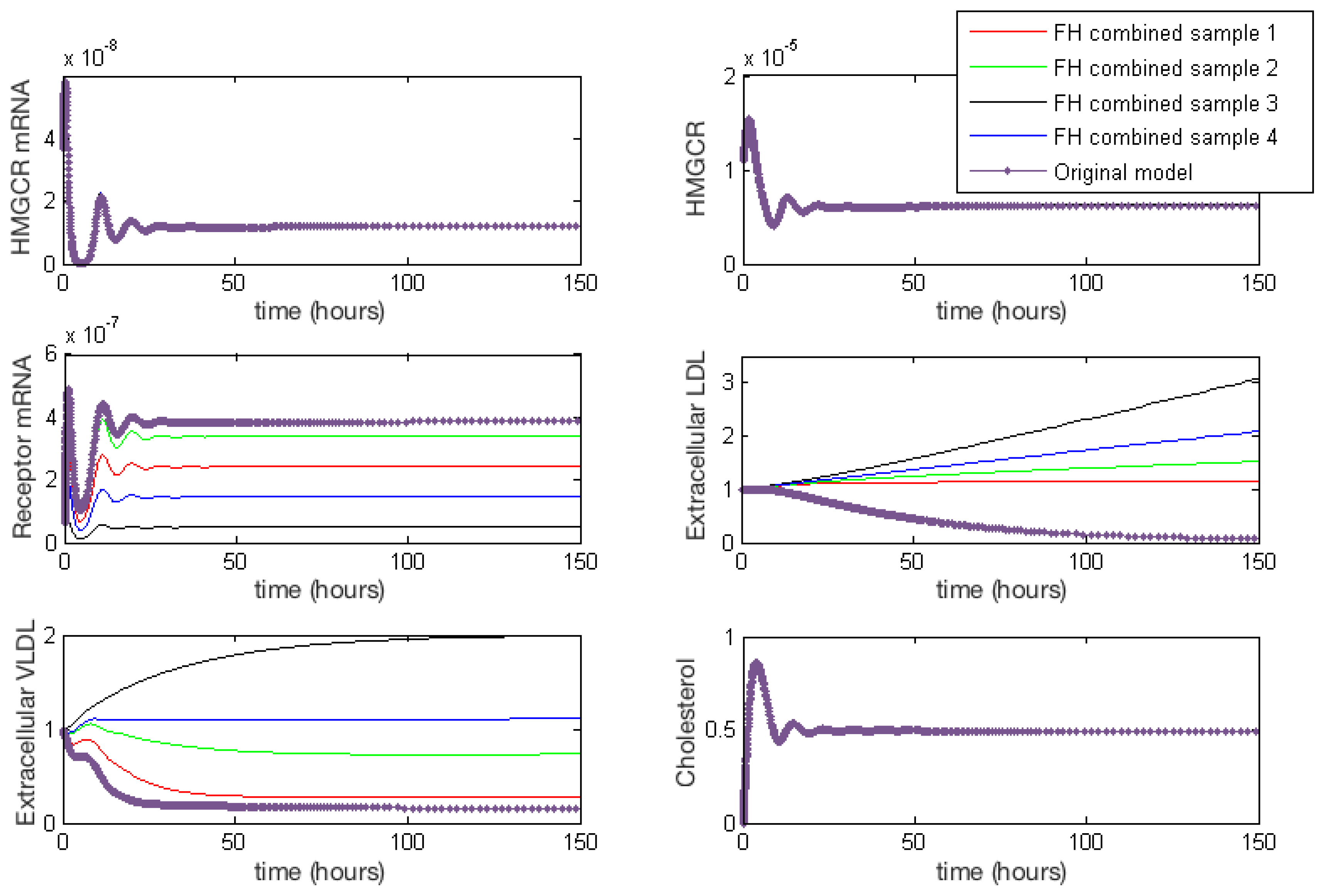
Figure 4 provides a summary of the effects of each of the combined cases of FH on HMGCR mRNA, HMGCR, LDLR mRNA, extracellular LDL and VLDL and cellular cholesterol levels. For completeness full model results are provided in Appendix E. These lead to a disruption in receptor production, free receptors, extracellular levels of VLDL and LDL and the binding and internalisation of VLDL and LDL that we would expect to see as a result of the disease. Our model shows that despite lipoprotein uptake being significantly reduced, the cell will keep intracellular cholesterol levels within a tightly controlled range as a result of genetic regulation via the SREBP-2 cholesterol feedback. This feedback ensures the cell responds to low levels of cholesterol by upregulating cholesterol biosynthesis, allowing it to produce around 80% of the cholesterol the cell needs, in spite of disruptions to receptor function. Whilst direct comparison with experimental values of intracellular cholesterol is not possible due to a lack of reported values in the literature, we postulate here that this effect could be tested experimentally via a series of VLDL and LDL uptake experiments. For populations of cells each affected by the different FH classes, the relative difference in the uptake of the lipoproteins could be compared to that of a control group of cells, in order to discern the differences detailed here.
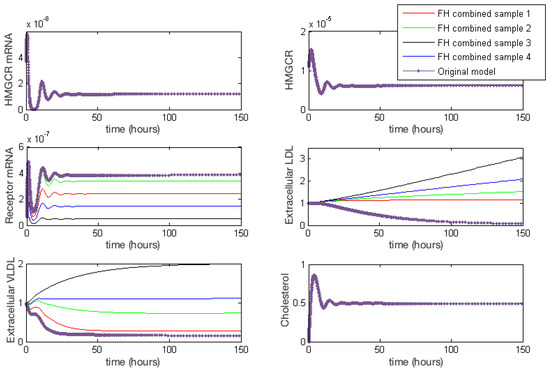
Figure 4.
Familial Hypercholesterolaemia combined effects on HMGCR mRNA, HMGCR, LDLR mRNA, extracellular LDL and VLDL and cellular cholesterol levels. Results for all model variables are given in Appendix E.
From these model results we can infer the increased susceptibility to CVD events, as a result of FH leading to increased plasma LDL levels concurs with the known biology. Furthermore, we can see from the samplings taken, Case 3 leads to the greatest rise in plasma LDL levels, due to low receptor synthesis and recycling combined with that of low LDL receptor binding affinity.
3.4. Modelling Statin Therapy
We can also consider if our model produces the known biological response to statins, globally the most commonly used pharmaceutical treatment for lowering plasma cholesterol levels. These drugs competitively bind to HMGCR preventing binding with HMGCoA and so inhibiting cholesterol biosynthesis. This reduces intracellular cholesterol concentrations thereby up-regulating receptor synthesis which clears more lipoproteins from the circulation. In this model, the effect of taking statins can be modelled by modifying the transcription of HMGCR mRNA, . We here show the numerical results for an idealised statin that instantaneously halts transcription of HMGCR mRNA, for 11 doses over a period of 7 days such that
where n is the number of dosage periods. Although this is a dramatic change in it is sufficient to show that the model replicates the expected dynamical behaviour. We have run the model to steady state and begun our simulation from that point. At time t = 9 h 45 m we set for the equivalent of approximately 13 h. Dosing in this way, the solutions give a 17.6% reduction in extracellular LDL. No further discernible differences in extracellular LDL levels were perceived after this period.
