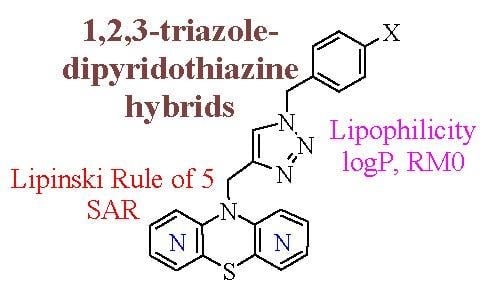
Evaluation of the Lipophilicity of New Anticancer 1,2,3-Triazole-Dipyridothiazine Hybrids Using RP TLC and Different Computational Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Chromatographic Procedure
2.3. Computational Programs
3. Results
4. Discussion
- the 2,7-diazaphenothiazine derivatives 1–5 RM0 = −57.811b − 0.0821 (r = 0.9936)
- the 3,6-diazaphenothiazine derivatives 6–10 RM0 = −63.632b − 0.327 (r = 0.9949)
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Arnott, J.A.; Planey, S.L. The influence of lipophilicity in drug discovery and design. Expert Opin. Drug Discov. 2012, 7, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Tsopelas, F.; Giaginis, C.; Tsantili, A. Lipophilicity and biomimetic properties to support drug discovery. Expert Opin. Drug Discov. 2017, 12, 885–896. [Google Scholar] [CrossRef]
- Waring, M.J. Lipophilicity in drug discovery. Expert Opin. Drug Discov. 2010, 5, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Lobo, S. Is there enough focus on lipophilicity in drug discovery? Expert Opin. Drug Discov. 2020, 15, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowska, M.; Komsta, Ł; Krzek, J.; Kokoszka, K. Lipophilicity study of eight cephalosporins by reversed-phase thin-layer chromatographic method. Biomed. Chromatogr. 2015, 29, 1759–1768. [Google Scholar] [CrossRef]
- Kulig, K.; Malawska, B. Estimation of the lipophilicity of antiarrhythmic and antihypertensive active 1-substituted pyrrolidin-2-one and pyrrolidine derivatives. Biomed. Chromatogr. 2003, 17, 318–324. [Google Scholar] [CrossRef]
- Dołowy, M.; Pyka, A. Evaluation of lipophilic properties of betamethasoneand related compounds. Acta Poloniae Pharm. Drug Res. 2015, 72, 671–681. [Google Scholar]
- Bajda, M.; Boryczka, S.; Wietrzyk, J.; Malawska, B. Investigation of the lipophilicity of anticancer-active tioquinoline derivatives. Biomed. Chromatogr. 2007, 21, 123–127. [Google Scholar] [CrossRef]
- Kadela-Tomanek, M.; Bober, K.; Bębenek, E.; Chrobak, E.; Boryczka, S. Application of thin-layer chromatography to evaluate the lipophilicity of 5,8-quinolinedione compounds. J. Planar Chromatogr. 2017, 30, 219–224. [Google Scholar] [CrossRef]
- Jeleń, M.; Pluta, K.; Morak-Młodawska, B. Lipophilicity estimation of anti-proliferative and anti-inflammatory 6-substituted 9-fluoroquino[3,2-b]benzo[1,4]thiazines. J. Liq. Chromatogr. Relat. Technol. 2019, 42, 563–569. [Google Scholar] [CrossRef]
