ACE Inhibitory Peptides from Bellamya bengalensis Protein Hydrolysates: In Vitro and In Silico Molecular Assessment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Materials
2.2. Preparation of B. bengalensis Protein Concentrates (BBPCs)
2.3. Preparation of B. bengalensis Protein Hydrolysates(BBPHs)
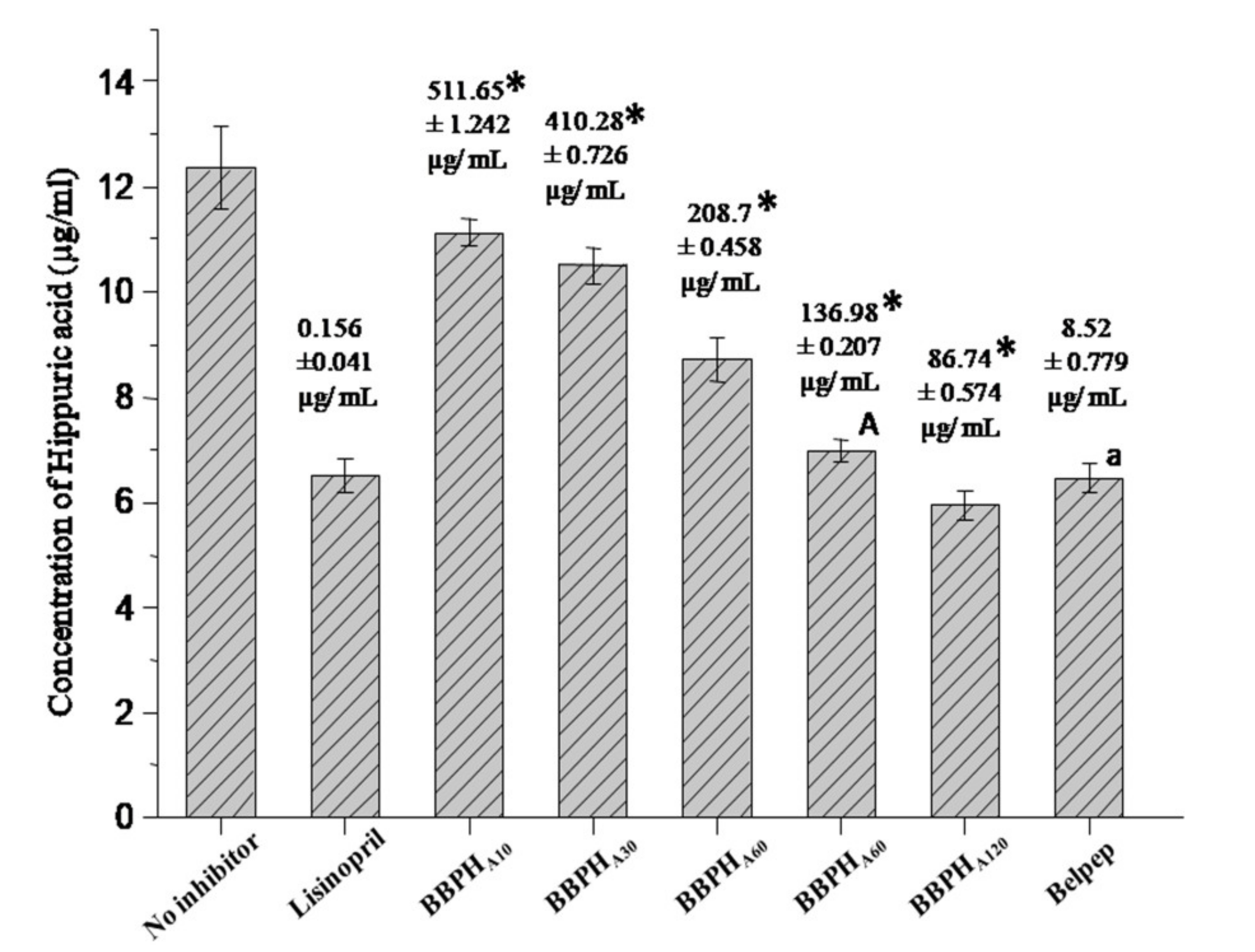
2.4. In Vitro ACE Inhibitory Assay
2.5. Identification and Preparation of ACE Inhibitory Peptides
2.6. Molecular Docking Studies
2.7. Determination of Kinetics of ACE Inhibitory Activity of the Peptide
2.8. Isothermal Titration Calorimetry (ITC)
2.9. Data Analysis
3. Results and Discussion
3.1. Degree of Hydrolysis of BBPCs and Their ACE Inhibitory Activities
3.2. Identification of the Peptide Sequence
3.3. Molecular Docking
3.4. Inhibitory Kinetics Study
3.5. Isothermal Titration Calorimetry
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwon, Y.I.; Apostolidis, E.; Kim, Y.C.; Shetty, K. Health benefits of traditional corn, beans, and pumpkin: In vitro studies for hyperglycemia and hypertension management. J. Med. Food 2007, 10, 266–275. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Chakraborty, M.; Bose, M.; Mukherjee, D.; Roychoudhury, A.; Dhar, P.; Mishra, R. Indian freshwater edible snail Bellamyabengalensis lipid extract prevents T cell mediated hypersensitivity and inhibits LPS induced macrophage activation. J. Ethnopharmacol. 2014, 157, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, A.K.; Roy, S.P. Ethno-medicinal uses of some Shell Fishes by people of Kosi River basin of North-Bihar, India. Stud. Ethno-Med. 2009, 3, 1–4. [Google Scholar] [CrossRef]
- Baby, R.L.; Hasan, I.; Kabir, K.A.; Naser, M.N. Nutrient analysis of some commercially important molluscs of Bangladesh. J. Sci. Res. 2010, 2, 390–396. [Google Scholar] [CrossRef] [Green Version]
- Hayes, M.; Mora, L. Alternative proteins as a source of bioactive peptides: The edible snail and generation of hydrolysates containing peptides with bioactive potential for use as functional foods. Foods 2021, 10, 276. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Mukherjee, A. Animal resources linked with the life of Birhor community settled in Ayodhya hills, Purulia District, West Bengal. Indian J. Appl. Pure Biol. 2012, 27, 31–36. [Google Scholar]
- Sajed Ali, S.; Acharyya, N.; Maiti, S. Promising anti-oxidative therapeutic potentials of edible freshwater snail Bellamyabengalensis extract against arsenic-induced rat hepatic tissue and DNA damage. Int. J. Aquat. Biol. 2016, 4, 239–255. [Google Scholar]
- Udenigwe, C.C.; Aluko, R.E. Food protein-derived bioactive peptides: Production, processing, and potential health benefits. J. Food Sci. 2012, 77, R11–R24. [Google Scholar] [CrossRef]