It is indicated that in general statins cause a 25%–55% decrease in LDL-cholesterol [44]. We were able to achieve a 25% reduction in extracellular concentrations by setting the rate of receptor mRNA transcription , with the same 11 doses over 7 days. The solutions in Figure 5 show that upon receiving a statin dose, levels of HMGCR mRNA and HMGCR decline to zero as transcription is inhibited. Solutions for all model variables are provided in Appendix F. Due to the lack of biosynthesis, cholesterol levels also decline dramatically which in turn up-regulates the transcription of receptor mRNA; a response by the cell to bring more cholesterol in to maintain healthy levels. Subsequently we see a rise in internal receptor levels and hence free receptors on the cell surface. Extracellular LDL and VLDL decrease as they bind to the abundant free receptors and are endocytosed. We see that with each statin dose extracellular LDL concentrations gradually decline for 7 days until they level out at a 25% decrease. Whilst of the same order of magnitude as that observed clinically we believe differences are a result of the short term duration of our statin application versus the longer term scale of measurements taken in patients (e.g. weeks or months). Furthermore, our model does not contain a detailed description of other elements of lipoprotein metabolism, for instance chylomicrons, or that of VLDL production by the hepatocyte. LDL levels are also directly linked to those of VLDL at present, whereas in vivo it is known they do not vary as much as other lipoproteins postprandially [45].
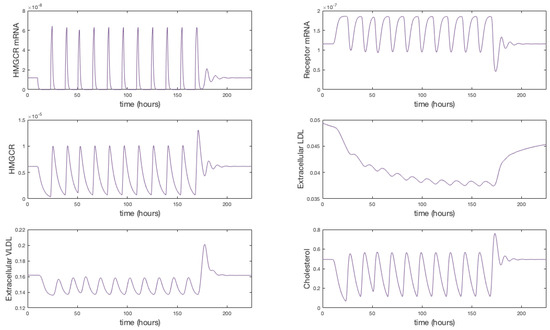
Figure 5.
A simulation showing the effect of statin therapy on our integrated model of cholesterol metabolism. Here 11 doses are applied over a seven day period starting at approximately 10 h. The inhibition of HMGCR mRNA transcription subsequently decreases HMGCR and cholesterol biosynthesis which up-regulates receptor mRNA transcription and receptor synthesis, which leads to a reduction in extracellular LDL and VLDL levels. A complete set of model solutions are provided in Appendix F.
After 168 h (7 days) we allow to return to its steady-state value of . In doing so concentrations of each of the two biosynthesis pathways (cholesterol and LDLR) exhibit periodic overshoot type behaviour as the system returns to its pre-stimulus steady-state; a result of the homoclinic Hopf bifurication behaviour that the cholesterol biosynthesis pathway exhibits [10]. Those entities directly affected by this change, for example receptor synthesis, also exhibit such behaviour, but this is dampened in the case of bound VLDL and LDL and considerably more so in the case of their extracellular levels.
4. Discussion and Conclusions
In this paper we have formulated, solved and analysed a nonlinear deterministic ODE model describing the key mechanisms of hepatocyte endocytosis of VLDL and LDL coupled with a description of cholesterol and receptor biosynthesis via the HMGCR pathway.
Parameterised with data from the relevant literature, the model was solved using the Matlab stiff differential equation solver ODE15s. Solutions showed the system synthesising cholesterol and receptors in response to mRNA transcription and translation of each entity, and uptake of extracellular VLDL and LDL as a result of receptor synthesis and the extraction of cholesterol from internalised lipoproteins.
Sensitivity analysis showed that the model qualitatively reproduced the known biology of lipoprotein uptake and receptor and cholesterol regulation [17,30,33]. It highlighted the competition effect between LDL and VLDL when binding to free receptors on the cell surface. VLDL have a greater binding affinity than LDL and so are removed from the lipoprotein rich medium faster than LDL. However, we found that when receptor numbers were reduced, LDL particles are able to bind in spaces that VLDL are not able to, given their differences in sizes, and thus extracellular LDL levels decrease faster than VLDL ones. We found the concentration of free receptors on the cell surface for lipoproteins to bind to have little effect on intracellular cholesterol levels but greatly impacted concentrations of extracellular LDL and VLDL which is a major risk factor for health problems such as CVD. Sensitivity analysis also demonstrated that periodic behaviour exhibited by the cholesterol biosynthesis system [12] becomes damped as the signal dissipates from transcription in the cell nucleus to translation in the cell cytoplasm.
Our work has highlighted that small changes in the rate of VLDL to LDL delipidation had significant effects on extracellular LDL concentrations but not the rest of the system. Hence this process could be a good candidate target for LDL reductive therapies. While modelling processes in this way is helpful to identifying possible new methods of treatments, the enzymes responsible for this reaction (lipoprotein lipase, hepatic lipase and cholesteryl ester transfer protein) have dual functions which are not represented by our model, so it is hard to predict the full extent of including this change without further work.
Our model demonstrates that cholesterol biosynthesis is the dominant source of cholesterol for the cell. Thus any major disruption of this pathway is likely to have a detrimental effect on human health. Thus whilst any therapy targeted at reducing intracellular cholesterol, such as statins, will favourably decrease plasma levels of circulating lipoproteins in time, our model results suggest the counteracting of any decrease in intracellular cholesterol is limited by the number of receptors available on the cell surface (a function of the cell size and receptor internationalisation and recycling) and their rate of internalisation. Hence in cases where cholesterol biosynthesis is compromised we speculate it will be difficult to supplement intracellular cholesterol levels via increases in plasma cholesterol levels alone.
Although the mathematical formulation of lipoprotein endocytosis used here assumes the surface of the cell is covered in a continuum of LDLR [15], the main outcome of the model (total cholesterol content) does not greatly differ to that of [12] in respect of predicting each class of Familial Hypercholesterolaemia and statin therapy. We, however, note the differences between each of [13,15] at the lower level detail of receptor occupancy levels and rate of lipoprotein uptake require further investigation.
We have been able to explore the effects of the genetic disease Familial Hypercholesterolaemia and statins using our integrated model of cholesterol metabolism. We found that Case I FH has the greatest effect on extracellular LDL concentrations and Case IV the least. The effects of combined cases were shown to affect receptor mRNA, free and internalised receptor levels and the extracellular concentrations of LDL and VLDL. We also found the model replicates the qualitative effects of statins very closely.
Quantitatively the model produced a 25% reduction in extracellular LDL levels for repeated statin dosing over a seven day period when the receptor mRNA transcription rate was decreased by 70%. Whilst our the model was able to reproduce the clinically reported lower bound of extracellular LDL reductions following statin therapy, we believe improvement in this result could be made by including a more thorough description of in vivo lipoprotein metabolism (e.g., chylomicrons, HDL) and longer dosing periods. Thus we believe further model extensions coupled with clinically informed parameterisation of this work are required to fully capture the quantitative regulation of lipoproteins and their responses to statin therapy.