- Bober, K.; Bębenek, E.; Boryczka, S. Application of TLC for Evaluation of the Lipophilicity of Newly Synthetized Esters: Betulin Derivatives. J. Anal. Methods Chem. 2019, 2019, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lipinski, C.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Chagas, C.M.; Moss, S.; Alisaraie, L. Drug metabolites and their effects on the development of adverse reactions: Revisiting Lipinski’s Rule of Five. Int. J. Pharm. 2018, 549, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Pluta, K.; Jeleń, M.; Morak-Młodawska, B.; Zimecki, M.; Artym, J.; Kocieba, M. Anticancer activity of newly synthesized azaphenothiazines from NCI’s anticancer screening bank. Pharmacol. Rep. 2010, 62, 319–332. Available online: http://www.if-pan.krakow.pl/pjp/pdf/2010/2_319.pdf (accessed on 8 February 2020). [CrossRef]
- Morak-Młodawska, B.; Pluta, K.; Matralis, A.N.; Kourounakis, A.P. Antioxidant Activity of Newly Synthesized 2,7-Diazaphenothiazines. Arch. Pharm. Chem. Life Sci. 2010, 343, 268–273. [Google Scholar] [CrossRef]
- Zimecki, M.; Artym, J.; Kocięba, M.; Pluta, K.; Morak-Młodawska, B.; Jeleń, M. Immunosupressive activities of newly synthesized azaphenothiazines in human and mouse models. Cell. Mol. Biol. Lett. 2009, 14, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Morak-Młodawska, B.; Pluta, K.; Zimecki, M.; Jeleń, M.; Artym, J.; Kocięba, M. Synthesis and selected immunological properties of 10-substituted 1,8-diazaphenothiazines. Med. Chem. Res. 2015, 24, 1408–1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Jeleń, M.; Kuśmierz, D. Synthesis, Anticancer Activity, and Apoptosis Induction of Novel 3,6-Diazaphenothiazines. Molecules 2019, 24, 267. [Google Scholar] [CrossRef] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Jelenń, M. Synthesis, spectroscopic characterization, and anticancer activity of new 10-substituted 1,6-diazaphenothiazines. Med. Chem. Res. 2016, 25, 2425–2433. [Google Scholar] [CrossRef] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Jeleń, M.; Kuśmierz, D. Synthesis and anticancer and lipophilic properties of 10-dialkylaminobutynyl derivatives of 1,8- and 2,7-diazaphenothiazines. J. Enzym. Inhib. Med. Chem. 2016, 31, 1132–1138. [Google Scholar] [CrossRef] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Suwińska, K.; Jeleń, M.; Kuśmierz, D. 3,6-Diazaphenothiazines as potential lead molecules–synthesis, characterization and anticancer activity. J. Enzyme Inhib. Med. Chem. 2016, 31, 1512–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Chen, M.; Wenzhi, Z.; Okechukwu, P.N.; Morak-Młodawska, B.; Pluta, K.; Jeleń, M.; Md Akim, A.; Ang, K.-P.; Ooi, K.K. 10H-3,6-Diazaphenothiazines Induce G2/M Phase Cell Cycle Arrest, Caspase-dependent Apoptosis and Inhibits Cell Invasion of A2780 Ovarian Carcinoma Cells through Regulation on NF-κB and [BIRC6-XIAP] Complexes. Drug Des. Develop. Ther. 2017, 11, 3045–3063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Jeleń, M.; Kuśmierz, D.; Suwinska, K.; Shkurenko, A.; Czuba, Z.; Jurzak, M. 10H-1,9-diazaphenothiazine and its 10-derivatives: Synthesis, characterisation and biological evaluation as potential anticancer agents. J. Enzyme Inhib. Med. Chem. 2019, 34, 1298–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Jeleń, M. Estimation of the Lipophilicity of New Anticancer and Immunosuppressive 1,8-Diazaphenothiazine Derivatives. J. Chromatogr. Sci. 2015, 53, 462–466. [Google Scholar] [CrossRef] [Green Version]