- Aluko, R.E. Antihypertensive peptides from food proteins. Annu. Rev. Food Sci. Technol. 2015, 6, 235–262. [Google Scholar] [CrossRef]
- Natesh, R.; Schwager, S.L.U.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Raghavan, S.; Kristinsson, H.G. ACE-inhibitory activity of tilapia protein hydrolysates. Food Chem. 2009, 117, 582–588. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Ye, R.; Wua, Y.; Xia, W. Comparison of analytical methods to assay inhibitors of angiotensin I-converting enzyme. Food Chem. 2013, 141, 3329–3334. [Google Scholar] [CrossRef]
- Ahn, C.B.; Jeon, Y.J.; Kim, Y.T.; Je, J.Y. Angiotensin i converting enzyme (ACE) inhibitory peptides from salmon byproduct protein hydrolysate by Alcalase hydrolysis. Process Biochem. 2012, 47, 2240–2245. [Google Scholar] [CrossRef]
- Mohammad, A.W.; Kumar, A.G.; Basha, R.K. Optimization of enzymatic hydrolysis of tilapia (Oreochromis spp.) scale gelatine. Int. Aquat. Res. 2015, 7, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, R.; Dey, T.K.; Ghosh, M.; Dhar, P. Enzymatic modification of sesame seed protein, sourced from waste resource for nutraceutical application. Food Bioprod. Process. 2015, 94, 70–81. [Google Scholar] [CrossRef]
- Li, G.H.; Liu, H.; Shi, Y.H.; Le, G.W. Direct spectrophotometric measurement of angiotensin I-converting enzyme inhibitory activity for screening bioactive peptides. J. Pharm. Biomed. Anal. 2005, 37, 219–224. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Sharma, M.; Nagpal, G.; Chauhan, J.S.; Singh, S.; Gautam, A.; Raghava, G.P.S. AHTPDB: A comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res. 2015, 43, D956–D962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeerschd, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero, L.; Castillo, J.; Quiñones, M.; Garcia-Vallvé, S.; Arola, L.; Pujadas, G.; Muguerza, B. Inhibition of angiotensin-converting enzyme activity by Flavonoids: Structure-Activity Relationship Studies. PLoS ONE 2012, 7, e49493. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Heo, L.; Lee, M.S.; Seok, C. GalaxyPepDock: A protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Res. 2015, 43, W431–W435. [Google Scholar] [CrossRef] [Green Version]
- Jimsheena, V.K.; Gowda, L.R. Arachin derived peptides as selective angiotensin I-converting enzyme (ACE) inhibitors: Structure-activity relationship. Peptides 2010, 31, 1165–1176. [Google Scholar] [CrossRef]
- Ni, H.; Li, L.; Liu, G.; Hu, S.Q. Inhibition mechanism and model of an angiotensin i-converting enzyme (ACE)-inhibitory hexapeptide from yeast (Saccharomyces cerevisiae). PLoS ONE 2012, 7, e37077. [Google Scholar] [CrossRef] [Green Version]
