The Expression of UGT46A1 Gene and Its Effect on Silkworm Feeding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Models
2.2. RNA Purification and cDNA Synthesis
2.3. Cloning, Expression, and Bioinformatics Analysis of UGT46A1
2.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.5. Double Strand RNA Synthesis and Injection
2.6. Determination of Feeding Behavior
3. Results
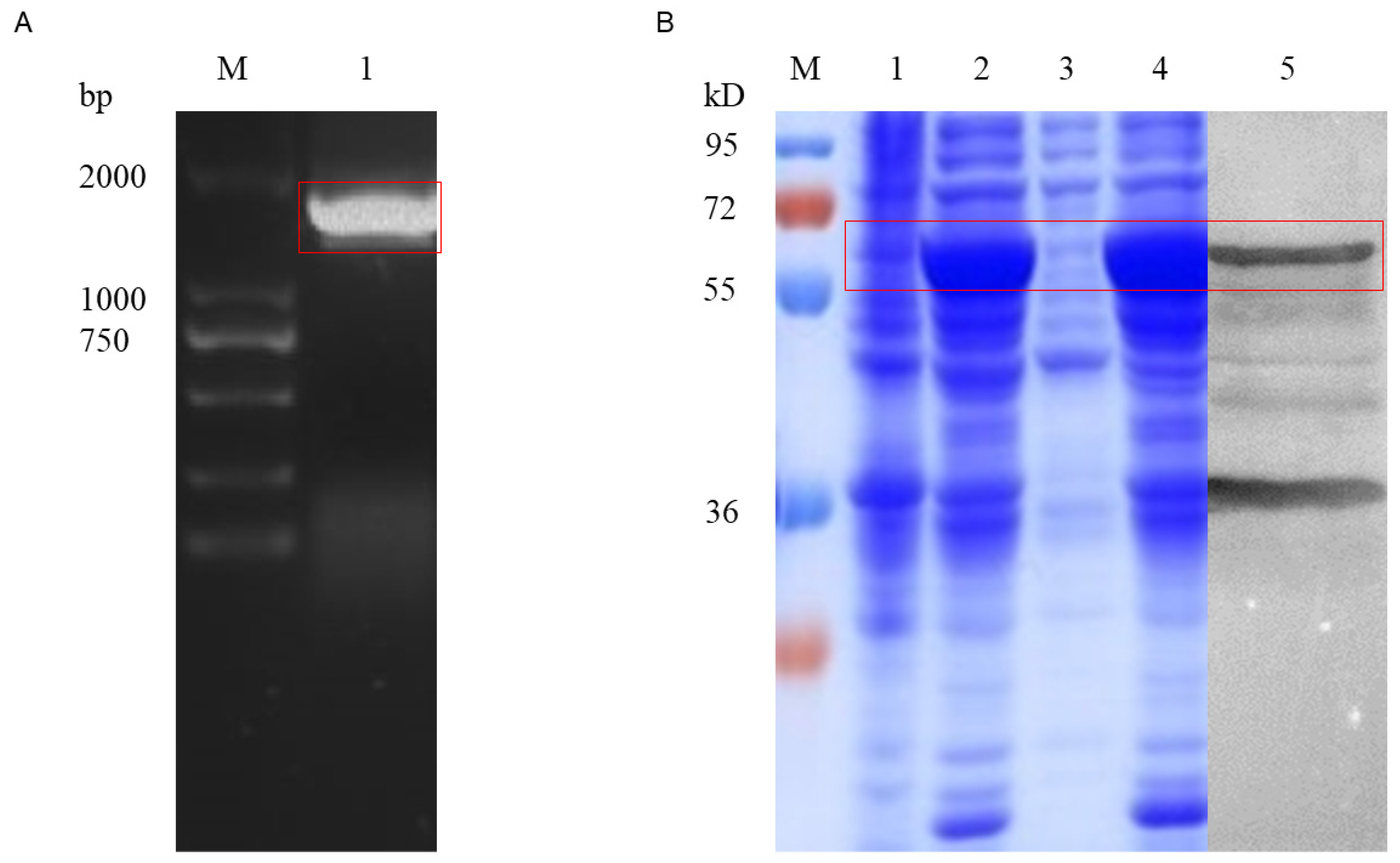
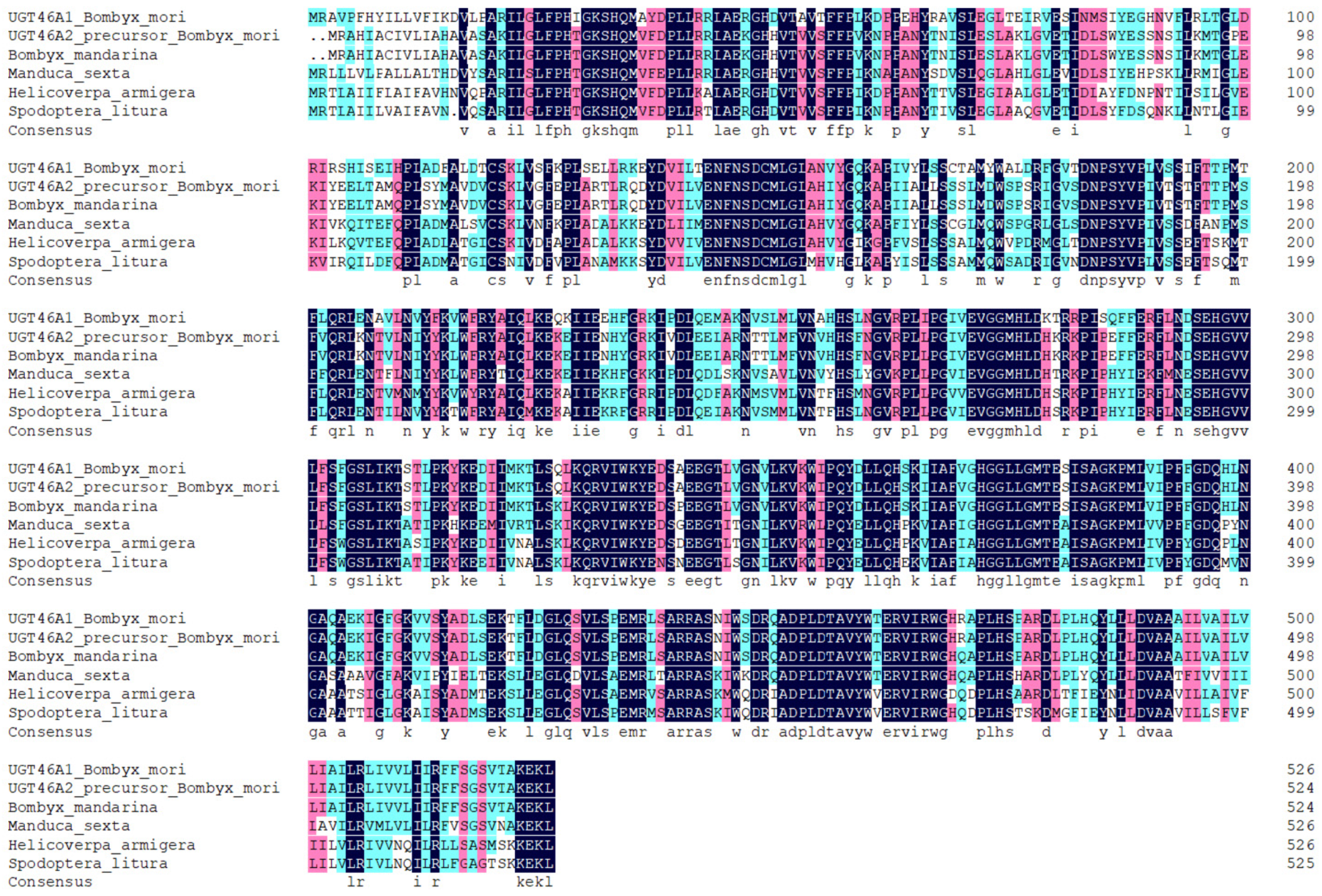
3.1. The Cloning, Expression, and Bioinformatics Analysis of UGT46A1
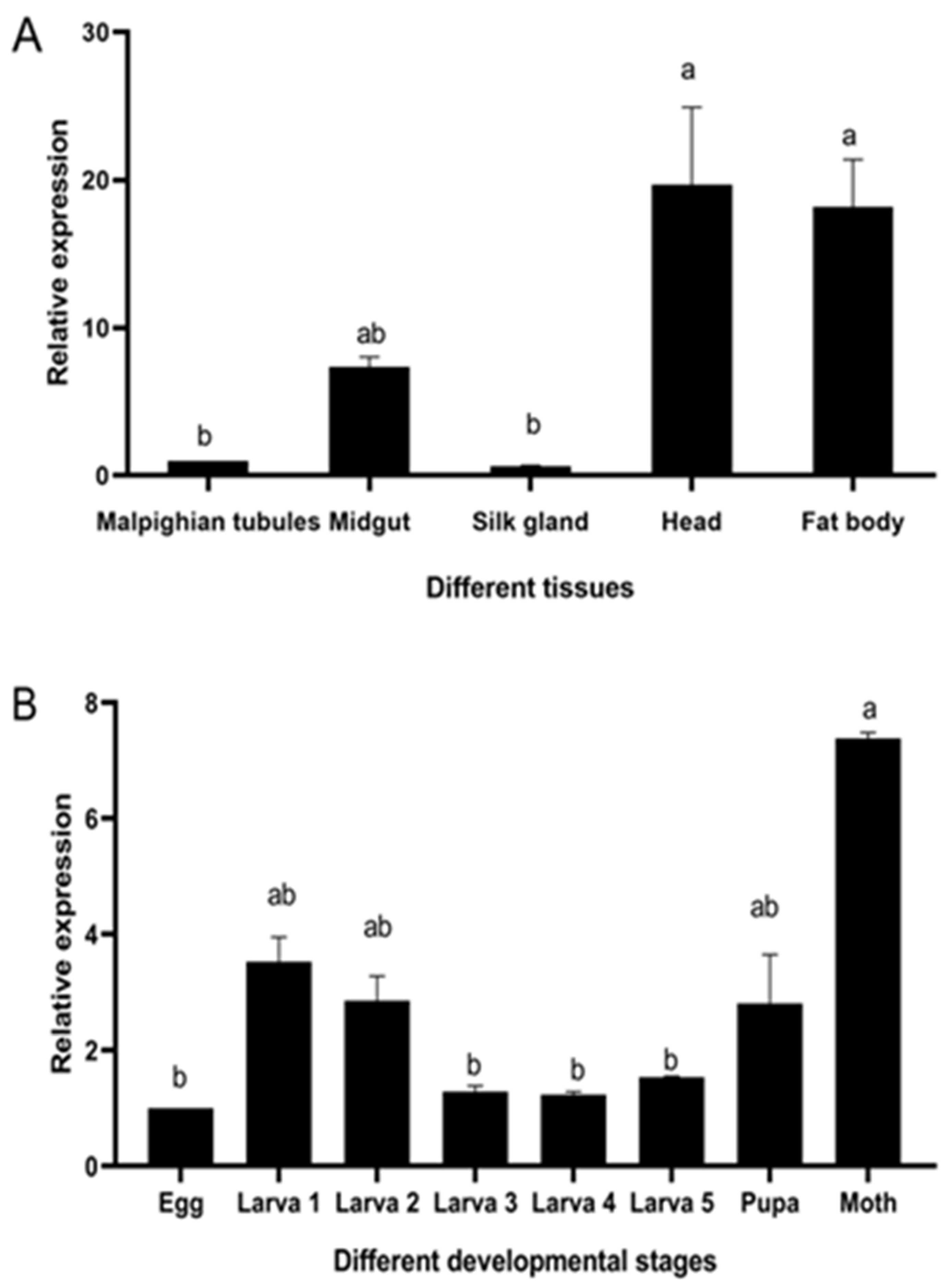
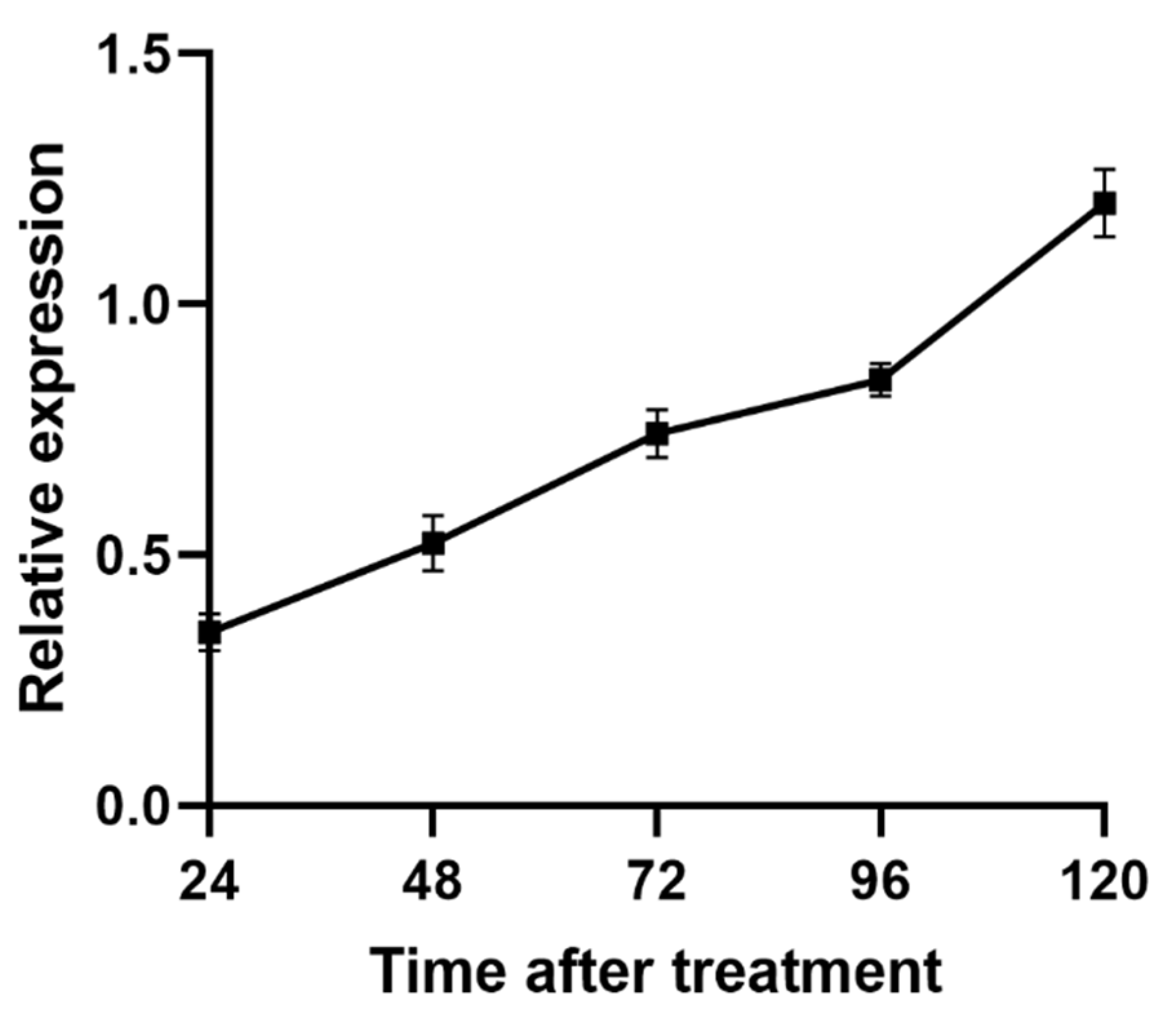
3.2. The Temporal Expression of UGT46A1 in B. mori
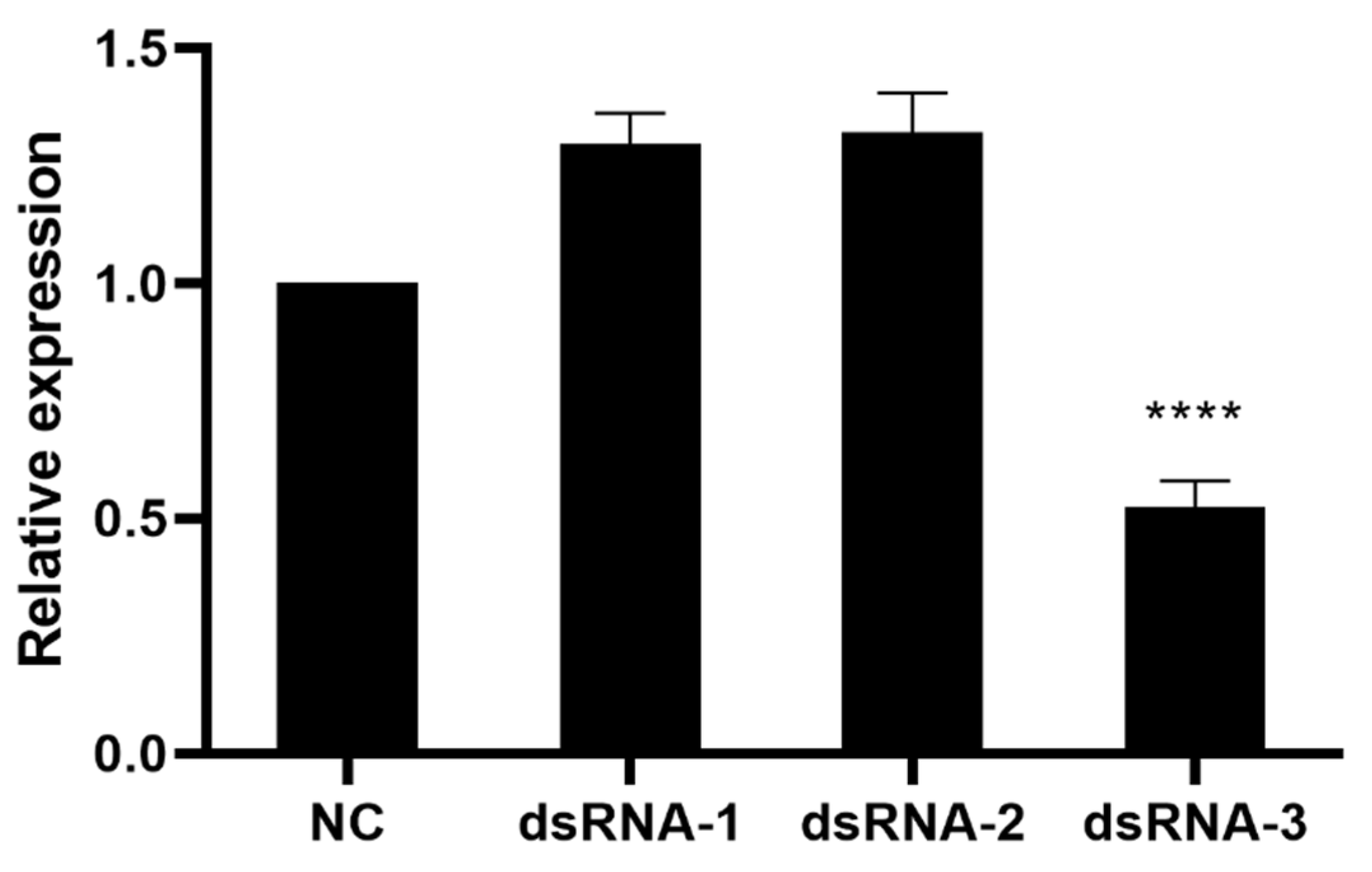
3.3. Knockdown Effect of dsRNA on UGT46A1 in B. mori
3.4. Effective Time of RNA Interference
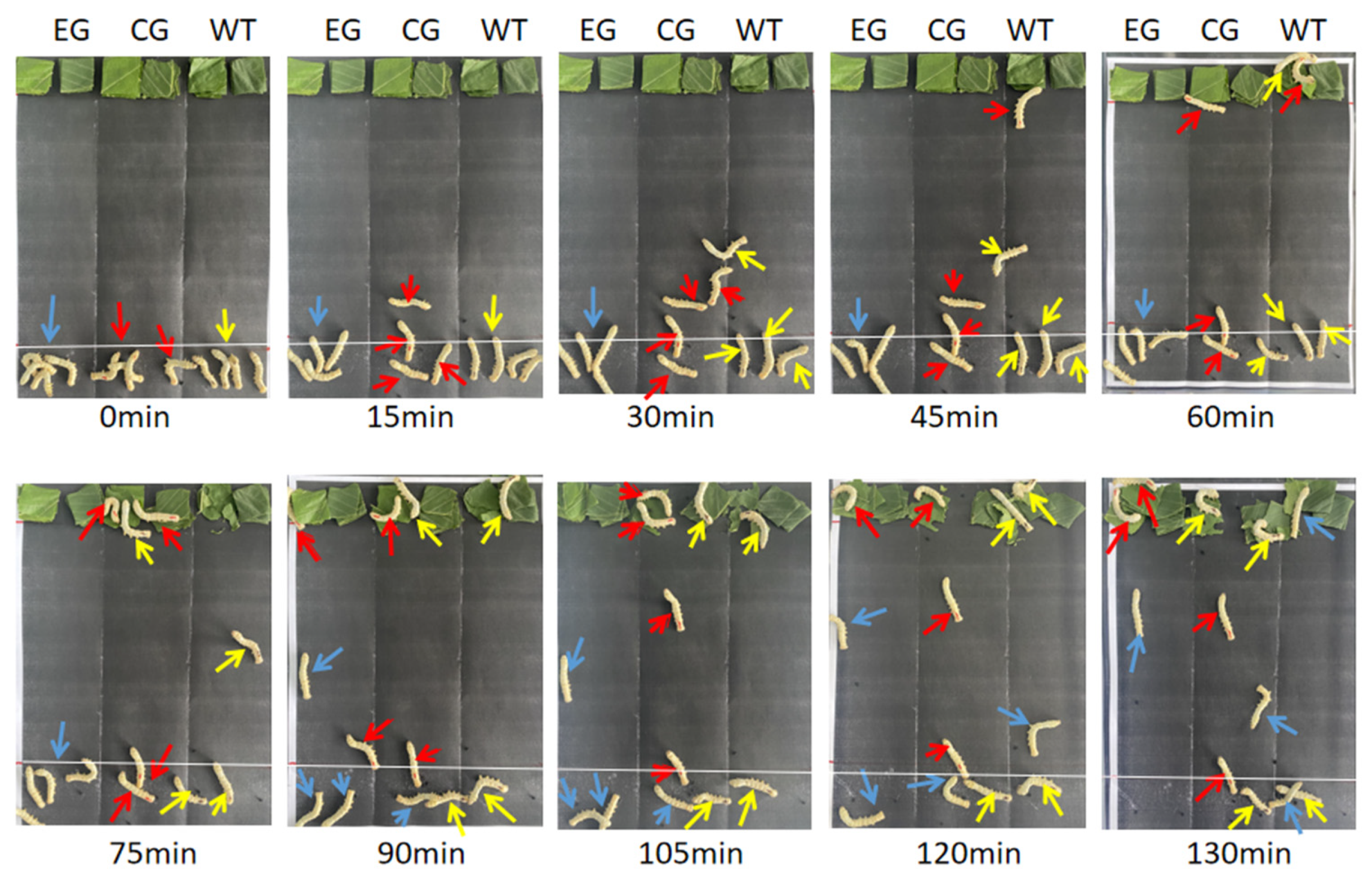
3.5. Results of Feeding Behavior Measurement
3.6. Determination of Food Selectivity of Silkworm
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fraenkel, G.S. The raison d’tre of secondary plant substances; these odd chemicals arose as a means of protecting plants from insects and now guide insects to food. Science 1959, 129, 1466. [Google Scholar] [CrossRef]
- Ishikawa, S.; Hirao, T.; Arai, N. Chemosensory basis of hostplant selection in the silkworm. Entomol. Exp. Appl. 2011, 12, 544–554. [Google Scholar] [CrossRef]
- Zhang, H.J.; Anderson, A.R.; Trowell, S.C.; Luo, A.R.; Xiang, Z.H.; Xia, Q.Y. Topological and Functional Characterization of an Insect Gustatory Receptor. PLoS ONE 2011, 6, e24111. [Google Scholar]
- Wilde, J. Host plant selection in the colorado beetle larva. Entomol. Exp. Appl. 2011, 1, 14–22. [Google Scholar] [CrossRef]
