Adaptive Control of Biomass Specific Growth Rate in Fed-Batch Biotechnological Processes. A Comparative Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biotechnological Process and Its Mathematical Model
2.2. Adaptive Control Algorithms
2.2.1. Adaptive PID Control Based on the Gain Scheduling Technique
2.2.2. Model-Free Adaptive ANN-Based Control
2.3. Numerical Simulation Techniques
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Boudreau, M.A.; McMillan, G.K. New Directions in Bioprocess Modelling and Control: Maximizing Process Analytical Technology Benefits; ISA: Durham, NC, USA, 2006; ISBN 978-1-556-17905-1. [Google Scholar]
- Dochain, D. Bioprocess Control; ISTE: London, UK, 2008; ISBN 978-0-470-61112-8. [Google Scholar]
- Food and Drug Administration. Guidance for Industry. PAT—A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance. 2004. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/pat-framework-innovative-pharmaceutical-development-manufacturing-and-quality-assurance (accessed on 1 October 2019).
- Simutis, R.; Lübbert, A. Bioreactor control improves bioprocess performance. Biotechnol. J. 2015, 10, 1115–1130. [Google Scholar] [CrossRef] [PubMed]
- Galvanauskas, V.; Simutis, R.; Levišauskas, D.; Urniežius, R. Practical solutions for specific growth rate control systems in industrial bioreactors. Processes 2019, 7, 693. [Google Scholar] [CrossRef]
- Schuler, M.M.; Marison, I.W. Real-time monitoring and control of microbial bioprocesses with focus on the specific growth rate: Current state and perspectives. Appl. Microbiol. Biotechnol. 2012, 94, 1469–1482. [Google Scholar] [CrossRef] [PubMed]
- Rocha, I.; Veloso, A.; Carneiro, S.; Costa, R.; Ferreira, E. Implementation of a specific rate controller in a fed-batch E. coli fermentation. IFAC Proc. Vol. 2008, 41, 15565–15570. [Google Scholar] [CrossRef]
- Puertas, J.; Ruiz, J.; Vega, M.R.; Lorenzo, J.; Caminal, G.; González, G. Influence of specific growth rate over the secretory expression of recombinant potato carboxypeptidase inhibitor in fed-batch cultures of Escherichia coli. Process. Biochem. 2010, 45, 1334–1341. [Google Scholar] [CrossRef]
