Gene Expression of Mouse Hippocampal Stem Cells Grown in a Galactose-Derived Molecular Gel Compared to In Vivo and Neurospheres
Abstract
:1. Introduction
2. Material and Methods
2.1. Mice
2.2. Dissection of Embryonic Hippocampus
2.3. Gender Detection of E19.5 Embryos
- DNA isolation
- b.
- Polymerase chain reaction (PCR)
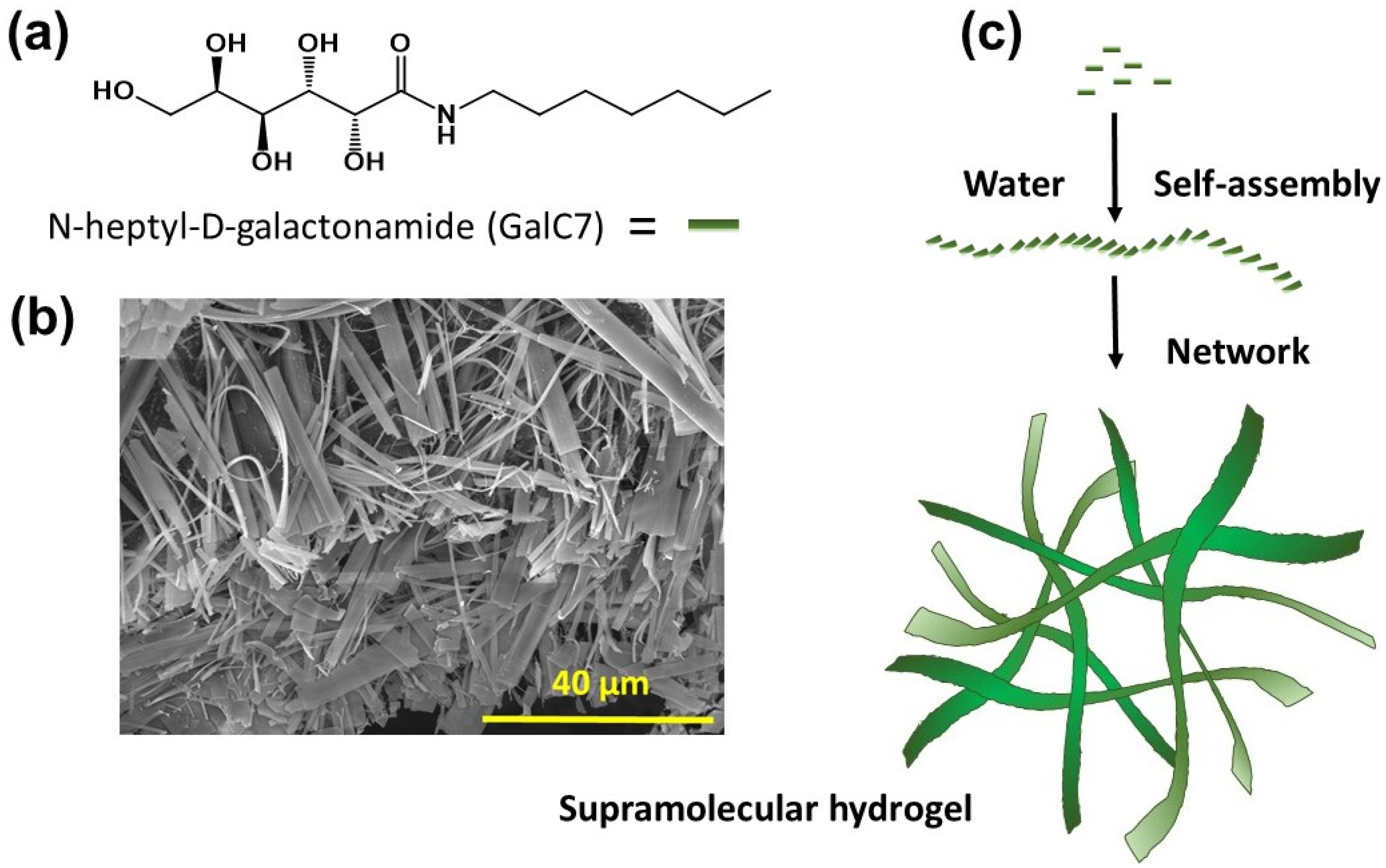
2.4. Preparation of the Hydrogel in Culture Plates
2.5. Primary Cell Culture
- Primary cell culture in culture plates coated with poly(HEMA)
- b.