- Devitt, B.D.; Smith, J. Morphology and fine structure of mouthpart sensilla in the dark-sided cutworm Euxoa messoria (Harris) (Lepidoptera: Noctuidae). Int. J. Insect Morphol. Embryol. 1982, 11, 255–270. [Google Scholar] [CrossRef]
- Hsiao, T.H. Chemical basis of host selection and plant resistance in oligophagous insects. Entomol. Exp. Appl. 2011, 12, 777–788. [Google Scholar] [CrossRef]
- Yoshizawa, Y.; Sato, R.; Tsuchihara, K.; Ozaki, K.; Mita, K.; Asaoka, K.; Taniai, K. Ligand carrier protein genes expressed in larval chemosensory organs of Bombyx mori. Insect Biochem. Mol. 2011, 41, 545–562. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.P.; Zhang, H.J.; Ping, Z.; Xia, Q.Y.; Xiang, Z.H. The Odorant Binding Protein Gene Family from the Genome of Silkworm, Bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef] [Green Version]
- Dani, F.R.; Michelucci, E.; Francese, S.; Mastrobuoni, G.; Cappellozza, S.; Marca, G.L.; Niccolini, A.; Felicioli, A.; Moneti, G. Odorant-Binding Proteins and Chemosensory Proteins in Pheromone Detection and Release in the Silkmoth Bombyx mori. Chem. Senses 2011, 36, 335–344. [Google Scholar] [CrossRef]
- Ban, L.; Scaloni, A.; Brandazza, S.; Angeli, Z. Chemosensory proteins of Locusta migratoria. Insect Mol. Biol. 2003, 12, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G. Biochemical diversity of odor detection: OBPs, ODEs and SNMPs—ScienceDirect. Insect Biochem. Mol. 2003, 391–445. [Google Scholar]