In conclusion, our work has demonstrated that mathematical modelling can provide a useful tool for understanding the cellular (lipoprotein endocytosis) and subcellular (biosynthesis and genetic regulation of cholesterol and receptors) processes that occur during lipoprotein metabolism. Whilst the level of abstraction of our mathematical model is quite high, this work demonstrates that such simplifications of a complex system can still reproduce the known biology of disease states and therapeutic interventions. Future work and extensions to the model presented here is thus needed to consider the effect of other aspects of the overall system, occurring at the subcellular and tissue level. There thus remains scope for further testing and application of such models and their extension to contexts that include a description of other dietary lipoproteins.
Author Contributions
F.P. undertook research and wrote the paper, P.K.S. and M.J.T. both supervised the research and wrote the paper.
Funding
F.P. acknowledges the support of an Engineering Physical Sciences Research Council (EPSRC) UK CASE studentship in collaboration with Syngenta (EP/P505682/1 & EP/J500501/1). M.J.T. is grateful for the support of a Research Council UK Fellowship (EP/C508777/1) during parts of the period in which this work was undertaken.
Conflicts of Interest
The atuhors declare no conflict of interest.
Appendix A. Model Reduction
Equations (3)–(20) were reduced as follows. We first observe that the total number of genes with a cell remains constant whether bound to SREBP-2 or otherwise. Adding Equations (3) and (6) for HMGCR synthesis, and (4) and (6) for LDLR synthesis, respectively, and integrating with respect to time leads to
The total amount of SREBP-2 in a cell is also constant such that
since .
We further reduce the system by assuming DNA-transcription factor binding is rapid in comparison to the reaction events in the cell [10,46] and so reaches a steady state faster than the rest of the system. This rapid equilibrium approximation applied to Equation (6) leads to
where .
Applying the same approximation to Equation (7) leads to
where .
Substitution of result (A3) into Equation (8) gives
and similarly for result (A4) into Equation (9) we find
Finally we assume that cholesterol-SREBP-2binding is rapid in comparison to other reaction events in the cell [10] such that
which upon rearranging yields
with .
Using result (A5) we can express Equation (5.19a) in terms of c such that
with .
Similarly for (5.20a) we have,
with .
Following the work in Tindall et al. [15] we seek to define the number of bound and occluded receptors. Quantities of are dependent on the average occupancy of surface receptors. In high levels of extracellular LDL and VLDL, average occupancy will be high and free receptors will be low, hence fewer free receptors will be internalised with each pit and will have a small value. Intuitively then, in low concentrations of extracellular LDL and VLDL, will have a larger value. To simplify, we can assume the total number of receptors is approximately constant over shorter time-scales, such as that of pit internalisation, then we can say the total number of receptors on the cell surface is
where represents bound receptors.
We can also assume, as pits are internalised, a fraction of all receptors are internalised also, given by
We assume the contribution of free receptor internalised with each particle is divided equitably between all bound particles, that is and with being the total density of free receptors. We can find by calculating the difference between Equations (A6) and (A7) giving
where, since the number of internalised receptors is small, a suitable value for is .
Appendix B. Parameter Estimation
—Initial value of receptor mRNA: Rudling et al. (2002) [38] details copy numbers of mRNA found in human liver cells under basal conditions. We take a value of 48 copies of receptor mRNA per cell, i.e., per mL which gives 48 molecules/ mL = 4.8 molecules/mL. This value was then refined using sensitivity analysis to give molecules/mL.
—Rate of receptor mRNA transcription: As for , one human LDLR mRNA transcript is 5265 bases long [21]. To transcribe one molecule of LDLR mRNA, from one gene, at a rate of 12 bases per second, takes
So per gene we have 1/438.75 s = 2.28 × molecules/s. There are two genes in a liver cell so LDLR mRNA molecules are synthesised per cell per second. With a cell volume equal to mL this gives 4.56 × molecules/s/1 × mL giving molecules/mL/s.
: Soutar and Knight (1990) [19] tell us a human LDLR mRNA transcript contains 839 amino acids. For one ribosome to transcribe one molecule of LDLR protein, from one LDLR mRNA, at 6 amino acids per second, it takes 839 amino acids/6 amino acids/s = 139.83 s. Then per ribosome this gives
The coding region of LDLR mRNA is 839 amino acids × 3 = 2517 nucleotides long, and a ribosome can attach every 35 nucleotides, so we have 71.91 ribosomes per mRNA molecule. Finally, 7.15 × molecules/s/ribosome × 71.91 ribosomes gives molecules/s.
: In the absence of further details we assume the rate of receptor mRNA degradation is equivalent to that HMGCR mRNA degradation and so take /s.
: Yang and Swartz (2011) [47] quantify DNA binding affinities to other transcription factors at nmol. We convert this value into units of molecules/mL by the use of Avogadro’s constant, such that
as an estimate we took . This value was then refined using sensitivity analysis to give molecules/mL.
—Rate of VLDL synthesis: We estimated a value for the rate of VLDL synthesis as particles/mL/s. This value was derived by a local sensitivity analysis such that model reproduced known intracellular cholesterol concentrations.
—Number of binding sites on LDLR gene for SREBP-2: The number of binding sites on the receptor gene available to SREBP-2 [12] is taken as being .
Appendix C. Sensitivity Analysis
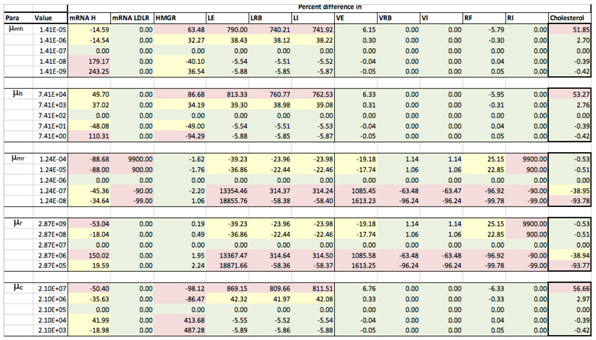
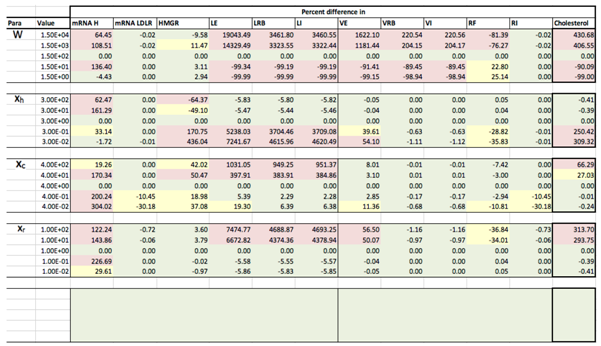
Results of the local sensitivity analysis of Equations (36)–(48) showing the relative percentage change from steady-state as each parameter was varied 100-fold below and above the values indicated in Table 2. Here “mRNA H” is HMGCR mRNA, “LE” is extracellular LDL, “LRB” is receptor bound LDL, “LI” is intenalised LDL, “VE” is extracellular VLDL, “VRB” is receptor bound VLDL, “VI” is internalised VLDL, “RF” is free receptors and “RI” is internalised receptors. Green indicates a less than 10% change from the steady-state value found when the model was solved using the values in Table 2, yellow greater than 10% but less than 50% and pink greater than 50%.