- Primary cell culture with GalC7 gel
- c.
- Cell culture of neurospheres with the GalC7 gel
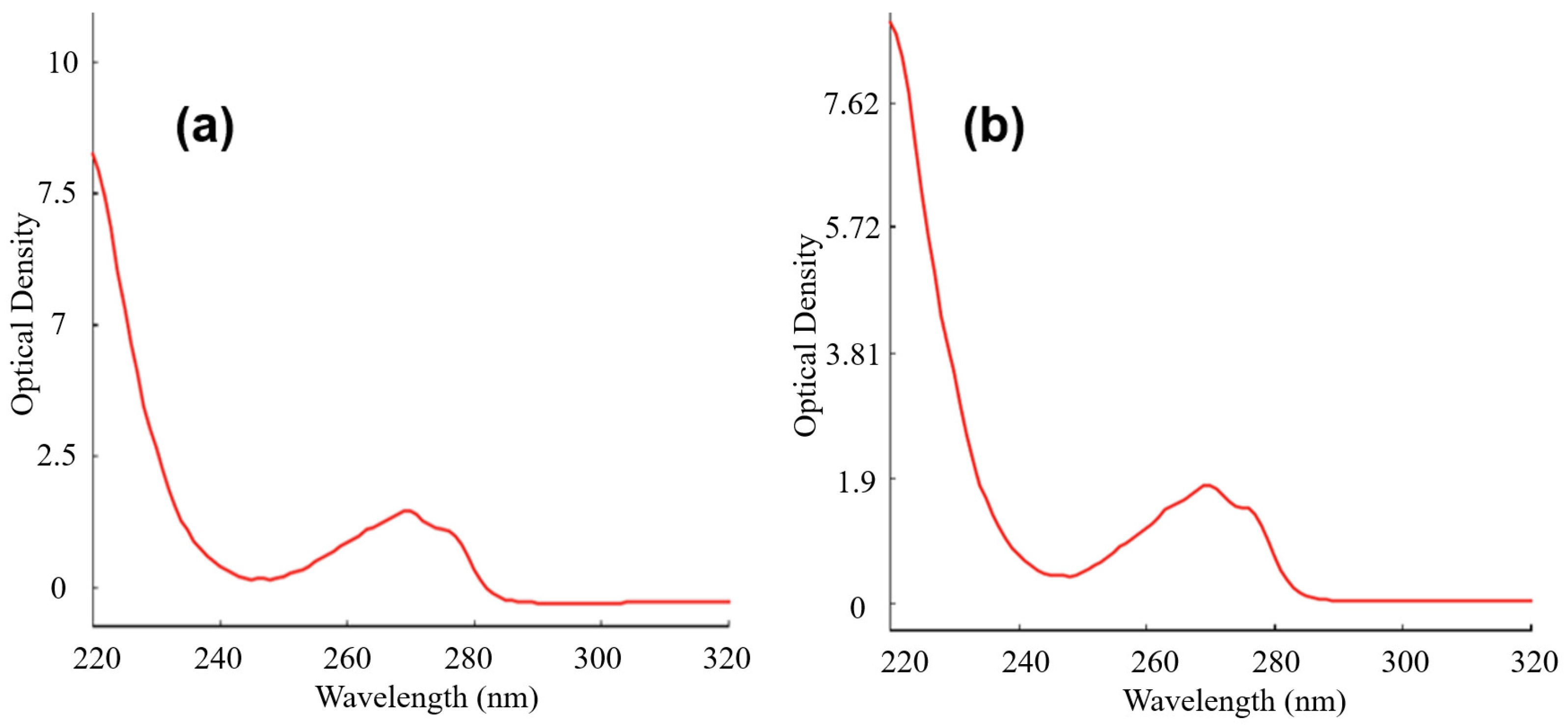
2.6. RNA Isolation
- Isolation of RNA from E19.5 embryonic hippocampi
- b.
