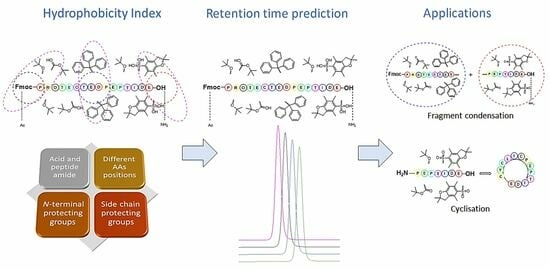
Determining the Hydrophobicity Index of Protected Amino Acids and Common Protecting Groups
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Peptide Synthesis
2.2.1. Incorporation Procedure
2.2.2. Peptide Assembly
2.3. Cleavage Protocols
2.3.1. Protected Fragments
2.3.2. Unprotected Fragments
3. Results and Discussions
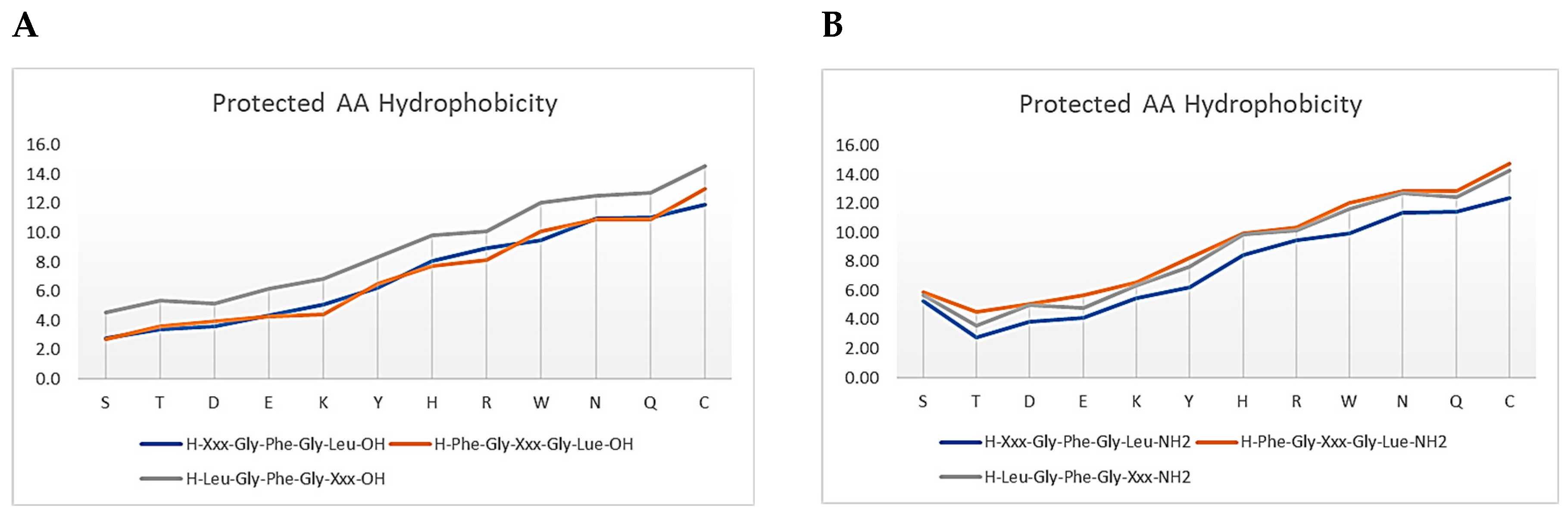
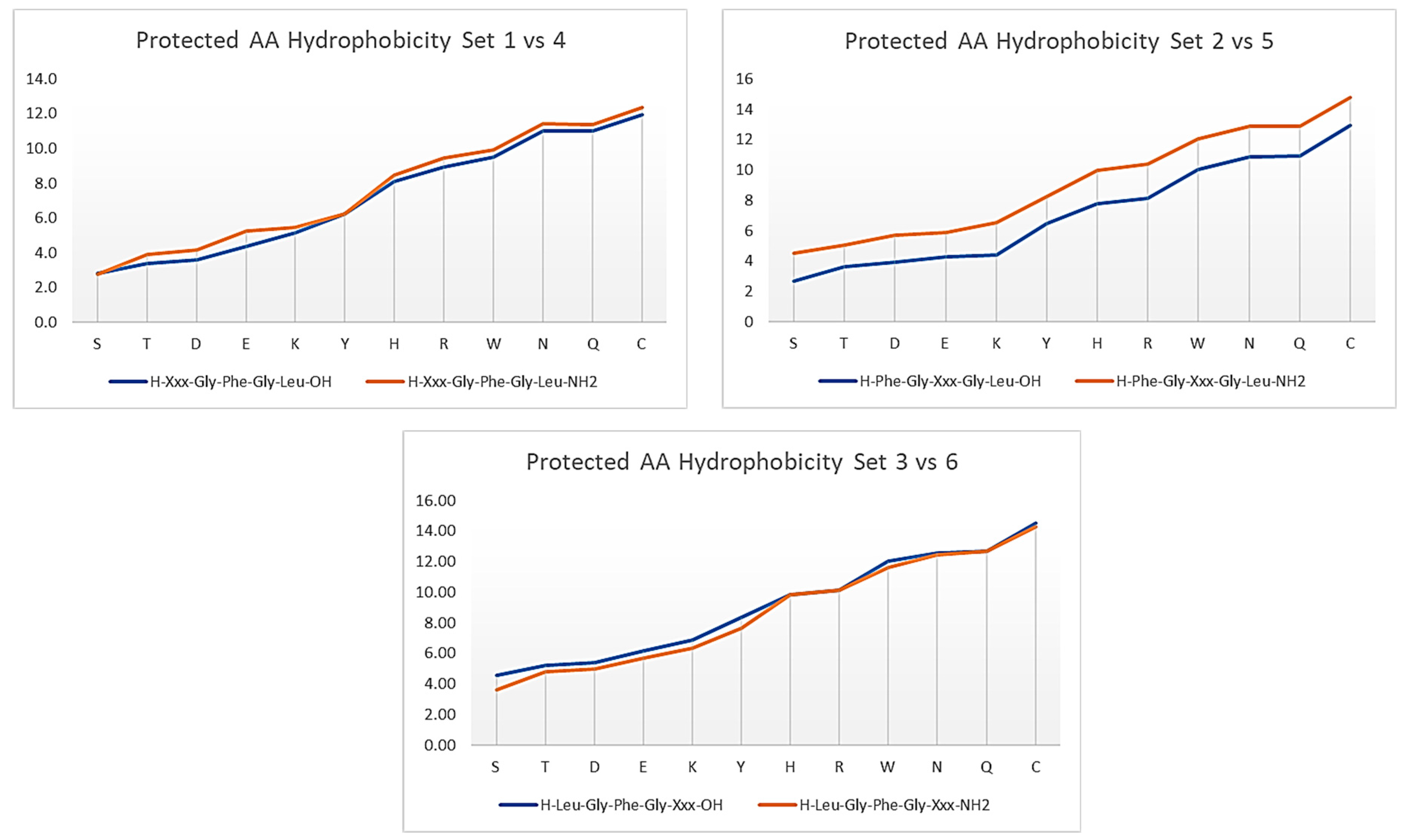
3.1. Amino Acids Hydrophobicity
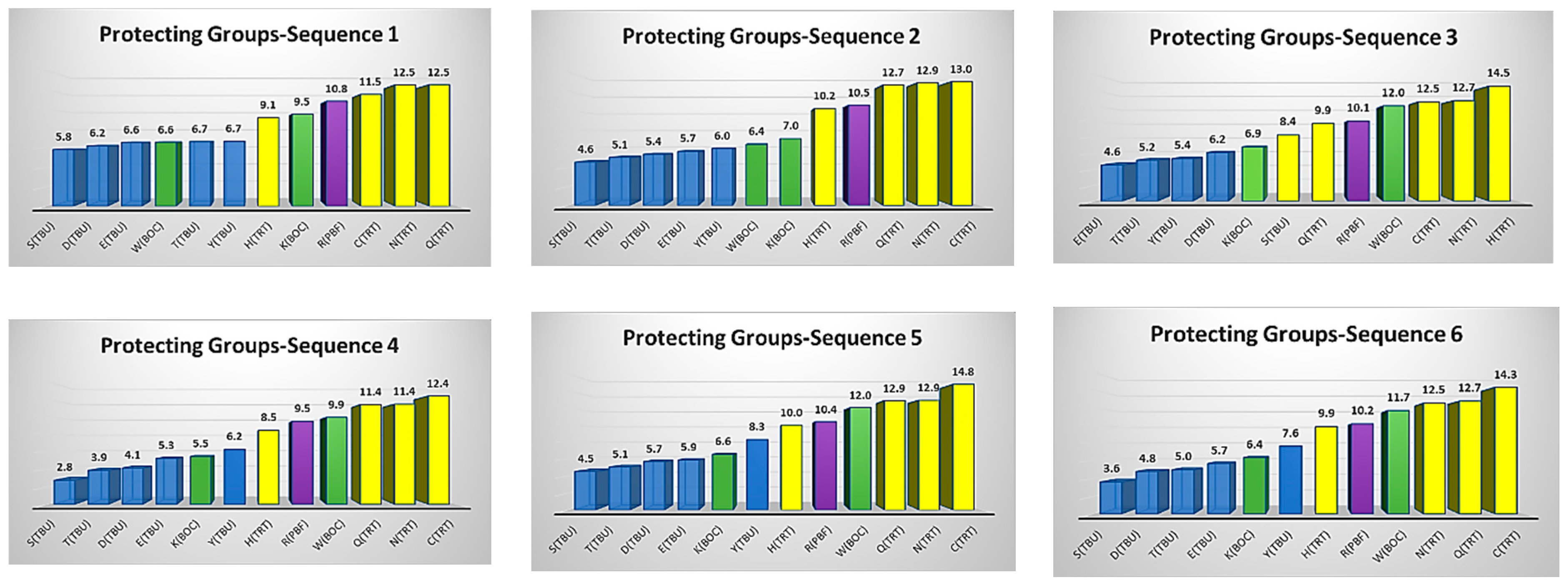
3.2. Protecting Groups’ Hydrophobicity
3.3. Retention Time Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Ac | acetyl group |
| Boc | tert-butyloxycarbonyl |
| CH2Cl2 | dichloromethane |
| CH3CN | acetonitrile |
| DIEA | N,N-diisopropylethylamine |
| DIC | N,N′-diisopropylcarbodiimide |
| DMF | N,N-dimethylformamide |
| GMP | good manufacturing practice |
| Pbf | 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl |
| RPLC | reversed-phase liquid chromatography |
| SPPS | solid-phase peptide synthesis |
| tBu | tert-butyl |
| TIS | triisopropylsilane |
| TFA | trifluoroacetic acid |
| Trt | trityl |
References
- Jaradat, D.M.M. Thirteen decades of peptide synthesis: Key developments in solid phase peptide synthesis and amide bond formation utilized in peptide ligation. Amino Acids 2018, 50, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, S. Peptides as ‘Drugs’: The Journey so Far. Int. J. Pept. Res. Ther. 2017, 23, 49–60. [Google Scholar] [CrossRef]
- Petrou, C.; Sarigiannis, Y. Peptide synthesis: Methods, trends, and challenges. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Sawston, UK, 2018; pp. 1–21. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Loffet, A. Peptides as Drugs: Is There a Market? J. Pept. Sci. 2002, 8, 1–7. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Al Shaer, D.; Albericio, F.; Torre, B.G.d.l. 2022 FDA TIDES (Peptides and Oligonucleotides) Harvest. Pharmaceuticals 2023, 16, 336. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Lombardi, L.; Williams, D.R.; Albericio, F. Strategies for Improving Peptide Stability and Delivery. Pharmaceuticals 2022, 15, 1283. [Google Scholar] [CrossRef]