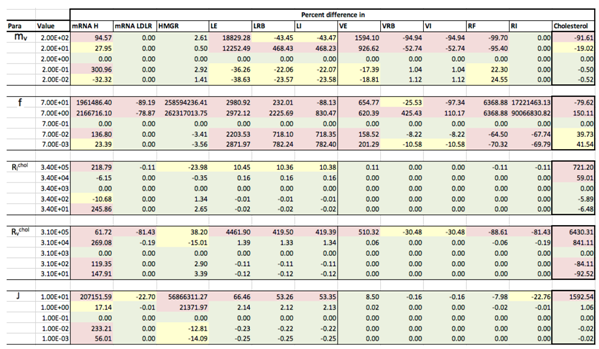
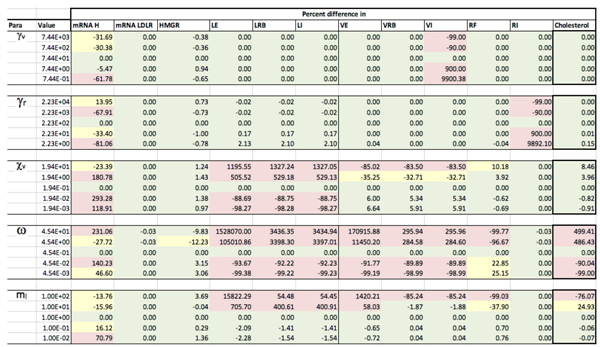
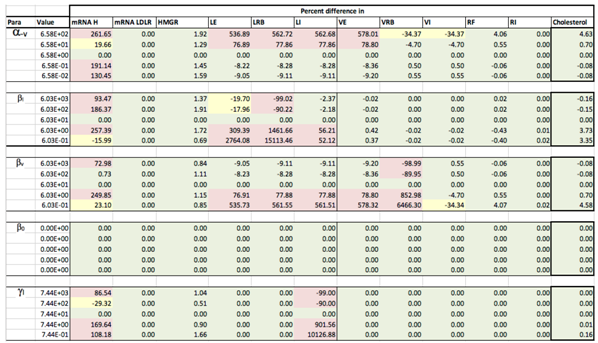
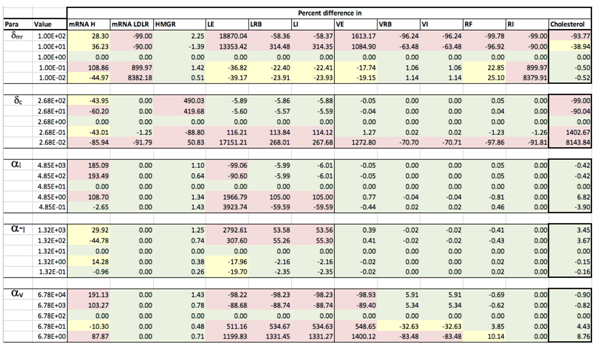
Appendix D. Familial Hypercholesterolaemia Analysis
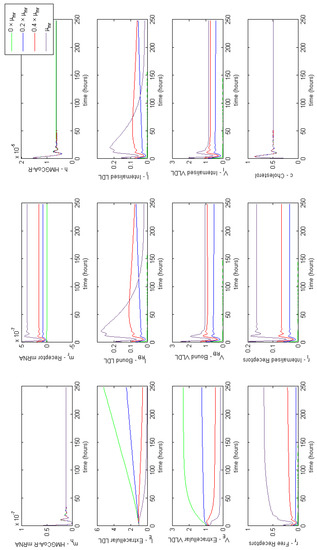
Figure A1.
Familial Hypercholesterolaemia Class I; LDLR are not synthesised correctly, affecting parameter . Results show a lack of receptors has a significant impact on extracellular LDL and VLDL concentrations.
Figure A1.
Familial Hypercholesterolaemia Class I; LDLR are not synthesised correctly, affecting parameter . Results show a lack of receptors has a significant impact on extracellular LDL and VLDL concentrations.
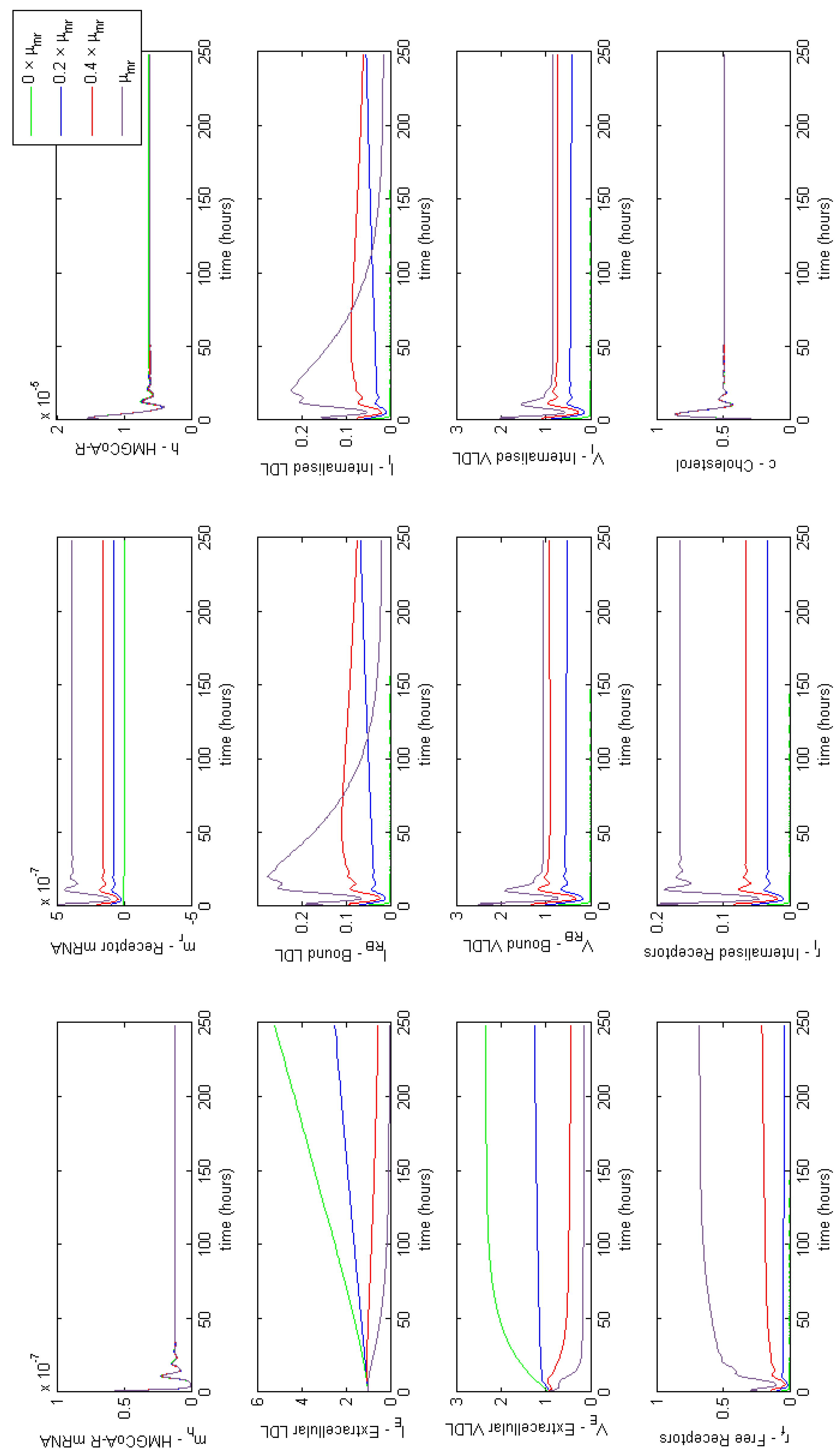
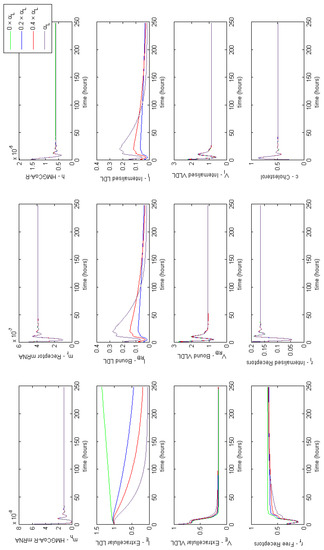
Figure A2.