- Isolation of RNA from primary cells cultured with pHEMA
- c.
- Isolation of RNA from primary cells cultured with GalC7 gel
2.7. Complementary DNA (cDNA) Synthesis
2.8. Preamplification Method
2.9. Measurement of mRNA Expression Levels by qPCR
3. Results and Discussion
3.1. Culture and Conditions
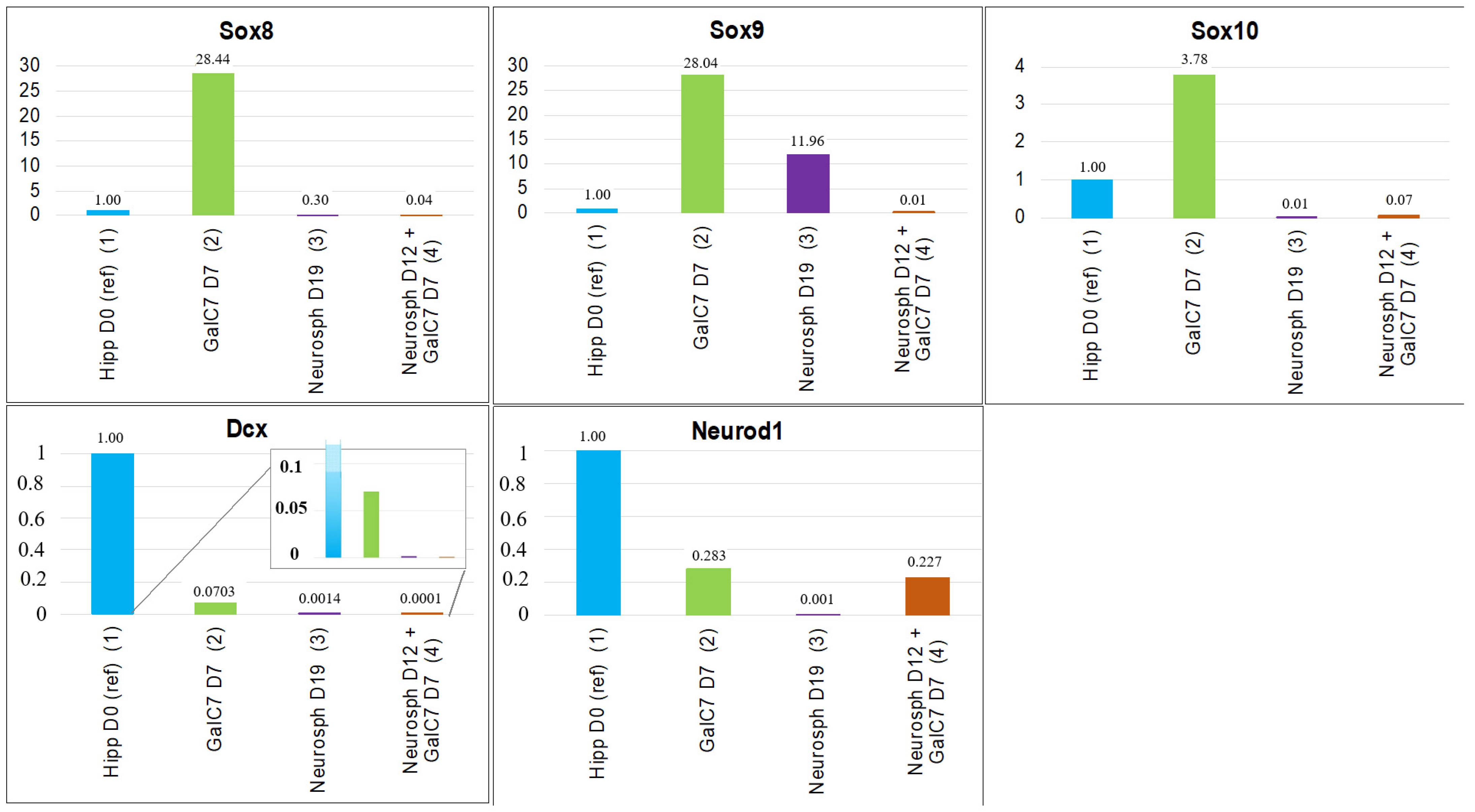
3.2. Gene Expression Studies with qPCR
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | Central nervous system |
| Dcx | Doublecortin |
| DMEM/F12 | Dulbecco’s modified Eagle medium/nutrient mixture F-12 |
| ECM | Extracellular matrix |
| GalC | Galactocerebrosides |
| GalC7 | N-heptyl-D-galactonamide |
| HBS | Hanks’ balanced salt |
| hNSC | Human neural stem cell |
| ITS | Insulin-Transferrin-Selenium-A |
| NFW | Nuclease-free water |
| PBS | Phosphate-buffered saline |
| pHEMA | Poly(hydroxyethylmethacrylate) |
| SAFIN | Self-assembling fibrillar network |
| SDS | Sodium dodecyl sulfate |
| Sox10 | SRY-box 10 |
| Sox8 | SRY-box 8 |
| Sox9 | SRY-box 9 |
| SRY | Sex Determining Region Y |
References
- Zhuang, P.; Sun, A.X.; An, J.; Chua, C.K.; Chew, S.Y. 3D neural tissue models: From spheroids to bioprinting. Biomaterials 2018, 154, 113–133. [Google Scholar] [CrossRef] [PubMed]
- Mahumane, G.D.; Kumar, P.; du Toit, L.C.; Choonara, Y.E.; Pillay, V. 3D scaffolds for brain tissue regeneration: Architectural challenges. Biomater. Sci. 2018, 6, 2812–2837. [Google Scholar] [CrossRef] [PubMed]
- Centeno, E.G.; Cimarosti, H.; Bithell, A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 2018, 13, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.J.; Do, J.T. Neural lineage differentiation from pluripotent stem cells to mimic human brain tissues. Front. Bioeng. Biotechnol. 2019, 7, 400. [Google Scholar] [CrossRef] [Green Version]
- Marton, R.M.; Miura, Y.; Sloan, S.A.; Li, Q.; Revah, O.; Levy, R.J.; Huguenard, J.R.; Pașca, S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019, 22, 484–491. [Google Scholar] [CrossRef]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Oliveira, B.; Yahya, A.Ç.; Novarino, G. Modeling cell-cell interactions in the brain using cerebral organoids. Brain Res. 2019, 1724, 146458. [Google Scholar] [CrossRef]
- Sood, D.; Cairns, D.M.; Dabbi, J.M.; Ramakrishnan, C.; Deisseroth, K.; Black, L.D.; Santaniello, S.; Kaplan, D.L. Functional maturation of human neural stem cells in a 3D bioengineered brain model enriched with fetal brain-derived matrix. Sci. Rep. 2019, 9, 1–15. [Google Scholar]
- Zychowicz, M.; Pietrucha, K.; Podobinska, M.; Kowalska-Wlodarczyk, M.; Lenart, J.; Augustyniak, J.; Buzanska, L. The collagen scaffold supports hiPSC-derived NSC growth and restricts hiPSC. Front. Biosci. (Sch. Ed.) 2019, 11, 105–121. [Google Scholar]