- Morak-Młodawska, B.; Pluta, K.; Jeleń, M. Lipophilicity of New Anticancer 1,6- and 3,6-diazaphenothiazines by of Use RP TLC and Different Computational Methods. J. Chromatogr. Sci. 2018, 56, 376–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeleń, M.; Pluta, K.; Morak-Młodawska, B. Determination of the lipophilicity parameters of new antiproliferative 8-10-substituted quinobenzothiazines by computational methods and RP TLC. J. Liq. Chromatogr. Relat. Technol. 2014, 37, 1373–1382. [Google Scholar] [CrossRef]
- Jeleń, M.; Pluta, K.; Morak-Młodawska, B. The Lipophilicity Parameters of New Antiproliferative 6,9-Disubstituted Quinobenzothiazines Determined by Computational Methods and RP TLC. J. Liq. Chromatogr. Relat. Technol. 2015, 38, 1577–1584. [Google Scholar] [CrossRef]
- Morak-Młodawska, B.; Pluta, K.; Latocha, M.; Jeleń, M.; Kusmierz, D. Design, Synthesis, and Structural Characterization of Novel Diazaphenothiazines with 1,2,3-Triazole Substituents as Promising Antiproliferative Agents. Molecules 2019, 24, 4388. [Google Scholar] [CrossRef] [Green Version]
- Bodor, N.; Gabanyi, Z.; Wong, C.K. A new method for the estimation of partition coefficient. J. Am. Chem. Soc. 1989, 111, 3783–3786. [Google Scholar] [CrossRef]
- Mannhold, R.; Cruciani, G.; Dross, K.; Rekker, R. Multivariate analysis of experimental and computational descriptors of molecular lipophilicity. J. Comput. Aided Mol. Des. 1998, 12, 573–581. [Google Scholar] [CrossRef]
- Tetko, I.V.; Tanchuk, V.Y. Application of Associative Neural Networks for Prediction of Lipophilicity in ALOGPS 2.1 Program. J. Chem. Inf. Comput. Sci. 2002, 42, 1136–1145. Available online: http://www.vcclab.org (accessed on 9 February 2020). [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. Available online: http://swissadme.ch (accessed on 9 February 2020). [CrossRef] [PubMed] [Green Version]
- ClogP (CS Chem 3D Ultra, Molecular Modeling and Analysis, version 7.0) distributed by CambridgeSoft.
- Available online: http://preadmet.bmdrc.org (accessed on 9 February 2020).
- Biaggi, G.L.; Barbaro, A.M.; Sapone, A. Determination of the lipophilicity by means of reversed-phase. I: Basic aspects and relationship between slope and intercept of TLC equitions. J. Chromatogr. A 1994, 662, 341–361. [Google Scholar]