- Nichols, Z.; Vogt, R.G. The SNMP/CD36 gene family in Diptera, Hymenoptera and Coleoptera: Drosophila melanogaster, D. pseudoobscura, Anopheles gambiae, Aedes aegypti, Apis mellifera, and Tribolium castaneum. Insect Biochem. Mol. 2008, 38, 398–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallem, E.A.; Dahanukar, A.; Carlson, J.R. Insect odor and taste receptors. Annu. Rev. Entomol. 2006, 51, 113–135. [Google Scholar] [CrossRef]
- Elmore, T.; Smith, D.P. Putative Drosophila odor receptor OR43b localizes to dendrites of olfactory neurons. Insect Biochem. Mol. 2001, 31, 791–798. [Google Scholar] [CrossRef]
- Mabèche-Coisne, M.; Merlin, C.; Rosell, G.; Carot, G.; Jacquin-Joly, E. Odorant-degrading enzymes of moths. In Proceedings of the 22th International Society of Chemical Ecology, Barcelona, Spain, 15–19 July 2006. [Google Scholar]
- Steiner, C.; Bozzolan, F.; Montagné, N.; Maïbèche, M.; Chertemps, T. Neofunctionalization of “Juvenile Hormone Esterase Duplication” in Drosophila as an odorant-degrading enzyme towards food odorants. Sci. Rep. 2017, 7, 12629. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.A.; Hopkins, T.L. β-Glucosylation of plant phenolics by phenol b-glucosyltransferase in larval tissues of the tobacco hornworm, Manduca sexta (L.). Insect Biochem. Mol. 1993, 23, 581–589. [Google Scholar] [CrossRef]
- Luque, T.; O’Reilly, D.R. Functional and phylogenetic analyses of a putative Drosophila melanogaster UDP-glycosyltransferase gene. Insect Biochem. Mol. 2002, 32, 1597–1604. [Google Scholar] [CrossRef]
- Ahmad, S.A.; Hopkins, T.L. Phenol β-glucosyltransferases in six species of insects: Properties and tissue localization. Comp. Biochem. Phys. B 1993, 104, 515–519. [Google Scholar] [CrossRef]
- Heydel, J.M.; Holsztynska, E.J.; Legendre, A.; Thiebaud, N.; Artur, Y.; Bon, A. UDP-glucuronosyltransferases (UGTs) in neuro-olfactory tissues: Expression, regulation, and function. Drug Metab. Rev. 2010, 42, 74–97. [Google Scholar] [CrossRef]