- Ferrazzano, L.; Catani, M.; Cavazzini, A.; Martelli, G.; Corbisiero, D.; Cantelmi, P.; Fantoni, T.; Mattellone, A.; De Luca, C.; Felletti, S.; et al. Sustainability in peptide chemistry: Current synthesis and purification technologies and future challenges. Green Chem. 2022, 24, 975–1020. [Google Scholar] [CrossRef]
- Kaspar, A.A.; Reichert, J.M. Future directions for peptide therapeutics development. Drug Discov. Today 2013, 18, 807–817. [Google Scholar] [CrossRef]
- D’Aloisio, V.; Dognini, P.; Hutcheon, G.A.; Coxon, C.R. PepTherDia: Database and structural composition analysis of approved peptide therapeutics and diagnostics. Drug Discov. 2021, 26, 1409–1419. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Albericio, F. Developments in peptide and amide synthesis. Curr. Opin. Chem. Biol. 2004, 8, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Zompra, A.A.; Galanis, A.S.; Werbitzky, O.; Albericio, F. Manufacturing peptides as active pharmaceutical ingredients. Future Med. Chem. 2009, 1, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Andersson, L.; Blomberg, L.; Flegel, M.; Lepsa, L.; Nilsson, B.; Verlander, M. Large-scale synthesis of peptides. Pept. Sci. 2000, 55, 227–250. [Google Scholar] [CrossRef]
- Merrifield, R.B. Solid Phase Peptide Synthesis. I. The Synthesis of a Tetrapeptide. J. Am. Chem. Soc. 1963, 85, 2149–2154. [Google Scholar] [CrossRef]
- Athanassopoulos, P.; Barlos, K.; Gatos, D.; Hatzi, O.; Tzavara, C. Application of 2-Chlorotrityl Chloride in Convergent Peptide Synthesis. Tetrahedron Lett. 1995, 36, 5645–5648. [Google Scholar] [CrossRef]
- Gongora-Benitez, M.; Tulla-Puche, J.; Albericio, F. Handles for Fmoc solid-phase synthesis of protected peptides. ACS Comb. Sci. 2013, 15, 217–228. [Google Scholar] [CrossRef]