Familial Hypercholesterolaemia Class III; binding of LDL to LDL receptors is ineffective, affecting parameter . Results show a lack of LDL binding prevents LDL being endocytosed.
Figure A2.
Familial Hypercholesterolaemia Class III; binding of LDL to LDL receptors is ineffective, affecting parameter . Results show a lack of LDL binding prevents LDL being endocytosed.
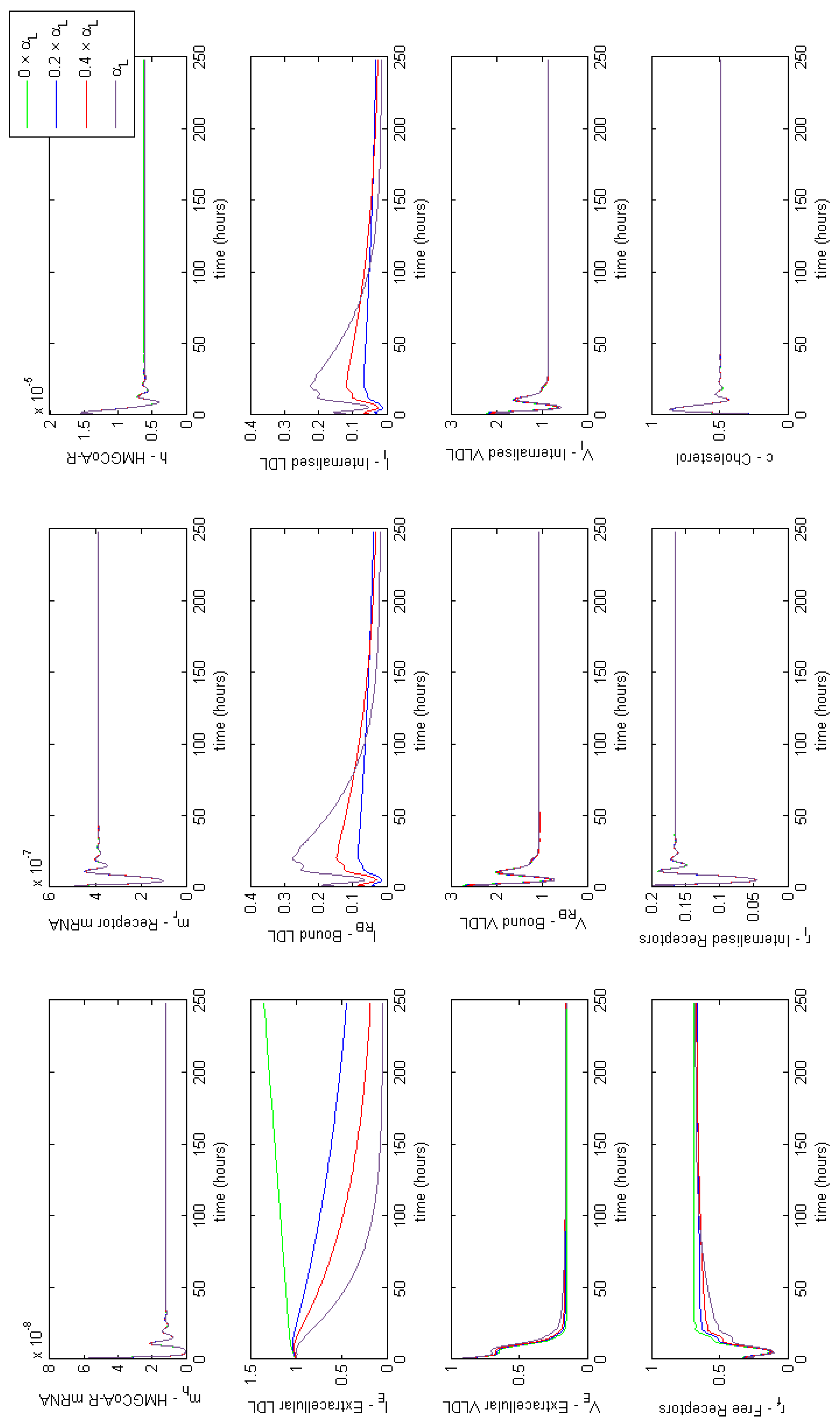
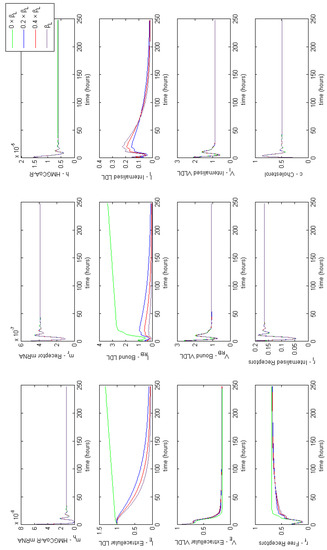
Figure A3.
Familial Hypercholesterolaemia Class IV; Bound LDL not internalised properly, affecting parameter . Results show the lack of internalisation of bound LDL particles blocking LDL binding.
Figure A3.
Familial Hypercholesterolaemia Class IV; Bound LDL not internalised properly, affecting parameter . Results show the lack of internalisation of bound LDL particles blocking LDL binding.