- Tephly, T.R.; Burchell, B. UDP-glucuronosyltransferases: A family of detoxifying enzymes. Trends Pharmacol. Sci. 1990, 11, 276–279. [Google Scholar] [CrossRef]
- Bozzolan, F.; Siaussat, D.; Maria, A.; Durand, N.; Maïbèche-Coisne, M. Antennal uridine diphosphate (UDP)-glycosyltransferases in a pest insect: Diversity and putative function in odorant and xenobiotics clearance. Insect Mol. Biol. 2015, 23, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.F.; Chai, C.L.; Zhang, Z.; Liu, Z.H.; Dai, F.Y.; Lu, C.; Xiang, Z.H. The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genom. 2008, 9, 563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brierley, C.H.; Burchell, B. Human UDP-glucuronosyl transferases: Chemical defence, jaundice and gene therapy. Bioessays 1993, 15, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.h.; Pelletier, Y.; Goyer, C. Identification of potential detoxification enzyme genes in Leptinotarsa decemlineata (Say) and study of their expression in insects reared on different plants. Can. J. Plant Sci. 2008, 88, 621–629. [Google Scholar] [CrossRef]
- Wang, Y.H.; Gu, Z.Y.; Wang, J.M.; Sun, S.S.; Wang, B.B.; Jin, Y.Q.; Shen, W.D.; Li, B. Changes in the activity and the expression of detoxification enzymes in silkworms (Bombyx mori) after phoxim feeding. Pestic. Biochem. Phys. 2013, 105, 13–17. [Google Scholar] [CrossRef]
- Bull, D.L.; Whitten, C.J. Factors influencing organophosphorus insecticide resistance in tobacco budworms. J. Agric. Food Chem. 1972, 20, 561. [Google Scholar] [CrossRef]
