Preparation, Antimicrobial Activity and Docking Study of Vanadium Mixed Ligand Complexes Containing 4-Amino-5-hydrazinyl-4H-1,2,4-triazole-3-thiol and Aminophenol Derivatives
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Physical Measurements
2.2. Elemental Analysis and Spectroscopy
2.3. Preparation of Mixed-Ligand Oxidovanadium (V) Complexes
2.4. Computational Study
2.4.1. Preparation of Ligands and Protein
2.4.2. Stepwise Docking Experiment
2.5. Assessment of Antimicrobial Capacity
3. Results
3.1. Synthesis of Oxidovanadium (V) Mixed Ligand Complexes
3.2. Molar Conductivity
3.3. IR Spectra
3.4. UV–Visible Spectra
3.5. 1H-NMR Spectra
3.6. Mass Spectra
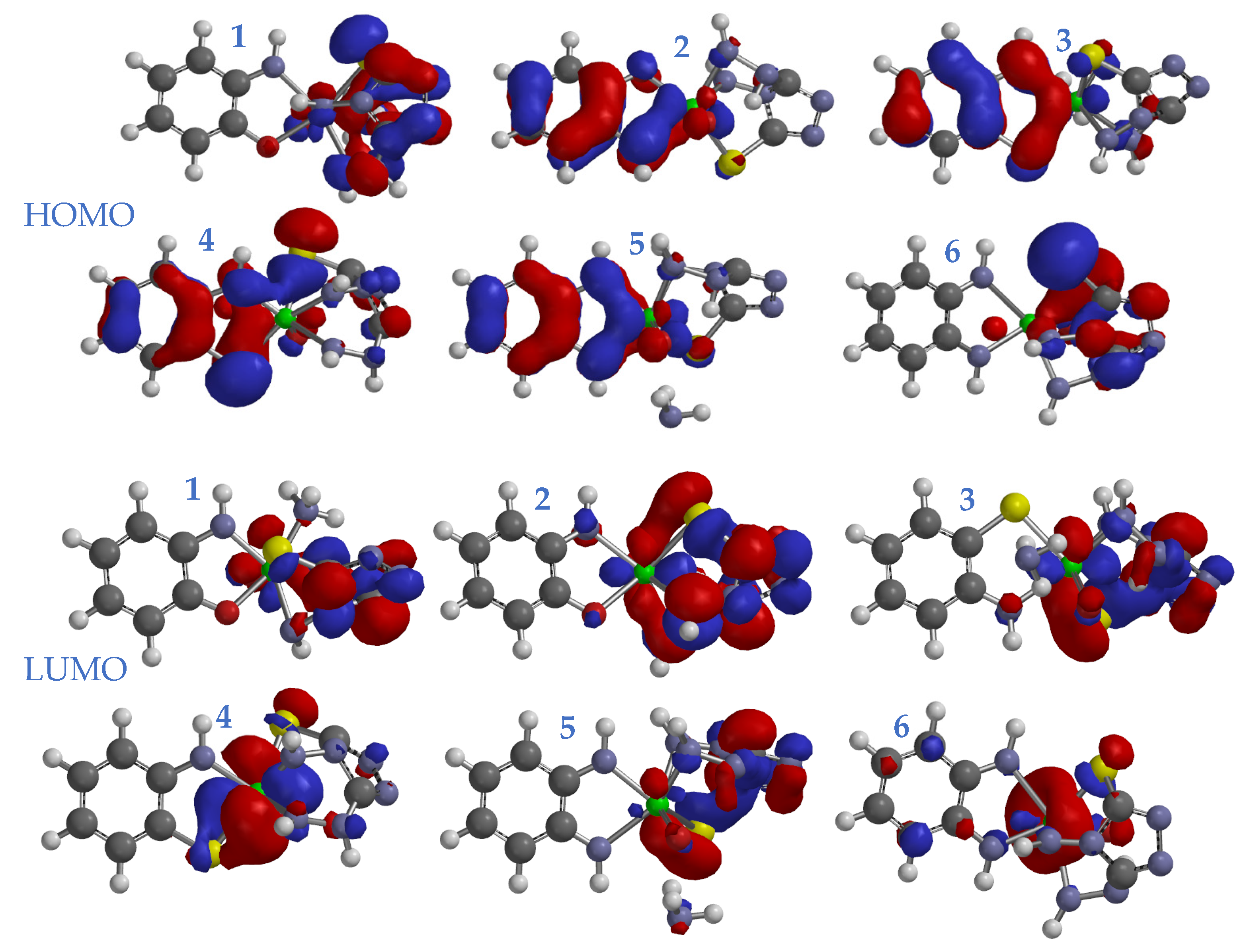
3.7. Molecular Modeling Studies
3.8. Docking Studies
3.9. Antimicrobial Tests
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alaraji, Y.H.; Shneine, J.K.; Ahmed, A.N.A.A. Synthesis, characterization, and antibacterial activity of new Schiff’s bases with 1,2,4-triazole moiety. J. Sci. 2015, 5, 293–299. [Google Scholar]
- Al-Khuzaie, M.G.; Al-Majidi, S.M. Synthesis, characterization and evaluation antimicrobial activity of some new substituted 2-mercapto-3-phenyl-4(3h)-quinazolinone. Iraqi J. Sci. 2014, 55, 582–593. [Google Scholar]
- Akhter, M.W.; Hassan, M.Z.; Amir, M. Synthesis and pharmacological evaluation of 3-diphenylmethyl-6-substituted-1,2,4-triazolo[3,4-b]-1,3,4-thiadiazoles: A condensed bridgehead nitrogen heterocyclic system. Arab. J. Chem. 2014, 7, 955–963. [Google Scholar] [CrossRef] [Green Version]
- Bekircan, O.; Menteşe, E.; Ülker, S.; Kucuk, C. Synthesis of some new 1,2,4-triazole derivatives starting from 3-(4-chlorophenyl)-5-(4-methoxybenzyl)-4H-1,2,4-triazol with anti-lipase and anti-urease activities. Arch. Pharm. 2014, 347, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.; Sumran, G. An insight on medicinal attributes of 1,2,4-triazoles. Eur. J. Med. Chem. 2020, 205. [Google Scholar] [CrossRef]
- Haasnoot, J.G. Mononuclear, oligonuclear and polynuclear metal coordination compounds with 1,2,4-triazole derivatives as ligands. Coord. Chem. Rev. 2000, 200, 131–185. [Google Scholar] [CrossRef]
- Fang, Z.Y.; Zhang, L.; Ma, J.P.; Zhao, L.; Wang, S.L.; Xie, N.H.; Liu, Y.Q.; Guo, X.Y.; Qin, J. Dinuclear cobalt and nickel complexes of a mercaptoacetic acid substituted 1,2,4-triazole ligand: Syntheses, structures and urease inhibitory studies. Acta Crystallogr. Sect. C Struct. Chem. 2019, 75, 1658–1665. [Google Scholar] [CrossRef]
- Althobiti, H.A.; Zabin, S.A. New Schiff bases of 2-(quinolin-8-yloxy)acetohydrazide and their Cu(ii), and Zn(ii) metal complexes: Their in vitro antimicrobial potentials and in silico physicochemical and pharmacokinetics properties. Open Chem. 2020, 18, 591–607. [Google Scholar] [CrossRef]