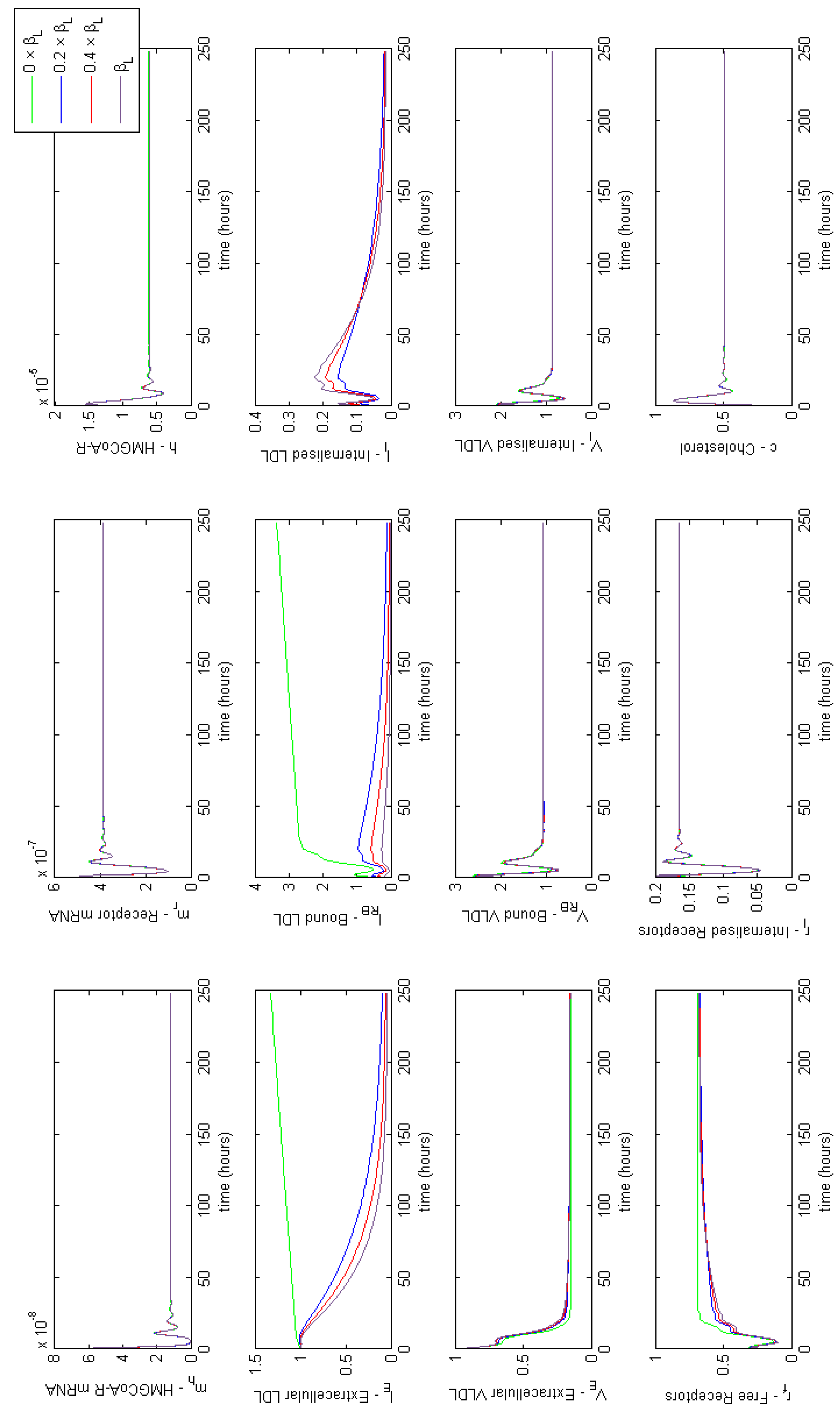
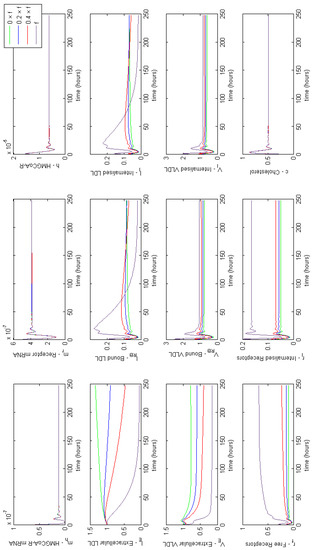
Figure A4.
Familial Hypercholesterolaemia Class V; receptors are not recycled properly, affecting model parameter f. Results show a decrease in free and internalised receptors, significantly affecting extracellular concentrations of both LDL and VLDL.
Figure A4.
Familial Hypercholesterolaemia Class V; receptors are not recycled properly, affecting model parameter f. Results show a decrease in free and internalised receptors, significantly affecting extracellular concentrations of both LDL and VLDL.
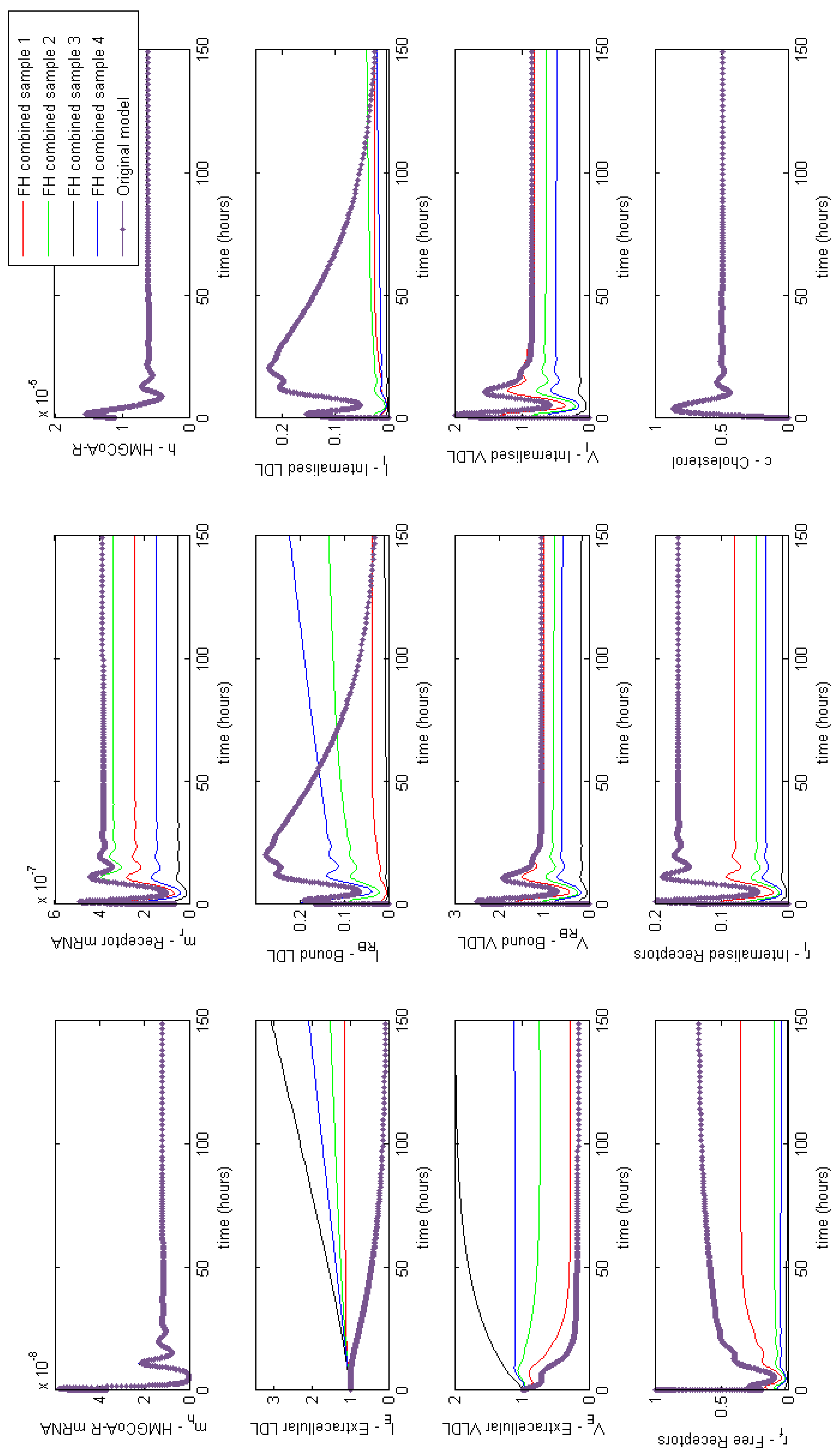
Appendix E. Full Model Results for Combined Cases of Familial Hypercholesterolaemia
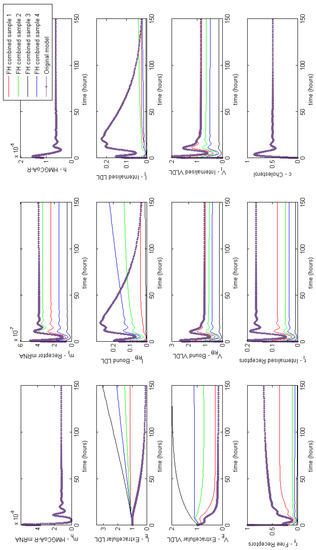
Figure A5.
Familial Hypercholesterolaemia combined effects. Showing a combination of reduced synthesis production and recycling and ineffective LDL binding and internalisation can dramatically increase levels of circulating LDL and VLDL.
Figure A5.
Familial Hypercholesterolaemia combined effects. Showing a combination of reduced synthesis production and recycling and ineffective LDL binding and internalisation can dramatically increase levels of circulating LDL and VLDL.
Appendix F. Full Model Results for Statin Application
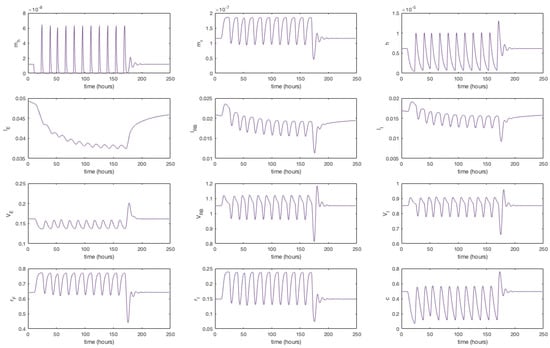
Figure A6.
A simulation showing the effect of statin therapy on our integrated model of cholesterol metabolism. Here 11 doses are applied over a seven day period starting at approximately 10 h.
Figure A6.
A simulation showing the effect of statin therapy on our integrated model of cholesterol metabolism. Here 11 doses are applied over a seven day period starting at approximately 10 h.
References
- O’Brien, J.; Sampson, E. Lipid composition of the normal human brain: Gray matter, white matter, and myelin. J. Lipid Res. 1965, 6, 537–544. [Google Scholar] [PubMed]
- Jousilahti, P.; Vartiainen, E.; Tuomilehto, J.; Puska, P. Sex, age, cardiovascular risk factors, and coronary heart disease A prospective follow-up study of 14,786 middle-aged men and women in Finland. Circulation 1999, 99, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Turley, S.; Dietschy, J. The intestinal absorption of biliary and dietary cholesterol as a drug target for lowering the plasma cholesterol level. Prev. Cardiol. 2003, 6, 29–64. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.; Brown, M.; Anderson, R.; Russell, D.; Schneider, W. Receptor-mediated endocytosis: Concepts emerging from the LDL receptor system. Annu. Rev. Cell Biol. 1985, 1, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Grundy, S. Inflammation, metabolic syndrome, and diet responsiveness. Circulation 2003, 108, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Gowdy, K.; Fessler, M. Emerging roles for cholesterol and lipoproteins in lung disease. Pulm. Pharmacol. Ther. 2013, 26, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Bermudes, A.; de Carvalho, W.; Dietist, P.; Muramoto, G.; Maranhão, R.; Delgado, A. Changes in lipid metabolism in pediatric patients with severe sepsis and septic shock. Nutrition 2018, 47, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Lagrost, G.; Grosjean, S.; Masson, D.; Deckert, V.; Gautier, T.; Debomy, F.; Vinault, S.; Jeannin, A.; Labbé, J.; Bonithon-Kopp, C. Low preoperative cholesterol level is a risk factor of sepsis and poor clinical outcome in patients undergoing cardiac surgery with cardiopulmonary bypass. Crit. Care Med. 2014, 42, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Paalvast, Y.; Kuivenhoven, J.; Groen, A. Evaluating computational models of cholesterol metabolism. Biochim. Biophys. Acta 2015, 1851, 1360–1376. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Sweby, P.; Minihane, A.M.; Jackson, K.; Tindall, M. A mathematical model of the sterol regulatory element binding protein 2 cholesterol biosynthesis pathway. J. Theor. Biol. 2014, 349, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Pool, F.; Currie, R.; Sweby, P.; Salazar, D.; Tindall, M. A mathematical model of the mevalonate cholesterol biosynthesis pathway. J. Theor. Biol. 2018, 443, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B. Mathematical Modelling of Low Density Lipoprotein Metabolism. Intracellular Cholesterol Regulation. Ph.D. Thesis, University of Reading, Reading, UK, 2011. [Google Scholar]
- Wattis, J.; O’Malley, B.; Blackburn, H.; Pickersgill, L.; Panovska, J.; Byrne, H.; Jackson, K. Mathematical model for low density lipoprotein (LDL) endocytosis by hepatocytes. Bull. Math. Biol. 2008, 70, 2303–2333. [Google Scholar] [CrossRef] [PubMed]
- Cromwell, W.; Otvos, J.; Keyes, M.; Pencina, M.; Sullivan, L.; Vasan, R.; Wilson, P.; D’Agostino, R. LDL particle number and risk of future cardiovascular disease in the Framingham Offspring Study implications for LDL management. J. Clin. Lipidol. 2007, 1, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Tindall, M.; Wattis, J.; O’Malley, B.; Pickersgill, L.; Jackson, K. A continuum receptor model of hepatic lipoprotein metabolism. J. Theor. Biol. 2009, 257, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. The Cell Cycle. In Molecular Biology of the Cell, 5th ed.; Garland Science: New York, NY, USA, 2008; pp. 1053–1114. [Google Scholar]
- Jackson, K.; Maitin, V.; Leake, D.; Yaqoob, P.; Williams, C. Saturated fat-induced changes in Sf 60–400 particle composition reduces uptake of LDL by HepG2 cells. J. Lipid Res. 2006, 47, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Lange, Y.; Ye, J.; Rigney, M.; Steck, T. Regulation of endoplasmic reticulum cholesterol by plasma membrane cholesterol. J. Lipid Res. 1999, 40, 2264–2270. [Google Scholar] [PubMed]
- Darzacq, X.; Shav-Tal, Y.; de Turris, V.; Brody, Y.; Shenoy, S.; Phair, R.; Singer, R. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007, 14, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.; Brown, M. Progress in understanding the LDL receptor and HMG-CoA reductase, two membrane proteins that regulate the plasma cholesterol. J. Lipid Res. 1984, 25, 1450–1461. [Google Scholar] [PubMed]
- National Center for Biotechnology Information. Genome. 2015. Available online: http://www.ncbi.nlm.nih.gov/genome (accessed on 31 July 2015).