- Rane, R.V.; Walsh, T.K.; Pearce, S.L.; Jermiin, L.S.; Oakeshott, J.G. Are feeding preferences and insecticide resistance associated with the size of detoxifying enzyme families in insect herbivores? Curr. Opin. Insect Sci. 2016, 13, 70–76. [Google Scholar] [CrossRef]
- Rane, R.V.; Ghodke, A.B.; Hoffmann, A.A.; Edwards, O.R.; Walsh, T.K.; Oakeshott, J.G. Detoxifying enzyme complements and host use phenotypes in 160 insect species. Curr. Opin. Insect Sci. 2019, 31, 131–138. [Google Scholar] [CrossRef]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Livak, K.J.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, L.C.; Okino, S.T.; Zhao, H.; Pookot, D.; Place, R.F.; Urakami, S.; Enokida, H.; DahiyaSmall, R. dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. USA 2006, 103, 17337–17342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhumika, B.; Kumar, S.A. Regulation of feeding behavior in Drosophila through the interplay of gustation, physiology and neuromodulation. Front. Biosci. 2018, 23, 2016–2027. [Google Scholar]
- Ahn, S.J.; Badenes-Pérez, F.; Reichelt, M.; Svato, A.; Schneider, B.; Gershenzon, J.; Heckel, D.G. Metabolic detoxification of capsaicin by UDP-glycosyltransferase in three Helicoverpa species. Arch. Insect Biochem. 2011, 78, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Vogel, H.; Heckel, D.G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 2012, 42, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Gao, X.; Zhang, S.; Zhang, Y.; Ma, D.; Cui, J. Dynamic changes of transcriptome of fifth-instar spodoptera litura larvae in response to insecticide. 3 Biotech 2021, 11, 98. [Google Scholar] [CrossRef]