- Kulkarni, A.; Han, Y.; Hopfinger, A.J. Predicting Caco-2 Cell Permeation Coefficients of Organic Molecules Using Membrane-Interaction QSAR Analysis. J. Chem. Inf. Comput. Sci. 2002, 42, 331–342. [Google Scholar] [CrossRef]
- Feher, M.; Schmidt, J.M. Property Distributions: Differences Between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. Chem. Inf. Comput. Sci. 2003, 34, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Irvine, J.D.; Takahashi, L.; Lockhart, K.; Cheong, J.; Tolan, J.W.; Selick, H.E.; Grove, J.R. MDCK (Madin-Darby Canine Kidney) Cells: A Tool for Membrane Permeability Screening. J. Pharm. Sci. 1999, 88, 28–33. [Google Scholar] [CrossRef]




| No | Alogps | AC_Logp | ALOGP | MLOGP | XLOGP2 | XLOGP3 | ILogP | XLogP | WlogP | MlogP | SILICOS-IT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.36 | 2.73 | 3.89 | 1.61 | 2.98 | 2.83 | 2.71 | 2.16 | 2.26 | 1.91 | 2.19 |
| 2 | 3.72 | 3.34 | 4.55 | 2.10 | 3.61 | 3.46 | 3.02 | 2.79 | 2.91 | 2.40 | 2.82 |
| 3 | 3.31 | 2.79 | 4.10 | 1.99 | 2.94 | 4.12 | 2.78 | 2.26 | 2.81 | 2.29 | 2.60 |
| 4 | 3.04 | 2.54 | 3.77 | 1.27 | 2.71 | 2.55 | 2.52 | 1.88 | 2.13 | 1.26 | 2.21 |
| 5 | 3.50 | 4.88 | 4.48 | 1.88 | 3.41 | 3.48 | 2.57 | 2.81 | 2.81 | 2.18 | 2.28 |
| 6 | 2.98 | 2.64 | 3.35 | 1.61 | 2.90 | 2.50 | 2.47 | 2.50 | 3.39 | 2.37 | 2.64 |
| 7 | 2.91 | 2.70 | 3.56 | 1.99 | 3.06 | 2.60 | 2.97 | 2.60 | 3.95 | 2.75 | 3.04 |
| 8 | 2.62 | 1.63 | 2.86 | 2.58 | 3.63 | 4.65 | 3.22 | 3.13 | 4.04 | 2.87 | 3.27 |
| 9 | 2.90 | 2.45 | 3.23 | 1.27 | 2.62 | 2.22 | 2.40 | 2.22 | 3.26 | 1.73 | 2.65 |
| 10 | 3.32 | 4.79 | 3.94 | 1.88 | 3.32 | 3.14 | 3.04 | 2.94 | 3.88 | 2.64 | 2.72 |
| No | −b | RM0 | r |
|---|---|---|---|
| 1 | 0.0227 | 1.229 | 0.9897 |
| 2 | 0.0198 | 1.150 | 0.9788 |
| 3 | 0.0215 | 1.155 | 0.9950 |
| 4 | 0.0253 | 1.274 | 0.9869 |
| 5 | 0.0426 | 2.407 | 0.9831 |
| 6 | 0.0416 | 2.217 | 0.9744 |
| 7 | 0.0341 | 1.867 | 0.9859 |
| 8 | 0.0492 | 2.823 | 0.9853 |
| 9 | 0.0261 | 1.332 | 0.9899 |
| 10 | 0.0417 | 2.388 | 0.9781 |
| No | Molecular Mass (M) | H-bond Acceptors | H-bond Donors | Rotatable Bonds | TPSA | Mol Refractivity (MR) |
|---|---|---|---|---|---|---|
| 1 | 372 | 4 | 0 | 4 | 85.03 | 108 |
| 2 | 390 | 4 | 0 | 4 | 85.03 | 109 |
| 3 | 406 | 4 | 0 | 4 | 85.03 | 113 |
| 4 | 397 | 5 | 0 | 4 | 108.8 | 114 |
| 5 | 404 | 4 | 0 | 5 | 110.3 | 116 |
| 6 | 372 | 4 | 0 | 4 | 85.03 | 108 |
| 7 | 390 | 4 | 0 | 4 | 85.03 | 109 |
| 8 | 406 | 4 | 0 | 4 | 85.03 | 113 |
| 9 | 397 | 5 | 0 | 4 | 108.8 | 114 |
| 10 | 404 | 4 | 0 | 5 | 110.3 | 116 |
| 11 | 286 | 2 | 0 | 4 | 44.6 | 86 |
| No | BBB | Caco2 | HIA | MDCK | PPB | SP |
|---|---|---|---|---|---|---|
| 1 | 0.547 | 24.769 | 98.110 | 94.808 | 88.062 | −3.742 |
| 2 | 0.283 | 26.146 | 98.558 | 3.203 | 73.476 | −4.184 |
| 3 | 0.982 | 50.735 | 97.663 | 34.206 | 89.739 | −3.795 |
| 4 | 0.196 | 22.382 | 99.752 | 30.308 | 87.208 | −3.682 |