- Lim, H.C.; Shin, H.S. Fed-batch Cultures: Principles and Applications of Semi-Batch Bioreactors; Cambridge Series in Chemical Engineering; Cambridge University Press: Cambridge, UK, 2013; ISBN 978-0-521-51336-4. [Google Scholar] [CrossRef]
- Mears, L.; Stocks, S.M.; Sin, G.; Gernaey, K.V. A review of control strategies for manipulating the feed rate in fed-batch fermentation processes. J. Biotechnol. 2017, 245, 34–46. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.Y.; Park, S.; Middelberg, A.P.J. Control of fed-batch fermentations. Biotechnol. Adv. 1999, 17, 29–48. [Google Scholar] [CrossRef]
- Åkesson, M.; Hagander, P. A gain-scheduling approach for control of dissolved oxygen in stirred bioreactors. IFAC Proc. Vol. 1999, 32, 7608–7613. [Google Scholar] [CrossRef]
- Kuprijanov, A.; Gnoth, S.; Simutis, R.; Lübbert, A. Advanced control of dissolved oxygen concentration in fed batch cultures during recombinant protein production. Appl. Microbiol. Biotechnol. 2009, 82, 221–229. [Google Scholar] [CrossRef]
- Gnoth, S.; Kuprijanov, A.; Simutis, R.; Lübbert, A. Simple adaptive pH control in bioreactors using gain-scheduling methods. Appl. Microbiol. Biotechnol. 2009, 85, 955–964. [Google Scholar] [CrossRef]
- Levišauskas, D. An algorithm for adaptive control of dissolved oxygen concentration in batch culture. Biotechnol. Tech. 1995, 9, 85–90. [Google Scholar] [CrossRef]
- Levišauskas, D.; Simutis, R.; Galvanauskas, V. Adaptive set-point control system for microbial cultivation processes. Nonlinear Anal. Model. Control. 2016, 21, 153–165. [Google Scholar] [CrossRef] [Green Version]
- Babuška, R.; Damen, M.R.; Hellinga, C.; Maarleveld, H. Intelligent adaptive control of bioreactors. J. Intell. Manuf. 2003, 14, 255–265. [Google Scholar] [CrossRef]
- Smets, I.Y.; Claes, J.E.; November, E.J.; Bastin, G.P.; Impe, J.F. Optimal adaptive control of (bio) chemical reactors: Past, present and future. J. Process. Contr. 2004, 14, 795–805. [Google Scholar] [CrossRef]
- Bastin, G.; Impe, J.F. Nonlinear and adaptive control in biotechnology: A tutorial. Eur. J. Contr. 1995, 1, 37–53. [Google Scholar] [CrossRef]