- Li, J.; Moghaddam, S.; Chen, X.; Chen, M.; Zhong, B. Shotgun strategy-based proteome profiling analysis on the head of silkworm Bombyx mori. Amino Acids 2010, 39, 751–761. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Chen, Q.; Hou, Y.; Xia, Q.; Ping, Z. Metabolomics Analysis of the Larval Head of the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2016, 17, 1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosquet, G. Nutritional and non nutritional stimulations of protein synthesis in the fat body of Bombyx mori. Insect Biochem. 1983, 13, 281–288. [Google Scholar] [CrossRef]
- Tojo, S.; Kiguchi, K.; Kimura, S. Hormonal control of storage protein synthesis and uptake by the fat body in the silkworm, Bombyx mori. J. Insect Physiol. 1981, 27, 491–497. [Google Scholar] [CrossRef]
- Bosquet, G. Occurrence of an active regulatory mechanism of protein synthesis during starvation and refeeding in Bombyx mori fat body. Biochimie 1979, 61, 165–170. [Google Scholar] [CrossRef]
- Nath, B.S.; Suresh, A.; Varma, B.M.; Kumar, R.P. Changes in protein metabolism in hemolymph and fat body of the silkworm, Bombyx mori (Lepidoptera: Bombycidae) in response to organophosphorus insecticides toxicity. Ecotoxicol. Environ. Saf. 1997, 74, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D. 100 years of pheromone research. Naturwissenschaften 1992, 79, 241–250. [Google Scholar] [CrossRef]
- Pansopha, P.; Ando, N.; Kanzaki, R. Dynamic use of optic flow during pheromone tracking by the male silkmoth, Bombyx mori. J. Exp. Biol. 2014, 217, 1811–1820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazard, D.; Zupko, K.; Poria, Y.; Net, P.; Lazarovits, J.; Horn, S.; Khen, M.; Lancet, D. Odorant