- Sutradhar, M.; Fernandes, A.R.; Silva, J.; Mahmudov, K.T.; Da Silva, M.F.C.G.; Pombeiro, A.J. Water soluble heterometallic potassium-dioxidovanadium (V) complexes as potential antiproliferative agents. J. Inorg. Biochem. 2016, 155, 17–25. [Google Scholar] [CrossRef]
- Levina, A.; Pires Vieira, A.; Wijetunga, A.; Kaur, R.; Koehn, J.T.; Crans, D.C.; Lay, P.A. A short-lived but highly cytotoxic vanadium (V) complex as a potential drug lead for brain cancer treatment by intratumoral injections. Angew. Chem. Int. Ed. 2020, 59, 15834–15838. [Google Scholar] [CrossRef]
- De Lima, L.M.; Belian, M.F.; Silva, W.E.; Postal, K.; Kostenkova, K.; Crans, D.C.; Rossiter, A.K.F.; da Silva Júnior, V.A. Vanadium (IV)-diamine complex with hypoglycemic activity and a reduction in testicular atrophy. J. Inorg. Biochem. 2021, 216. [Google Scholar] [CrossRef]
- Frank, P.; Carlson, R.M.K.; Carlson, E.J.; Hodgson, K.O. Medium-dependence of vanadium K-edge X-ray absorption spectra with application to blood cells from phlebobranch tunicates. Coord. Chem. Rev. 2003, 237, 31–39. [Google Scholar] [CrossRef]
- Munawar, K.S.; Ali, S.; Tahir, M.N.; Khalid, N.; Abbas, Q.; Qureshi, I.Z.; Shahzadi, S. Investigation of derivatized Schiff base ligands of 1,2,4-triazole amine and their oxovanadium (IV) complexes: Synthesis, structure, DNA binding, alkaline phosphatase inhibition, biological screening, and insulin mimetic properties. Russ. J. Gen. Chem. 2015, 85, 2183–2197. [Google Scholar] [CrossRef]
- Sutradhar, M.; Alegria, E.C.; Ferretti, F.; Raposo, L.R.; da Silva, M.F.C.G.; Baptista, P.V.; Fernandes, A.R.; Pombeiro, A.J. Antiproliferative activity of heterometallic sodium and potassiumdioxidovanadium (V) polymers. J. Inorg. Biochem. 2019, 200. [Google Scholar] [CrossRef]
- Biswas, N.; Patra, D.; Mondal, B.; Drew, M.G.B.; Ghosh, T. Synthesis of mixed-ligand complexes of VO2+ and VO3+ incorporating hydrazone, 1,10-phenanthroline and 8-hydroxyquinoline. J. Coord. Chem. 2016, 69, 318–329. [Google Scholar] [CrossRef]
- Sutradhar, M.; Pombeiro, A.J. Coordination chemistry of non-oxido, oxido and dioxidovanadium (IV/V) complexes with azine fragment ligands. Coord. Chem. Rev. 2014, 265, 89–124. [Google Scholar] [CrossRef]
- Benítez, J.; Guggeri, L.; Tomaz, I.; Arrambide, G.; Navarro, M.; Pessoa, J.C.; Garat, B.; Gambino, D. Design of vanadium mixed-ligand complexes as potential anti-protozoa agents. J. Inorg. Biochem. 2009, 103, 609–616. [Google Scholar] [CrossRef] [PubMed]
- Adeniyi, A.A.; Ajibade, P.A. Comparing the suitability of autodock, gold and glide for the docking and predicting the possible targets of Ru (II)-based complexes as anticancer agents. Molecules 2013, 18, 3760–3778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alikhani, M.; Hakimi, M.; Moeini, K.; Mashreghi, M.; Eigner, V.; Dusek, M. Spectral, structural, biological and molecular docking studies of a new mixed-valence V (IV)/V (V) ofloxacin complex. J. Mol. Struct. 2020, 1216. [Google Scholar] [CrossRef]