- Bray, B.L. Large-scale manufacture of peptide therapeutics by chemical synthesis. Nat. Rev. Drug Discov. 2003, 2, 587–593. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Morse, S.V.; Lombardi, L.; Serban, S.; Basso, A.; Williams, D.R. Successful synthesis of a glial-specific blood–brain barrier shuttle peptide following a fragment condensation approach on a solid-phase resin. J. Pept. Sci. 2022, 29, e3448. [Google Scholar] [CrossRef]
- White, C.J.; Yudin, A.K. Contemporary strategies for peptide macrocyclization. Nat. Chem. 2011, 3, 509–524. [Google Scholar] [CrossRef]
- Snyder, L.R.; Kirkland, J.J.; Dolan, J.W. Introduction to Modern Liquid Chromatography, 3rd ed.; Wiley-Interscience: New York, NY, USA, 2010. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Valenzo, O.M.M.; Williams, D.R. Prediction of peptides retention behavior in reversed-phase liquid chromatography based on their hydrophobicity. J. Sep. Sci. 2023, 46, 2200743. [Google Scholar] [CrossRef]
- De Luca, C.; Lievore, G.; Bozza, D.; Buratti, A.; Cavazzini, A.; Ricci, A.; Macis, M.; Cabri, W.; Felletti, S.; Catani, M. Downstream Processing of Therapeutic Peptides by Means of Preparative Liquid Chromatography. Molecules 2021, 26, 4688. [Google Scholar] [CrossRef] [PubMed]
- Musaimi, O.A.; Mercado-Valenzo, O.M.; Williams, D.R. Factors Influencing the Prediction Accuracy of Model Peptides in Reversed-Phase Liquid Chromatography. Separations 2023, 10, 81. [Google Scholar] [CrossRef]
- Tripet, B.; Cepeniene, D.; Kovacs, J.M.; Mant, C.T.; Krokhin, O.V.; Hodges, R.S. Requirements for prediction of peptide retention time in reversed-phase high-performance liquid chromatography: Hydrophilicity/hydrophobicity of side-chains at the N- and C-termini of peptides are dramatically affected by the end-groups and location. J. Chromatogr. A 2007, 1141, 212–225. [Google Scholar] [CrossRef] [PubMed]