signal termination by olfactory UDP glucuronosyl transferase. Nature 1991, 349, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Eubanks, M.D.; Styrsky, J.D.; Denno, R.F. The evolution of omnivory in heteropteran insects. Ecology 2003, 84, 2521. [Google Scholar] [CrossRef]
- Tanaka, K.; Uda, Y.; Ono, Y.; Nakagawa, T.; Suwa, M.; Yamaoka, R.; Touhara, K. Highly Selective Tuning of a Silkworm Olfactory Receptor to a Key Mulberry Leaf Volatile. Curr. Biol. 2009, 19, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.J.; Zhang, S.S.; Niu, B.L.; Ji, D.F.; Liu, X.J.; Li, M.W.; Bai, H.; Palli, S.R.; Wang, C.Z.; Tan, A.J. A determining factor for insect feeding preference in the silkworm, Bombyx mori. PLoS Biol. 2019, 17, e3000162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mang, D.; Shu, M.; Endo, H.; Yoshizawa, Y.; Nagata, S.; Kikuta, S.; Sato, R. Expression of a sugar clade gustatory receptor, bmgr6, in the oral sensory organs, midgut, and central nervous system of larvae of the silkworm bombyx mori. Insect Biochem. Mol. 2016, 70, 85–98. [Google Scholar] [CrossRef]
- Bock, H.E.; Hartl, W.; Genth, E. In vitro binding of complement on leukocytes. contribution to the immunopathogenesis of schultz’s drug allergic agranulocytosis. Schweiz. Med. Wochr. 1970, 100, 617–622. [Google Scholar]
- Zhang, S.X. Expression of general odorant binding proteins in silkworm bombyx mori and molecular evaluation in lepidoteran insects. Sci. Seric. 2010, 36, 610–618. [Google Scholar]