- Gómez, V.; Benet-Buchholz, J.; Martin, E.; Galán-Mascarós, J.R. Hysteretic spin crossover above room temperature and magnetic coupling in trinuclear transition-metal complexes with anionic 1,2,4-triazole ligands. Chem. A Eur. J. 2014, 20, 5369–5379. [Google Scholar] [CrossRef]
- Sumrra, S.H.; Anees, M.; Asif, A.; Zafar, M.N.; Mahmood, K.; Nazar, M.F.; Khalid, M.; Nadeem, M.A.; Khan, M.U. Synthesis, structural, spectral and biological evaluation of metals endowed 1,2,4-Triazole. Bull. Chem. Soc. Ethiop. 2020, 34, 335–351. [Google Scholar] [CrossRef]
- Sener, S.; Kul, I.; Bhat, K. Photochemical reactions of metal carbonyls [M(CO)6 (M = Cr, Mo, W), Mn(CO)3 Cp] with 4-amino-3-hydrazino-5-mercapto-1,2,4-triazole (AHMT) and 3-amino-5-mercapto-1,2,4-triazole (AMT). Synth. React. Inorg. Met. Nano-Met. Chem. 2015, 45, 495–501. [Google Scholar] [CrossRef]
- Sanoja, W.; Martínez, J.D.; Araujo, M.L.; Brito, F.; Hernández, L.; Del Carpio, E.; Lubes, V. Stability constants of mixed ligand complexes of vanadium (III) with 8-hydroxyquinoline and the amino acids glycine, proline, α-alanine and β-alanine. J. Mol. Liq. 2014, 197, 223–225. [Google Scholar] [CrossRef]
- Dales, G.E.; Kostrewa, D.; Gsell, B.; Stieger, M.; D’Arcy, A. Crystal engineering: Deletion mutagenesis of the 24 kDa fragment of the DNA gyrase B subunit from Staphylococcus aureus. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 1626–1629. [Google Scholar] [CrossRef]
- Gellibert, F.; Woolven, J.; Fouchet, M.-H.; Mathews, N.; Goodland, H.; Lovegrove, V.; Laroze, A.; Nguyen, V.-L.; Sautet, S.; Wang, R.; et al. Identification of 1,5-naphthyridine derivatives as a novel series of potent and selective TGF-β type I receptor inhibitors. J. Med. Chem. 2004, 47, 4494–4506. [Google Scholar] [CrossRef]
- Rana, R.; Sharma, R.; Kumar, A. Repurposing of fluvastatin against Candida albicans CYP450 lanosterol 14 α-demethylase, a target enzyme for antifungal therapy: An in silico and in vitro study. Curr. Mol. Med. 2019, 19, 506–524. [Google Scholar] [CrossRef]
- Jacob, K.S.; Ganguly, S.; Kumar, P.; Poddar, R.; Kumar, A. Homology model, molecular dynamics simulation and novel pyrazole analogs design of Candida albicans CYP450 lanosterol 14 α-demethylase, a target enzyme for antifungal therapy. J. Biomol. Struct. Dyn. 2017, 35, 1446–1463. [Google Scholar] [CrossRef]
- Kant, K.; Lal, U.R.; Kumar, A.; Ghosh, M. A merged molecular docking, ADME-T and dynamics approaches towards the genus of Arisaema as herpes simplex virus type 1 and type 2 inhibitors. Comput. Biol. Chem. 2019, 78, 217–226. [Google Scholar] [CrossRef]