- Salaris, F.; Colosi, C.; Brighi, C.; Soloperto, A.; de Turris, V.; Benedetti, M.C.; Ghirga, S.; Rosito, M.; Di Angelantonio, S.; Rosa, A. 3D bioprinted human cortical neural constructs derived from induced pluripotent stem cells. J. Clin. Med. 2019, 8, 1595. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves-Pimentel, C.; Moreno, G.M.M.; Trindade, B.S.; Isaac, A.R.; Rodrigues, C.G.; Savariradjane, M.; de Albuquerque, A.V.; de Andrade Aguiar, J.L.; da Silveira Andrade-da, B.L. Cellulose exopolysaccharide from sugarcane molasses as a suitable substrate for 2D and 3D neuron and astrocyte primary cultures. J. Mater. Sci. Mater. Med. 2018, 29, 1–12. [Google Scholar] [CrossRef]
- Moxon, S.R.; Corbett, N.J.; Fisher, K.; Potjewyd, G.; Domingos, M.; Hooper, N.M. Blended alginate/collagen hydrogels promote neurogenesis and neuronal maturation. Mater. Sci. Eng. C 2019, 104, 109904. [Google Scholar] [CrossRef]
- Palazzolo, G.; Broguiere, N.; Cenciarelli, O.; Dermutz, H.; Zenobi-Wong, M. Ultrasoft alginate hydrogels support long-term three-dimensional functional neuronal networks. Tissue Eng. Part A 2015, 21, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Accardo, A.; Blatché, M.-C.; Courson, R.; Loubinoux, I.; Vieu, C.; Malaquin, L. Direct laser fabrication of free-standing PEGDA-hydrogel scaffolds for neuronal cell growth. Mater. Today 2018, 21, 315–316. [Google Scholar] [CrossRef]
- Haring, A.P.; Thompson, E.G.; Tong, Y.; Laheri, S.; Cesewski, E.; Sontheimer, H.; Johnson, B.N. Process-and bio-inspired hydrogels for 3D bioprinting of soft free-standing neural and glial tissues. Biofabrication 2019, 11, 025009. [Google Scholar] [CrossRef]
- Berns, E.J.; Sur, S.; Pan, L.; Goldberger, J.E.; Suresh, S.; Zhang, S.; Kessler, J.A.; Stupp, S.I. Aligned neurite outgrowth and directed cell migration in self-assembled monodomain gels. Biomaterials 2014, 35, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skilling, K.J.; Citossi, F.; Bradshaw, T.D.; Ashford, M.; Kellam, B.; Marlow, M. Insights into low molecular mass organic gelators: A focus on drug delivery and tissue engineering applications. Soft Matter 2014, 10, 237–256. [Google Scholar] [CrossRef]
- Du, X.; Zhou, J.; Shi, J.; Xu, B. Supramolecular hydrogelators and hydrogels: From soft matter to molecular biomaterials. Chem. Rev. 2015, 115, 13165–13307. [Google Scholar] [CrossRef] [PubMed]
- Chalard, A.; Vaysse, L.; Joseph, P.; Malaquin, L.; Souleille, S.; Lonetti, B.; Sol, J.-C.; Loubinoux, I.; Fitremann, J. Simple synthetic molecular hydrogels from self-assembling alkylgalactonamides as scaffold for 3D neuronal cell growth. ACS Appl. Mater. Interfaces 2018, 10, 17004–17017. [Google Scholar] [CrossRef] [PubMed]
- Latxague, L.; Ramin, M.A.; Appavoo, A.; Berto, P.; Maisani, M.; Ehret, C.; Chassande, O.; Barthélémy, P. Control of stem-cell behavior by fine tuning the supramolecular assemblies of low-molecular-weight gelators. Angew. Chem. 2015, 127, 4600–4604. [Google Scholar] [CrossRef]