- Forstner, M.; Gohl, T.; Breer, H.; Krieger, J. Candidate pheromone binding proteins of the silkmoth bombyx mori. Invertebr. Neurosci. 2006, 6, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Wanner, K.W.; Anderson, A.R.; Trowell, S.C.; Theilmann, D.A.; Robertson, H.M.; Newcomb, R.D. Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, bombyx mori. Insect Mol. Biol. 2007, 16, 107–119. [Google Scholar] [CrossRef] [PubMed]


| Genes | Primer Names | Sequences |
|---|---|---|
| UGT46A1 | UGT46A1-F UGT46A1-R | GCGTGCCGTGCCATTTCATTAC CATCGTGTCCTCTCTCAGCCAATC |
| Actin A3 | Actin A3-F Actin A3-R | AACGGAATCCACGAAACCAC ACAATACGGTGTTGGCGTAC |
| Name | Target Location | Sense Strand | Antisense Strand |
|---|---|---|---|
| dsRNA-1 | UGT46A1-337 | GCAGACUUCGCUUUAGAUATT | UAUCUAAAGCGAAGUCUGCTT |
| dsRNA-2 | UGT46A1-987 | GCAGAGAGUUAUCUGGAAATT | UUUCCAGAUAACUCUCUGCTT |
| dsRNA-3 | UGT46A1-1550 | GCGGUAGUGUCACUGCUAATT | UUAGCAGUGACACUACCGCTT |
| Negative control | control | UUCUCCGAACGUGUCACGUTT | ACGUGACACGUUCGGAGAATT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, W.; Fan, Y.; Zhu, F.; Taha, R.H.; Chen, K. The Expression of UGT46A1 Gene and Its Effect on Silkworm Feeding. Processes 2021, 9, 1473. https://doi.org/10.3390/pr9081473
Song W, Fan Y, Zhu F, Taha RH, Chen K. The Expression of UGT46A1 Gene and Its Effect on Silkworm Feeding. Processes. 2021; 9(8):1473. https://doi.org/10.3390/pr9081473
Chicago/Turabian StyleSong, Wenting, Yixuan Fan, Feifei Zhu, Rehab Hosny Taha, and Keping Chen. 2021. "The Expression of UGT46A1 Gene and Its Effect on Silkworm Feeding" Processes 9, no. 8: 1473. https://doi.org/10.3390/pr9081473
APA StyleSong, W., Fan, Y., Zhu, F., Taha, R. H., & Chen, K. (2021). The Expression of UGT46A1 Gene and Its Effect on Silkworm Feeding. Processes, 9(8), 1473. https://doi.org/10.3390/pr9081473