- Trachsel, H. Translation in Eukaryotes; CRC Press: Boca Raton, FL, USA, 1991; pp. 149–176. [Google Scholar]
- Istvan, E.; Palnitkar, M.; Buchanan, S.; Deisenhofer, J. Crystal structure of the catalytic portion of human HMG-CoA reductase: Insights into regulation of activity and catalysis. EMBO J. 2000, 19, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Segal, I. Enzyme Kinetics Behaviour and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems; A Wiley-Interscience Publication; Wiley: New York, NY, USA, 1975; pp. 60–100. [Google Scholar]
- Tanaka, R.; Edwards, P.; Lan, S.; Knöppel, E.; Fogelman, A. Purification of 3-hydroxy-3-methylglutaryl coenzyme A reductase from human liver. J. Lipid Res. 1982, 23, 523–530. [Google Scholar] [PubMed]
- Soutar, A.; Knight, B. Structure and regulation of the LDL-receptor and its gene. Br. Med. Bull. 1990, 46, 891–916. [Google Scholar] [CrossRef] [PubMed]
- Wilson, G.; Deeley, R. An episomal expression vector system for monitoring sequence-specific effects on mRNA stability in human cell lines. Plasmid 1995, 33, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Vargas, N.; Brewer, B.; Rogers, T.; Wilson, G. Protein kinase C activation stabilizes LDL receptor mRNA via the JNK pathway in HepG2 cells. J. Lipid Res. 2009, 50, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Dana, S.; Goldstein, J. Regulation of 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase Activity in Cultured Human Fibroblasts comparison of cells from a normal subject and from a patient with homozygous familial hypercholesterolemia. J. Biol. Chem. 1974, 249, 789–796. [Google Scholar] [PubMed]
- Harwood, J.R.H.; Pellarin, L. Kinetics of low-density lipoprotein receptor activity in Hep-G2 cells: Derivation and validation of a Briggs–Haldane-based kinetic model for evaluating receptor-mediated endocytotic processes in which receptors recycle. Biochem. J. 1997, 323, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Mahley, R.; Innerarity, T. Lipoprotein receptors and cholesterol homeostasis. Biochim. Biophys. Acta-Rev. Biomembr. 1983, 737, 197–222. [Google Scholar] [CrossRef]
- Goldstein, J.; Anderson, R.; Brown, M. Coated pits, coated vesicles, and receptor-mediated endocytosis. Nature 1979, 279, 679–685. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Goldstein, J. Receptor-mediated endocytosis: Insights from the lipoprotein receptor system. Proc. Natl. Acad. Sci. USA 1979, 76, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Goldstein, J.; Brown, M. Characterization of the low density lipoprotein receptor in membranes prepared from human fibroblasts. J. Biol. Chem. 1978, 253, 3852–3856. [Google Scholar] [PubMed]
- Shearer, K. The Regulation and Uptake of Dietary Fats. Ph.D. Thesis, University of Reading, Reading, UK, 2010. [Google Scholar]
- Dunn, K.; McGraw, T.; Maxfield, F. Iterative fractionation of recycling receptors from lysosomally destined ligands in an early sorting endosome. J. Cell Biol. 1989, 109, 3303–3314. [Google Scholar] [CrossRef] [PubMed]
- Panovska, J.; Tindall, M.; Wattis, J.; Byrne, H. Mathematical models of hepatic lipoprotein metabolism. In Proceedings of the 5th Mathematics in Medicine Study Group, Oxford, UK, 12–16 September 2005. [Google Scholar]
- Rudling, M.; Angelin, B.; Ståhle, L.; Reihnér, E.; Sahlin, S.; Olivecrona, H.; Björkhem, I.; Einarsson, C. Regulation of hepatic low-density lipoprotein receptor, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and cholesterol 7α-hydroxylase mRNAs in human liver. J. Clin. Endocrinol. Metab. 2002, 87, 4307–4313. [Google Scholar] [CrossRef] [PubMed]
- Go, G.; Mani, A. Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J. Biol. Med. 2012, 85, 19–28. [Google Scholar] [PubMed]
- Foundation, F.H. Familial Hypercholesterolaemia. 2018. Available online: https://thefhfoundation.org (accessed on 6 June 2018).
- Dammerman, M.; Breslow, J. Genetic basis of lipoprotein disorders. Circulation 1995, 91, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, H.; Russell, D.; Brown, M.; Goldstein, J. The LDL receptor locus in familial hypercholesterolemia: Mutational analysis of a membrane protein. Annu. Rev. Genet. 1990, 24, 133–170. [Google Scholar] [CrossRef] [PubMed]
- Iman, R. Latin Hypercube Sampling. In Encyclopedia of Quantitative Risk Analysis and Assessment; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2008. [Google Scholar]
- Golan, D.; Tashjian, A.; Armstrong, E. Principles of Pharmacology: The Pathophysiologic Basis of Drug Therapy; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2011. [Google Scholar]
- Balasubramaniam, S.; Szanto, A.; Roach, P. Circadian rhythm in hepatic low-density-lipoprotein (LDL)-receptor expression and plasma LDL levels. Biochem. J. 1994, 298, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Lickwar, C.R.; Mueller, F.; Hanlon, S.E.; McNally, J.G.; Lieb, J.D. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 2012, 484, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Swartz, J. A filter microplate assay for quantitative analysis of DNA binding proteins using fluorescent DNA. Anal. Biochem. 2011, 415, 168–174. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).