- Klee, W.A.; Nirenberg, M. Mode of action of endogenous opiate peptides. Nature 1976, 263, 609–612. [Google Scholar] [CrossRef]
- Kovacs, J.M.; Mant, C.T.; Hodges, R.S. Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side chains in peptides in the absence of nearest-neighbor or conformational effects. J. Pept. Sci. 2006, 84, 283–297. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, M.; Al Musaimi, O.; Collins, J.M.; Albericio, F.; de la Torre, B.G. Cleaving protected peptides from 2-chlorotrityl chloride resin. Moving away from dichloromethane. Green Chem. 2020, 22, 2840–2845. [Google Scholar] [CrossRef]
- Al Musaimi, O.; Gavva, V.; Williams, D.R. Greener Cleavage of Protected Peptide Fragments from Sieber Amide Resin. ChemistryOpen 2022, 11, e202200236. [Google Scholar] [CrossRef]

| Peptide Fragment | Predicted τ (Minute) | Actual (Minute) | Difference (Minute) |
|---|---|---|---|
| Fmoc-APPPS(tBu)-OH | 16.9 | 17.5 | −0.6 |
| Fmoc-GPS(tBu)S(tBu)G-OH | 20.5 | 20.3 | +0.2 |
| Fmoc-E(tBu)FIE(tBu)-OH | 30.7 | 30.9 | −0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gavva, V.; Al Musaimi, O.; Bent, C.; Williams, D.R. Determining the Hydrophobicity Index of Protected Amino Acids and Common Protecting Groups. Separations 2023, 10, 456. https://doi.org/10.3390/separations10080456
Gavva V, Al Musaimi O, Bent C, Williams DR. Determining the Hydrophobicity Index of Protected Amino Acids and Common Protecting Groups. Separations. 2023; 10(8):456. https://doi.org/10.3390/separations10080456
Chicago/Turabian StyleGavva, Varshitha, Othman Al Musaimi, Colin Bent, and Daryl R. Williams. 2023. "Determining the Hydrophobicity Index of Protected Amino Acids and Common Protecting Groups" Separations 10, no. 8: 456. https://doi.org/10.3390/separations10080456
APA StyleGavva, V., Al Musaimi, O., Bent, C., & Williams, D. R. (2023). Determining the Hydrophobicity Index of Protected Amino Acids and Common Protecting Groups. Separations, 10(8), 456. https://doi.org/10.3390/separations10080456