- Demirhan, E.; Apar, D.K.; Özbek, B. Sesame cake protein hydrolysis by alcalase: Effects of process parameters on hydrolysis, solubilisation, and enzyme inactivation. Korean J. Chem. Eng. 2011, 28, 195–202. [Google Scholar] [CrossRef]
- Bhaskar, N.; Benila, T.; Radha, C.; Lalitha, R.G. Optimization of enzymatic hydrolysis of visceral waste proteins of Catla (Catlacatla) for preparing protein hydrolysate using a commercial protease. Bioresour. Technol. 2008, 99, 335–343. [Google Scholar] [CrossRef]
- Wu, Q.; Jia, J.; Yan, H.; Du, J.; Gui, Z. A novel angiotensin-I converting enzyme (ACE) inhibitory peptide from gastrointestinal protease hydrolysate of silkworm pupa (Bombyx mori) protein: Biochemical characterization and molecular docking study. Peptides 2015, 68, 17–24. [Google Scholar] [CrossRef]
- Wu, Q.; Du, J.; Jia, J.; Kuang, C. Production of ACE inhibitory peptides from sweet sorghum grain protein using alcalase: Hydrolysis kinetic, purification and molecular docking study. Food Chem. 2016, 199, 140–149. [Google Scholar] [CrossRef]
- Forghani, B.; Zarei, M.; Ebrahimpour, A.; Philip, R.; Bakar, J.; Hamid, A.A.; Saari, N. Purification and characterization of angiotensin converting enzyme-inhibitory peptides derived from Stichopushorrens: Stability study against the ACE and inhibition kinetics. J. Funct. Foods 2016, 20, 276–290. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. The scientific evidence for the role of milk protein-derived bioactive peptides in humans: A Review. J. Funct. Foods 2015, 17, 640–656. [Google Scholar] [CrossRef] [Green Version]
- Sagardia, I.; Roa-Ureta, R.H.; Bald, C. A new QSAR model, for angiotensin I-converting enzyme inhibitory oligopeptides. Food Chem. 2013, 136, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Puchalska, P.; Marina Alegre, M.L.; García López, M.C. Isolation and characterization of peptides with antihypertensive activity in foodstuffs. Crit. Rev. Food Sci. Nutr. 2015, 55, 521–551. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Suzuki, K.; Funatsu, G. Isolation from α-zein of thermolysin peptides with angiotensin i-converting enzyme inhibitory activity. Biosci. Biotechnol. Biochem. 1996, 60, 661–663. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, J.; Škalič, M.; Martínez-Rosell, G.; De Fabritiis, G. KDEEP: Protein-ligand absolute binding affinity prediction via 3D-Convolutional neural networks. J. Chem. Inf. Model. 2018, 58, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Brew, K. Structure of human ACE gives new insights into inhibitor binding and design. Trends Pharmacol. Sci. 2003, 24, 391–394. [Google Scholar] [CrossRef]
- Jao, C.L.; Huang, S.L.; Hsu, K.C. Angiotensin I-converting enzyme inhibitory peptides: Inhibition mode, bioavailability, and antihypertensive effects. Biomedicine 2012, 2, 130–136. [Google Scholar] [CrossRef]