- Moore, A.N.; Silva, T.L.L.; Carrejo, N.C.; Marmolejo, C.A.O.; Li, I.-C.; Hartgerink, J.D. Nanofibrous peptide hydrogel elicits angiogenesis and neurogenesis without drugs, proteins, or cells. Biomaterials 2018, 161, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Álvarez, Z.; Edelbrock, A.N.; Sato, K.; Stupp, S.I. Bioactive nanofibers induce neural transdifferentiation of human bone marrow mesenchymal stem cells. Acs Appl. Mater. Interfaces 2018, 10, 41046–41055. [Google Scholar] [CrossRef]
- Edelbrock, A.N.; Àlvarez, Z.; Simkin, D.; Fyrner, T.; Chin, S.M.; Sato, K.; Kiskinis, E.; Stupp, S.I. Supramolecular nanostructure activates TrkB receptor signaling of neuronal cells by mimicking brain-derived neurotrophic factor. Nano Lett. 2018, 18, 6237–6247. [Google Scholar] [CrossRef] [PubMed]
- Frick, C.; Müller, M.; Wank, U.; Tropitzsch, A.; Kramer, B.; Senn, P.; Rask-Andersen, H.; Wiesmüller, K.-H.; Löwenheim, H. Biofunctionalized peptide-based hydrogels provide permissive scaffolds to attract neurite outgrowth from spiral ganglion neurons. Colloids Surf. B Biointerfaces 2017, 149, 105–114. [Google Scholar] [CrossRef]
- Martin, A.D.; Chua, S.W.; Au, C.G.; Stefen, H.; Przybyla, M.; Lin, Y.; Bertz, J.; Thordarson, P.; Fath, T.; Ke, Y.D. Peptide nanofiber substrates for long-term culturing of primary neurons. ACS Appl. Mater. Interfaces 2018, 10, 25127–25134. [Google Scholar] [CrossRef]
- Kim, H.N.; Choi, N. Consideration of the Mechanical Properties of Hydrogels for Brain Tissue Engineering and Brain-on-a-chip. BioChip J. 2019, 13, 8–19. [Google Scholar] [CrossRef]
- Koser, D.E.; Thompson, A.J.; Foster, S.K.; Dwivedy, A.; Pillai, E.K.; Sheridan, G.K.; Svoboda, H.; Viana, M.; da F Costa, L.; Guck, J. Mechanosensing is critical for axon growth in the developing brain. Nat. Neurosci. 2016, 19, 1592–1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Yoshitomi, T.; Yoshimoto, K. Analysis of chirality effects on stem cell fate using three-dimensional fibrous peptide hydrogels. ACS Appl. Bio Mater. 2018, 1, 538–543. [Google Scholar] [CrossRef]
- Liu, G.F.; Zhang, D.; Feng, C.L. Control of three-dimensional cell adhesion by the chirality of nanofibers in hydrogels. Angew. Chem. Int. Ed. 2014, 53, 7789–7793. [Google Scholar] [CrossRef] [PubMed]
- Franze, K.; Gerdelmann, J.; Weick, M.; Betz, T.; Pawlizak, S.; Lakadamyali, M.; Bayer, J.; Rillich, K.; Gögler, M.; Lu, Y.-B. Neurite branch retraction is caused by a threshold-dependent mechanical impact. Biophys. J. 2009, 97, 1883–1890. [Google Scholar] [CrossRef] [Green Version]
- Chalard, A.; Mauduit, M.; Souleille, S.; Joseph, P.; Malaquin, L.; Fitremann, J. 3D printing of a biocompatible low molecular weight supramolecular hydrogel by dimethylsulfoxide water solvent exchange. Addit. Manuf. 2020, 33, 101162. [Google Scholar] [CrossRef]