- Ginkel, S.Z.; Dooley, T.P.; Suling, W.J.; Barrow, W.W. Identification and cloning of the Mycobacterium avium folA gene, required for dihydrofolate reductase activity. FEMS Microbiol. Lett. 2006, 156, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Levišauskas, D.; Galvanauskas, V.; Henrich, S.; Wilhelm, K.; Volk, N.; Lübbert, A. Model-based optimization of viral capsid protein production in fed-batch culture of recombinant Escherichia coli. Bioproc. Biosyst. Eng. 2003, 25, 255–262. [Google Scholar] [CrossRef]
- Galvanauskas, V.; Volk, N.; Simutis, R.; Lübbert, A. Design of recombinant protein production processes. Chem. Eng. Comm. 2004, 191, 732–748. [Google Scholar] [CrossRef]
- Gasser, B.; Saloheimo, M.; Rinas, U.; Dragosits, M.; Rodríguez-Carmona, E.; Baumann, K.; Villaverde, A. Protein folding and conformational stress in microbial cells producing recombinant proteins: A host comparative overview. Microb. Cell. Fact. 2008, 7, 11. [Google Scholar] [CrossRef]
- Gnoth, S.; Jenzsch, M.; Simutis, R.; Lübbert, A. Product formation kinetics in genetically modified E. coli bacteria: Inclusion body formation. Bioproc. Biosyst. Eng. 2007, 31, 41–46. [Google Scholar] [CrossRef]
- Gnoth, S.; Simutis, R.; Lübbert, A. Selective expression of the soluble product fraction in Escherichia coli cultures employed in recombinant protein production processes. Appl. Microbiol. Biotechnol. 2010, 87, 2047–2058. [Google Scholar] [CrossRef] [PubMed]
- Luedeking, R.; Piret, E.L. A kinetic study of the lactic acid fermentation. Batch process at controlled pH. Biotechnol. Bioeng. 2000, 67, 636–644. [Google Scholar] [CrossRef]
- Pirt, S.J. Principles of Microbe and Cell Cultivation; Blackwell Scientific Publications: Oxford, UK, 1985; ISBN 978-0-632-01455-2. [Google Scholar]
- Rivera, D.E.; Morari, M.; Skogestad, S. Internal model control: PID controller design. Ind. Eng. Chem. Process. Des. Dev. 1986, 25, 252–265. [Google Scholar] [CrossRef]
- Simutis, R.; Lübbert, A. Hybrid approach to state estimation for bioprocess control. Bioengineering 2017, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Urniezius, R.; Survyla, A.; Paulauskas, D.; Bumelis, V.A.; Galvanauskas, V. Generic estimator of biomass concentration for Escherichia coli and Saccharomyces cerevisiae fed-batch cultures based on cumulative oxygen consumption rate. Microb. Cell Factories 2019, in press. [Google Scholar] [CrossRef]