- Andújar-Sánchez, M.; Cámara-Artigas, A.; Jara-Pérez, V. A calorimetric study of the binding of lisinopril, enalaprilat and captopril to angiotensin-converting enzyme. Biophys. Chem. 2004, 111, 183–189. [Google Scholar] [CrossRef]
- Brautigam, C.A. Fitting two- and three-site binding models to isothermal titration calorimetric data. Methods 2015, 76, 124–136. [Google Scholar] [CrossRef] [Green Version]
- Freiburger, L.A.; Auclair, K.; Mittermaier, A.K. Elucidating protein binding mechanisms by variable-c ITC. ChemBioChem 2009, 10, 2871–2873. [Google Scholar] [CrossRef]
- Fuchs, S.; Xiao, H.D.; Cole, J.M.; Adams, J.W.; Frenzel, K.; Michaud, A.; Zhao, H.; Keshelava, G.; Capecchi, M.R.; Corvol, P.; et al. Role of the N-terminal catalytic domain of angiotensin-converting enzyme investigated by targeted inactivation in mice. J. Biol. Chem. 2004, 279, 15946–15953. [Google Scholar] [CrossRef] [Green Version]




| Sequence | Mol. wt. (g/mol) | Hydrophobicity (%) | pKd | ∆G (kcal/mol) |
|---|---|---|---|---|
| LTPVPGSPF | 914.608 | 66.67 | 2.86 | −3.86 |
| IIAPTPVPAAH | 1086.736 | 81.82 | 6.47 | −8.49 |
| TIGAPDGIPSAPR | 1251.721 | 53.85 | 3.57 | −5.87 |
| HEFPGVVVGANDD | 1374.808 | 46.15 | 3.64 | −4.92 |
| LNPGAGLPRGPNGADTF | 1653.991 | 47.06 | 6.53 | −8.82 |
| Km | Vmax | |
|---|---|---|
| ACE+HHL+ (no inhibitor) | 3.008 µM | 0.001 µM/min |
| ACE+HHL+Belpep | 1.55625 µM | 0.0005 µM/min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, T.K.; Chatterjee, R.; Mandal, R.S.; Roychoudhury, A.; Paul, D.; Roy, S.; Pateiro, M.; Das, A.K.; Lorenzo, J.M.; Dhar, P. ACE Inhibitory Peptides from Bellamya bengalensis Protein Hydrolysates: In Vitro and In Silico Molecular Assessment. Processes 2021, 9, 1316. https://doi.org/10.3390/pr9081316
Dey TK, Chatterjee R, Mandal RS, Roychoudhury A, Paul D, Roy S, Pateiro M, Das AK, Lorenzo JM, Dhar P. ACE Inhibitory Peptides from Bellamya bengalensis Protein Hydrolysates: In Vitro and In Silico Molecular Assessment. Processes. 2021; 9(8):1316. https://doi.org/10.3390/pr9081316
Chicago/Turabian StyleDey, Tanmoy Kumar, Roshni Chatterjee, Rahul Shubhra Mandal, Anadi Roychoudhury, Debjyoti Paul, Souvik Roy, Mirian Pateiro, Arun K. Das, Jose M. Lorenzo, and Pubali Dhar. 2021. "ACE Inhibitory Peptides from Bellamya bengalensis Protein Hydrolysates: In Vitro and In Silico Molecular Assessment" Processes 9, no. 8: 1316. https://doi.org/10.3390/pr9081316
APA StyleDey, T. K., Chatterjee, R., Mandal, R. S., Roychoudhury, A., Paul, D., Roy, S., Pateiro, M., Das, A. K., Lorenzo, J. M., & Dhar, P. (2021). ACE Inhibitory Peptides from Bellamya bengalensis Protein Hydrolysates: In Vitro and In Silico Molecular Assessment. Processes, 9(8), 1316. https://doi.org/10.3390/pr9081316