- Chalard, A.; Joseph, P.; Souleille, S.; Lonetti, B.; Saffon-Merceron, N.; Loubinoux, I.; Vaysse, L.; Malaquin, L.; Fitremann, J. Wet spinning and radial self-assembly of a carbohydrate low molecular weight gelator into well organized hydrogel filaments. Nanoscale 2019, 11, 15043–15056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnescu, T.; Arter, J.; Reiprich, S.; Tamm, E.R.; Waisman, A.; Wegner, M. Sox8 and Sox10 jointly maintain myelin gene expression in oligodendrocytes. Glia 2018, 66, 279–294. [Google Scholar] [CrossRef]
- Singh, A.; Harada, S.; Mishina, Y. Downstream genes of Sox8 that would affect adult male fertility. Sex. Dev. 2009, 3, 16–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kordes, U.; Cheng, Y.-C.; Scotting, P.J. Sox group E gene expression distinguishes different types and maturational stages of glial cells in developing chick and mouse. Dev. Brain Res. 2005, 157, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Weider, M.; Wegner, M. SoxE factors: Transcriptional regulators of neural differentiation and nervous system development. In Proceedings of the Semin. Semin. Cell Dev. Biol. 2017, 63, 35–42. [Google Scholar] [CrossRef]
- Wittstatt, J.; Reiprich, S.; Küspert, M. Crazy little thing called Sox—new insights in oligodendroglial Sox protein function. Int. J. Mol. Sci. 2019, 20, 2713. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Cornwell, A.; Li, J.; Peng, S.; Osorio, M.J.; Aalling, N.; Wang, S.; Benraiss, A.; Lou, N.; Goldman, S.A. SOX9 is an astrocyte-specific nuclear marker in the adult brain outside the neurogenic regions. J. Neurosci. 2017, 37, 4493–4507. [Google Scholar] [CrossRef] [Green Version]
- Pompolo, S.; Harley, V. Localisation of the SRY-related HMG box protein, SOX9, in rodent brain. Brain Res. 2001, 906, 143–148. [Google Scholar] [CrossRef]
- Lovatt, D.; Sonnewald, U.; Waagepetersen, H.S.; Schousboe, A.; He, W.; Lin, J.H.-C.; Han, X.; Takano, T.; Wang, S.; Sim, F.J. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J. Neurosci. 2007, 27, 12255–12266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnabé-Heider, F.; Göritz, C.; Sabelström, H.; Takebayashi, H.; Pfrieger, F.W.; Meletis, K.; Frisén, J. Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell 2010, 7, 470–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molofsky, A.V.; Glasgow, S.M.; Chaboub, L.S.; Tsai, H.H.; Murnen, A.T.; Kelley, K.W.; Fancy, S.P.; Yuen, T.J.; Madireddy, L.; Baranzini, S. Expression profiling of Aldh1l1-precursors in the developing spinal cord reveals glial lineage-specific genes and direct Sox9-Nfe2l1 interactions. Glia 2013, 61, 1518–1532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Farmer, W.T.; Abrahamsson, T.; Chierzi, S.; Lui, C.; Zaelzer, C.; Jones, E.V.; Bally, B.P.; Chen, G.G.; Théroux, J.-F.; Peng, J. Neurons diversify astrocytes in the adult brain through sonic hedgehog signaling. Science 2016, 351, 849–854. [Google Scholar] [CrossRef]