- Cheng, G.S. Model Free Adaptive control with CYBOCON. In Techniques for Adaptive Control; VanDoren, V.J., Ed.; Butterworth-Heinemann: Amsterdam, The Netherlands, 2003; pp. 145–202. [Google Scholar]
- Press, W.H.; Vetterling, W.T. Numerical Recipes; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Fogel, D.B. Evolutionary Computation toward a New Philosophy of Machine Intelligence; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]

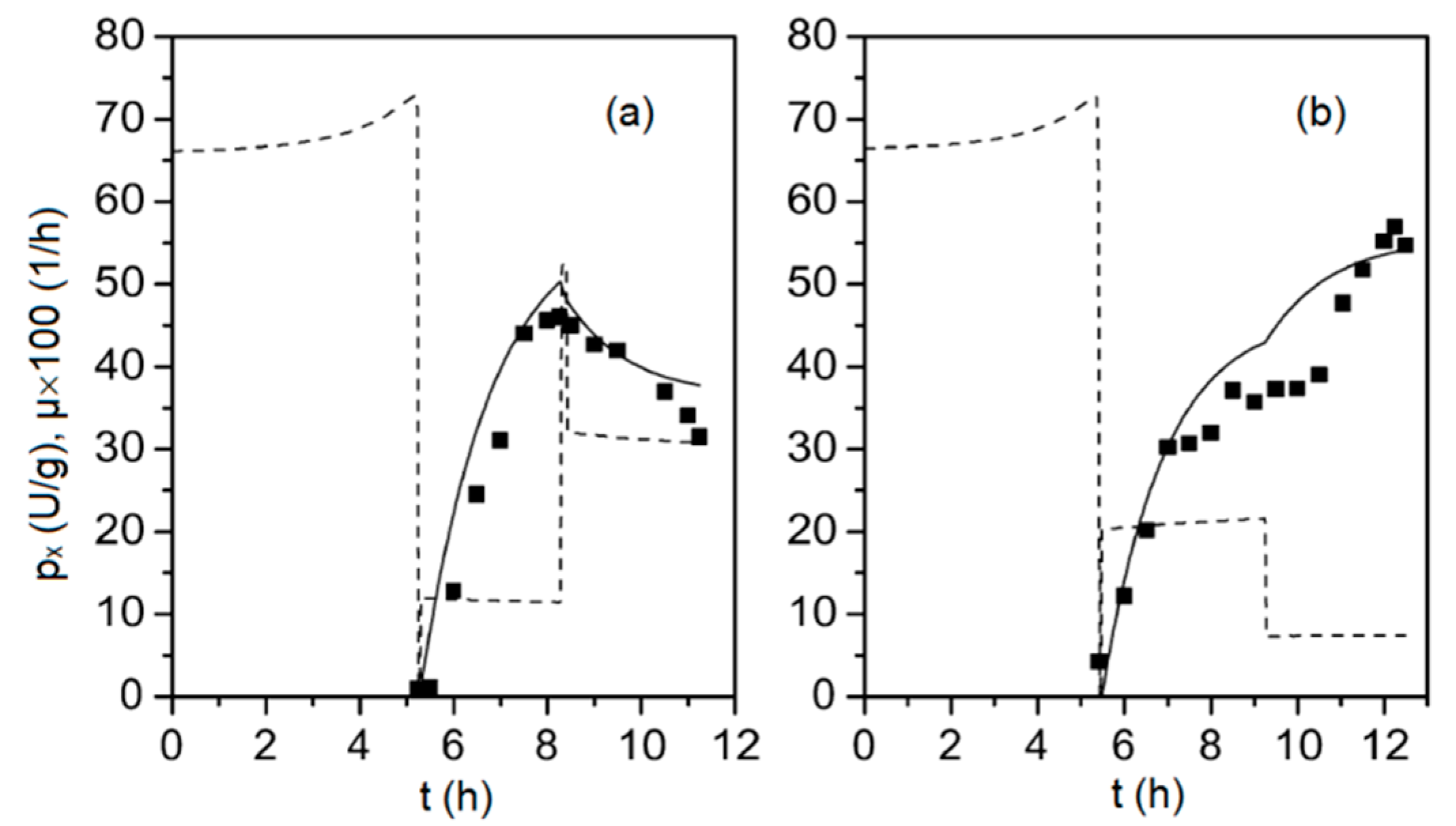


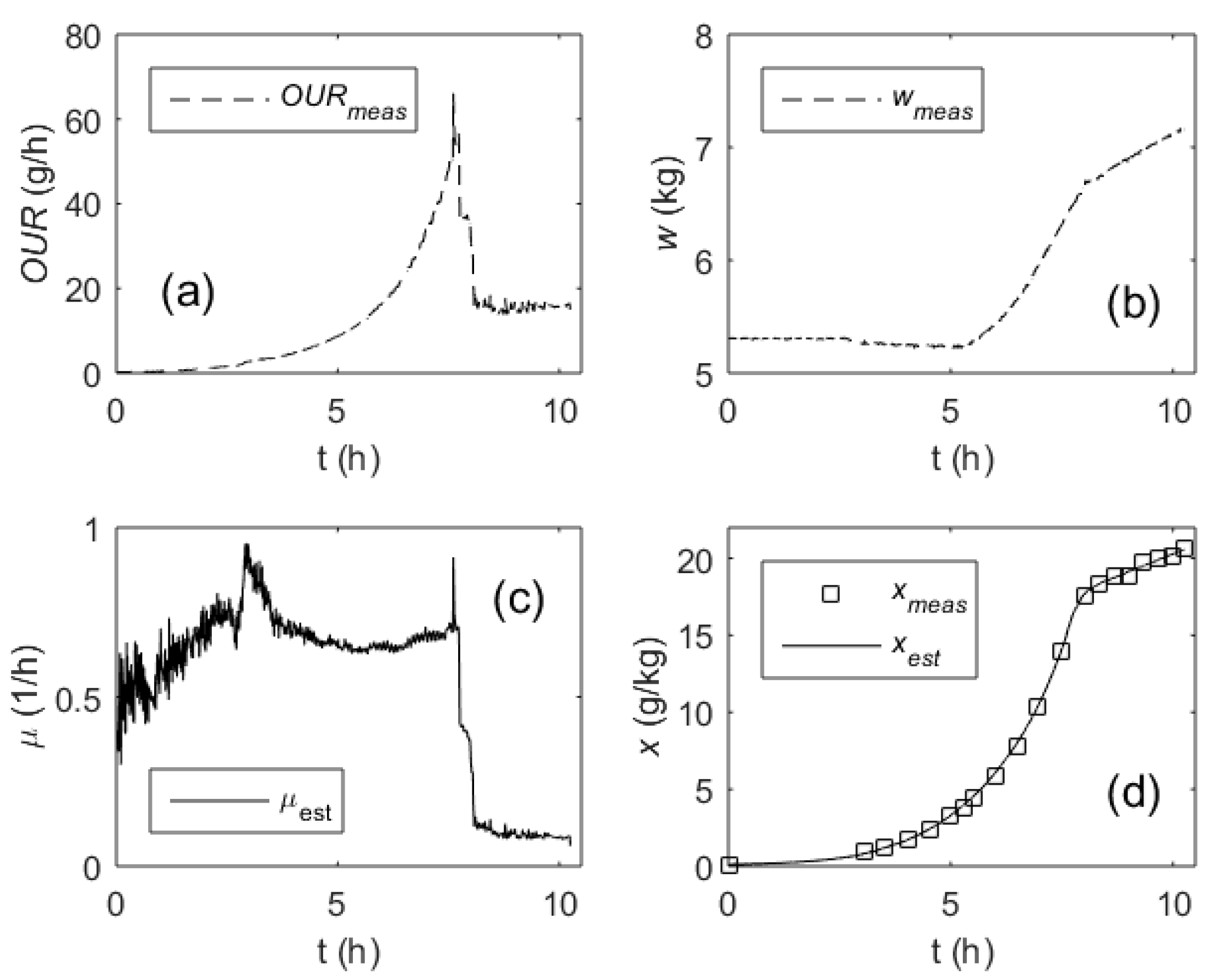







| Model Parameter | Value | Units |
|---|---|---|
| Ki | 93.8 ± 12.7 | g/kg |
| Kiµ | 0.0174 ± 0.0012 | 1/h |
| Km | 751 ± 27 | U/g |
| Ks | 0.01 ± 0.005 | g/kg |
| Kµ | 0.61 ± 0.03 | 1/h |
| m | 0.0242 ± 0.004 | g/(gh) |
| mox | 0.05 ± 0.0025 | g/(gh) |
| sf | 151 | g/kg |
| Tpx | 1.5 ± 0.1 | h |
| Tref | 37 | °C |
| Yox | 0.7 ± 0.01 | g/g |
| Yxs | 0.46 ± 0.01 | g/g |
| α | 0.0495 ± 0.0025 | 1/°C |
| µmax | 0.737 ± 0.01 | 1/h |
| Model Parameter | Value | Units |
|---|---|---|
| KC | 0.4 | kg |
| η | 1.5 | – |
| κ0 | 0.14 | kg |
| κ1 | 0.008 | g/kg |
| κ2 | 0.8 | g/(kg h) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galvanauskas, V.; Simutis, R.; Vaitkus, V. Adaptive Control of Biomass Specific Growth Rate in Fed-Batch Biotechnological Processes. A Comparative Study. Processes 2019, 7, 810. https://doi.org/10.3390/pr7110810
Galvanauskas V, Simutis R, Vaitkus V. Adaptive Control of Biomass Specific Growth Rate in Fed-Batch Biotechnological Processes. A Comparative Study. Processes. 2019; 7(11):810. https://doi.org/10.3390/pr7110810
Chicago/Turabian StyleGalvanauskas, Vytautas, Rimvydas Simutis, and Vygandas Vaitkus. 2019. "Adaptive Control of Biomass Specific Growth Rate in Fed-Batch Biotechnological Processes. A Comparative Study" Processes 7, no. 11: 810. https://doi.org/10.3390/pr7110810
APA StyleGalvanauskas, V., Simutis, R., & Vaitkus, V. (2019). Adaptive Control of Biomass Specific Growth Rate in Fed-Batch Biotechnological Processes. A Comparative Study. Processes, 7(11), 810. https://doi.org/10.3390/pr7110810