- Navyashree, V.; Kant, K.; Kumar, A. Natural chemical entities from Arisaema genus might be a promising break-through against Japanese encephalitis virus infection: A molecular docking and dynamics approach. J. Biomol. Struct. Dyn. 2020, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Sharma, R.; Kumar, A. Comparative potential of simvastatin, rosuvastatin and fluvastatin against bacterial infection: An in silico and in vitro study. Orient. Pharm. Exp. Med. 2019, 19, 259–275. [Google Scholar] [CrossRef]
- Frisch, M.J.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Chemical Computing Group ULC. Molecular Operating Environment (MOE) 1010 Handbook; Chemical Computing Group ULC: Montreal, QC, Canada, 2017. [Google Scholar]
- Laurie, A.T.R.; Jackson, R.M. Q-SiteFinder: An energy-based method for the prediction of protein–ligand binding sites. Bioinformatics 2005, 21, 1908–1916. [Google Scholar] [CrossRef]
- Zabin, S.A.; Abdelbaset, M. Oxo/dioxo-vanadium (V) complexes with Schiff base ligands derived from 4-amino-5-mercapto-3-phenyl-1,2,4-triazole. Eur. J. Chem. 2016, 7, 322–328. [Google Scholar] [CrossRef]
- Yaul, A.; Pethe, G.; Deshmukh, R.; Aswar, A. Vanadium complexes with quadridentate Schiff bases: Synthesis, characterization, thermal and catalytic studies. J. Therm. Anal. Calorim. 2013, 113, 745–752. [Google Scholar] [CrossRef]
- Kareem, A.; Khan, M.S.; Nami, S.A.A.; Bhat, S.A.; Mirza, A.U.; Nishat, N. Curcumin derived Schiff base ligand and their transition metal complexes: Synthesis, spectral characterization, catalytic potential and biological activity. J. Mol. Struct. 2018, 1167, 261–273. [Google Scholar] [CrossRef]
- Singh, K.; Kumar, Y.; Puri, P.; Singh, G. Spectroscopic, thermal, and antimicrobial studies of Co(II), Ni(II), Cu(II), and Zn(II) complexes derived from bidentate ligands containing N and S donor atoms. Bioinorg. Chem. Appl. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, N.V.; Sathisha, M.P.; Budagumpi, S.; Kurdekar, G.S.; Revankar, V.K. Binuclear transition metal complexes of bicompartmental SNO donor ligands: Synthesis, characterization, and electrochemistry. J. Coord. Chem. 2010, 63, 1451–1461. [Google Scholar] [CrossRef]
- Abdelbaset, M.; Zabin, S. Vanadium (V) complexes containing 1,2,4-triazole moiety and their antimicrobial activity. Int. J. Adv. Res. 2016, 4, 1861–1871. [Google Scholar] [CrossRef]
- Barakat, A.S.; Gaballa, A.S.; Mohammed, S.F.; Teleb, S.M. Vibrational and thermal studies of the complexes (NH4)[VO(O2)2(phen)]·2H2O and (NH 4)[V(O2)3(phen)]·2H2O. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2005, 62, 814–818. [Google Scholar] [CrossRef] [PubMed]
- Alghamdi, I.A.; Abdelbaset, M.; El Mannoubi, I. Mixed ligand complexes of copper(II) and cobalt(II) with hydrazones derivatives and ortho-vanillin: Syntheses, characterizations and antimicrobial activity. Orient. J. Chem. 2019, 35, 1722–1730. [Google Scholar] [CrossRef]
- Thakur, S.; Sarkar, N.; Drew, M.G.B.; Bauzá, A.; Frontera, A.; Chattopadhyay, S. Estimating the energy of noncovalent interactions in a dioxovanadium (V) Schiff base complex: Exploration of its phenoxazinone synthase like activity. Polyhedron 2018, 142, 83–92. [Google Scholar] [CrossRef]