- Nagao, M.; Ogata, T.; Sawada, Y.; Gotoh, Y. Zbtb20 promotes astrocytogenesis during neocortical development. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.; Li, G. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef] [Green Version]
- Azar, S.; Leventoux, N.; Ripoll, C.; Rigau, V.; Gozé, C.; Lorcy, F.; Bauchet, L.; Duffau, H.; Guichet, P.; Rothhut, B. Cellular and molecular characterization of IDH1-mutated diffuse low grade gliomas reveals tumor heterogeneity and absence of EGFR/PDGFRα activation. Glia 2018, 66, 239–255. [Google Scholar] [CrossRef]
- Souza, D.G.; Bellaver, B.; Terra, S.R.; Guma, F.C.R.; Souza, D.O.; Quincozes-Santos, A. In vitro adult astrocytes are derived from mature cells and reproduce in vivo redox profile. J. Cell. Biochem. 2017, 118, 3111–3118. [Google Scholar] [CrossRef]
- Stolt, C.C.; Lommes, P.; Friedrich, R.P.; Wegner, M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 2004, 131, 2349–2358. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.P.; Couillard-Després, S.; Cooper-Kuhn, C.M.; Winkler, J.; Aigner, L.; Kuhn, H.G. Transient expression of doublecortin during adult neurogenesis. J. Comp. Neurol. 2003, 467, 1–10. [Google Scholar] [CrossRef]
- Couillard-Despres, S.; Winner, B.; Schaubeck, S.; Aigner, R.; Vroemen, M.; Weidner, N.; Bogdahn, U.; Winkler, J.; Kuhn, H.G.; Aigner, L. Doublecortin expression levels in adult brain reflect neurogenesis. Eur. J. Neurosci. 2005, 21, 1–14. [Google Scholar] [CrossRef]
- Gleeson, J.G.; Lin, P.T.; Flanagan, L.A.; Walsh, C.A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron 1999, 23, 257–271. [Google Scholar] [CrossRef] [Green Version]
- Klempin, F.; Kronenberg, G.; Cheung, G.; Kettenmann, H.; Kempermann, G. Properties of doublecortin-(DCX)-expressing cells in the piriform cortex compared to the neurogenic dentate gyrus of adult mice. PLoS ONE 2011, 6, e25760. [Google Scholar] [CrossRef] [Green Version]
- Corbo, J.C.; Deuel, T.A.; Long, J.M.; LaPorte, P.; Tsai, E.; Wynshaw-Boris, A.; Walsh, C.A. Doublecortin is required in mice for lamination of the hippocampus but not the neocortex. J. Neurosci. 2002, 22, 7548–7557. [Google Scholar] [CrossRef]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef]
- Aprea, J.; Nonaka-Kinoshita, M.; Calegari, F. Generation and characterization of Neurod1-CreERT2 mouse lines for the study of embryonic and adult neurogenesis. Genesis 2014, 52, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.H.; Stein, G.H.; Lee, J.E. NeuroD: The predicted and the surprising. Mol. Cells 2004, 18, 271–288. [Google Scholar]
- Miyata, T.; Maeda, T.; Lee, J.E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999, 13, 1647–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roybon, L.; Hjalt, T.; Stott, S.; Guillemot, F.; Li, J.-Y.; Brundin, P. Neurogenin2 directs granule neuroblast production and amplification while NeuroD1 specifies neuronal fate during hippocampal neurogenesis. PLoS ONE 2009, 4, e4779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.K.; Cho, J.H.; Hwang, W.S.; Lee, Y.D.; Reu, D.S.; Suh-Kim, H. Expression of neuroD/BETA2 in mitotic and postmitotic neuronal cells during the development of nervous system. Dev. Dyn. 2000, 217, 361–367. [Google Scholar] [CrossRef]