| 5 | 0.273 | 25.465 | 99.026 | 4.763 | 94.100 | −3.491 |
| 6 | 0.836 | 27.476 | 98.110 | 74.714 | 91.502 | −3.508 |
| 7 | 1.061 | 29.803 | 98.099 | 12.723 | 91.532 | −3.881 |
| 8 | 1.439 | 51.402 | 97.663 | 33.067 | 93.582 | −3.634 |
| 9 | 0.224 | 23.546 | 99.752 | 25.266 | 89.391 | −3.517 |
| 10 | 0.406 | 27.037 | 99.026 | 4.277 | 98.907 | −3.320 |
| 11 | 3.103 | 22.684 | 97.476 | 18.983 | 75.453 | −3.100 |
| No | Molecular Descriptor or ADME Activities | Equation | r |
|---|---|---|---|
| 1–5 6–10 | M | RM0 = 69.511M2 − 240.09M + 579.21 RM0 = 30.107M2 − 118.14M + 501.3 | 0.4987 0.6546 |
| 1–5 6–10 | TPSA | RM0 = 17.351TPSA + 69.8 RM0 = −9.699TPSA + 115.45 | 0.6989 0.4051 |
| 1–5 6–10 | MR | RM0 = 4.235MR + 105.89 RM0 = 6.099M2 − 24.708M + 135.43 | 0.6761 0.5020 |
| 1–10 | BBB | BBB = 8.8792RM03 − 22.208RM02 + 16.225RM0 − 0.9889 | 0.8769 |
| 1–10 | Caco-2 | Caco-2 = 0.002RM03 − 0.2097RM02 + 6.9362RM0 − 71.729 | 0.6545 |
| 1–10 | HIA | HIA = −1.1492RM03 + 340.02RM02 − 335.35RM0 + 0.01 | 0.5377 |
| 1–10 | MDCK | MDCK = 0.0005RM03 + 0.0018RM02 − 0.0583RM0 + 2.1802 | 0.4144 |
| 1–10 | PPB | PPB = −0.0012RM03 + 0.3263RM02 − 28.461RM0 + 820.96 | 0.9145 |
| 1–10 | SP | SP = −1.5107RM03 – 15.909RM02 − 53.876RM0 − 56.38 | 0.5870 |
| No | −b | RM0 | r | logPTLC |
|---|---|---|---|---|
| I | 0.018 | 1.001 | 0.9979 | 1.21 (29) |
| II | 0.019 | 1.501 | 0.9974 | 1.58 (29) |
| III | 0.033 | 2.231 | 0.9960 | 2.43 (30) |
| IV | 0.034 | 2.886 | 0.9944 | 3.18 (29) |
| V | 0.044 | 3.488 | 0.9964 | 4.45 (29) |
| Compound | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| logPTLC | 1.408 | 1.330 | 1.335 | 1.452 | 2.569 | 2.382 | 2.037 | 2.979 | 1.509 | 2.551 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morak-Młodawska, B.; Pluta, K.; Jeleń, M. Evaluation of the Lipophilicity of New Anticancer 1,2,3-Triazole-Dipyridothiazine Hybrids Using RP TLC and Different Computational Methods. Processes 2020, 8, 858. https://doi.org/10.3390/pr8070858
Morak-Młodawska B, Pluta K, Jeleń M. Evaluation of the Lipophilicity of New Anticancer 1,2,3-Triazole-Dipyridothiazine Hybrids Using RP TLC and Different Computational Methods. Processes. 2020; 8(7):858. https://doi.org/10.3390/pr8070858
Chicago/Turabian StyleMorak-Młodawska, Beata, Krystian Pluta, and Małgorzata Jeleń. 2020. "Evaluation of the Lipophilicity of New Anticancer 1,2,3-Triazole-Dipyridothiazine Hybrids Using RP TLC and Different Computational Methods" Processes 8, no. 7: 858. https://doi.org/10.3390/pr8070858
APA StyleMorak-Młodawska, B., Pluta, K., & Jeleń, M. (2020). Evaluation of the Lipophilicity of New Anticancer 1,2,3-Triazole-Dipyridothiazine Hybrids Using RP TLC and Different Computational Methods. Processes, 8(7), 858. https://doi.org/10.3390/pr8070858