- Maurya, R.C.; Rajput, S. Neutral dioxovanadium (V) complexes of biomimetic hydrazones ONO donor ligands of bioinorganic and medicinal relevance: Synthesis via air oxidation of bis(acetylaceto-nato)oxovanadium (IV), characterization, biological activity and 3D molecular modeling. J. Mol. Struct. 2007, 833. [Google Scholar] [CrossRef]
- Zabin, S.A.; Abdelbaset, M. Vanadium (V) complexes containing 1,2,4-triazole moiety and their antimicrobial activity. Eur. J. Chem. 2016, 7, 322–328. [Google Scholar] [CrossRef]
- Singh, K.; Singh, D.P.; Singh Barwa, M.; Tyagi, P.; Mirza, Y. Some bivalent metal complexes of Schiff bases containing N and S donor atoms. J. Enzym. Inhib. Med. Chem. 2006, 21, 749–755. [Google Scholar] [CrossRef]
- Haddad, R.; Yousif, E.; Ahmed, A. Synthesis and characterization of transition metal complexes of 4-Amino-5-pyridyl-4H-1,2,4-triazole-3-thiol. Springerplus 2013, 2, 510. [Google Scholar] [CrossRef] [Green Version]
- Kumaran, J.S.; Priya, S.; Jayachandramani, N.; Mahalakshmi, S. Synthesis, spectroscopic characterization and biological activities of transition metal complexes derived from a tridentate schiff base. J. Chem. 2013. [Google Scholar] [CrossRef] [Green Version]
- Wildman, S.A.; Crippen, G.M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Al-Harbi, L.M.; Nassar, H.S.; Moustfa, A.; Alosaimi, A.M.; Mohamed, H.M.; Khowdiary, M.M.; El-Gazzar, M.A.; Elhenawy, A.A. Novel coumarin amino acid derivatives: Design, synthesis, docking, absorption, distribution, metabolism, elimination, toxicity (admet), quantitative structure–activity relationship (qsar) and anticancer studies. Mater. Express 2020, 10, 1375–1394. [Google Scholar] [CrossRef]
- Aldred, K.J.; McPherson, S.A.; Turnbough, C.L.; Kerns, R.J.; Osheroff, N. Topoisomerase IV-quinolone interactions are mediated through a water-metal ion bridge: Mechanistic basis of quinolone resistance. Nucleic Acids Res. 2013, 41, 4628–4639. [Google Scholar] [CrossRef]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- Corbeil, C.R.; Williams, C.I.; Labute, P. Variability in docking success rates due to dataset preparation. J. Comput. Mol. Des. 2012, 26, 775–786. [Google Scholar] [CrossRef] [Green Version]
- Amro, N.A.; Kotra, L.P.; Wadu-Mesthrige, K.; Bulychev, A.; Mobashery, S.; Liu, G.Y. High-resolution atomic force microscopy studies of the Escherichia coli outer membrane: Structural basis for permeability. Langmuir 2000, 16, 2789–2796. [Google Scholar] [CrossRef]
- Pellieux, C.; Dewilde, A.; Pierlot, C.; Aubry, J.M. Bactericidal and virucidal activities of singlet oxygen generated by thermolysis of naphthalene endoperoxides. Methods Enzymol. 2000, 319, 197–207. [Google Scholar] [CrossRef]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef]