- Liu, M.-H.; Li, W.; Zheng, J.-J.; Xu, Y.-G.; He, Q.; Chen, G. Differential neuronal reprogramming induced by NeuroD1 from astrocytes in grey matter versus white matter. Neural Regen. Res. 2020, 15, 342. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, Y.; Iwai, L.; Toda, T.; Lu, Q.R.; Kawasaki, H. Immunostaining for oligodendrocyte-specific galactosphingolipids in fixed brain sections using the cholesterol-selective detergent digitonin. J. Neurosci. Methods 2009, 178, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Marcus, J.; Popko, B. Galactolipids are molecular determinants of myelin development and axo–glial organization. Biochim. Biophys. Acta 2002, 1573, 406–413. [Google Scholar] [CrossRef]
- Ozgen, H.; Baron, W.; Hoekstra, D.; Kahya, N. Oligodendroglial membrane dynamics in relation to myelin biogenesis. Cell Mol. Life Sci. 2016, 73, 3291–3310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignoles, R.; Lentini, C.; d’Orange, M.; Heinrich, C. Direct lineage reprogramming for brain repair: Breakthroughs and challenges. Trends Mol. Med. 2019, 25, 897–914. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Primer Sequences (5′–3′) | Product Size (bp) | Primer Concentration (µM) |
|---|---|---|---|
| Sry | Forward: TGCACAATTGTCTAGAGAGC Reverse: ACTGCAGAAGGTTGTACAGT | 329 | 10 µM |
| Pax6 | Forward: CTTTCTCCAGAGCCTCAAT Reverse: GCAACAGGAAGGAGGGGGAGA | 150 | 10 µM |
| Reaction buffer (5X) | 4 µL |
| Template RNA (2.5 µg/20 µL) | * |
| NFW | ** |
| kept on ice for 5 min. | |
| Enzyme mixture | 2 µL |
| 5 min at 42 °C | |
| 5 min at 85 °C | |
| 15 min at 65 °C | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayram, K.K.; Fitremann, J.; Bayram, A.; Yılmaz, Z.; Mehmetbeyoğlu, E.; Özkul, Y.; Rassoulzadegan, M. Gene Expression of Mouse Hippocampal Stem Cells Grown in a Galactose-Derived Molecular Gel Compared to In Vivo and Neurospheres. Processes 2021, 9, 716. https://doi.org/10.3390/pr9040716
Bayram KK, Fitremann J, Bayram A, Yılmaz Z, Mehmetbeyoğlu E, Özkul Y, Rassoulzadegan M. Gene Expression of Mouse Hippocampal Stem Cells Grown in a Galactose-Derived Molecular Gel Compared to In Vivo and Neurospheres. Processes. 2021; 9(4):716. https://doi.org/10.3390/pr9040716
Chicago/Turabian StyleBayram, Keziban Korkmaz, Juliette Fitremann, Arslan Bayram, Zeynep Yılmaz, Ecmel Mehmetbeyoğlu, Yusuf Özkul, and Minoo Rassoulzadegan. 2021. "Gene Expression of Mouse Hippocampal Stem Cells Grown in a Galactose-Derived Molecular Gel Compared to In Vivo and Neurospheres" Processes 9, no. 4: 716. https://doi.org/10.3390/pr9040716