- Mahmoud, W.H.; Deghadi, R.G.; Mohamed, G.G. Metal complexes of novel Schiff base derived from iron sandwiched organometallic and 4-nitro-1,2-phenylenediamine: Synthesis, characterization, DFT studies, antimicrobial activities and molecular docking. Appl. Organometal. Chem. 2018, 32, e4289. [Google Scholar] [CrossRef]
- Aziz, A.A.A.; Salem, A.N.M.; Sayed, M.A.; Aboaly, M.M. Synthesis, structural characterization, thermal studies, catalytic efficiency and antimicrobial activity of some M(II) complexes with ONO tridentate Schiff base N-salicylidene-o-aminophenol (saphH2). J. Mol. Struct. 2012, 1010, 130–138. [Google Scholar] [CrossRef]
- Bharti, S.; Choudhary, M.; Mohan, B.; Rawat, S.P.; Sharma, S.R.; Ahmad, K. Syntheses, spectroscopic characterization, SOD-like properties and antibacterial activities of dimer copper (II) and nickel (II) complexes based on imine ligands containing 2-aminothiophenol moiety: X-ray crystal structure determination of disulfide Schiff bases. J. Mol. Struct. 2018, 1164, 137–154. [Google Scholar]





| “Antibacterial Activity 200 μg/disc” | “Antifungal Activity” | ||||
|---|---|---|---|---|---|
| Ligand and Complexes | “Gram-Positive Bacteria” | Gram-Negative Bacteria | Yeast | ||
| S.a | S.p | E.c | P.a | C.a | |
| NH4[VO(L1)(L2)] 2.5H2O(1) | 0 | 20 | 0 | 0 | 12 |
| K[VO(L1)(L2)] 1.5H2O (2) | 12 | 23 | 0 | 0 | 17 |
| NH4[VO(L1)(L3)] 4H2O(3) | 12 | 26 | 7 | 6 | 11 |
| K[VO(L1)(L3)] H2O (4) | 16 | 21 | 0 | 0 | 14 |
| NH4[VO(L1)(L4)] H2O (5) | 0 | 24 | 0 | 0 | 13 |
| K[VO(L1)(L4)]2 H2O (6) | 15 | 21 | 9 | 7 | 18 |
| Amoxicillin | 28 | 35 | 21 | 23 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domyati, D.; Zabin, S.A.; Elhenawy, A.A.; Abdelbaset, M. Preparation, Antimicrobial Activity and Docking Study of Vanadium Mixed Ligand Complexes Containing 4-Amino-5-hydrazinyl-4H-1,2,4-triazole-3-thiol and Aminophenol Derivatives. Processes 2021, 9, 1008. https://doi.org/10.3390/pr9061008
Domyati D, Zabin SA, Elhenawy AA, Abdelbaset M. Preparation, Antimicrobial Activity and Docking Study of Vanadium Mixed Ligand Complexes Containing 4-Amino-5-hydrazinyl-4H-1,2,4-triazole-3-thiol and Aminophenol Derivatives. Processes. 2021; 9(6):1008. https://doi.org/10.3390/pr9061008
Chicago/Turabian StyleDomyati, Doaa, Sami A. Zabin, Ahmed A. Elhenawy, and Mohamed Abdelbaset. 2021. "Preparation, Antimicrobial Activity and Docking Study of Vanadium Mixed Ligand Complexes Containing 4-Amino-5-hydrazinyl-4H-1,2,4-triazole-3-thiol and Aminophenol Derivatives" Processes 9, no. 6: 1008. https://doi.org/10.3390/pr9061008
APA StyleDomyati, D., Zabin, S. A., Elhenawy, A. A., & Abdelbaset, M. (2021). Preparation, Antimicrobial Activity and Docking Study of Vanadium Mixed Ligand Complexes Containing 4-Amino-5-hydrazinyl-4H-1,2,4-triazole-3-thiol and Aminophenol Derivatives. Processes, 9(6), 1008. https://doi.org/10.3390/pr9061008






