Plant Sample Preparation for Metabolomics, Lipidomics, Ionomics, Fluxomics, and Peptidomics
Abstract
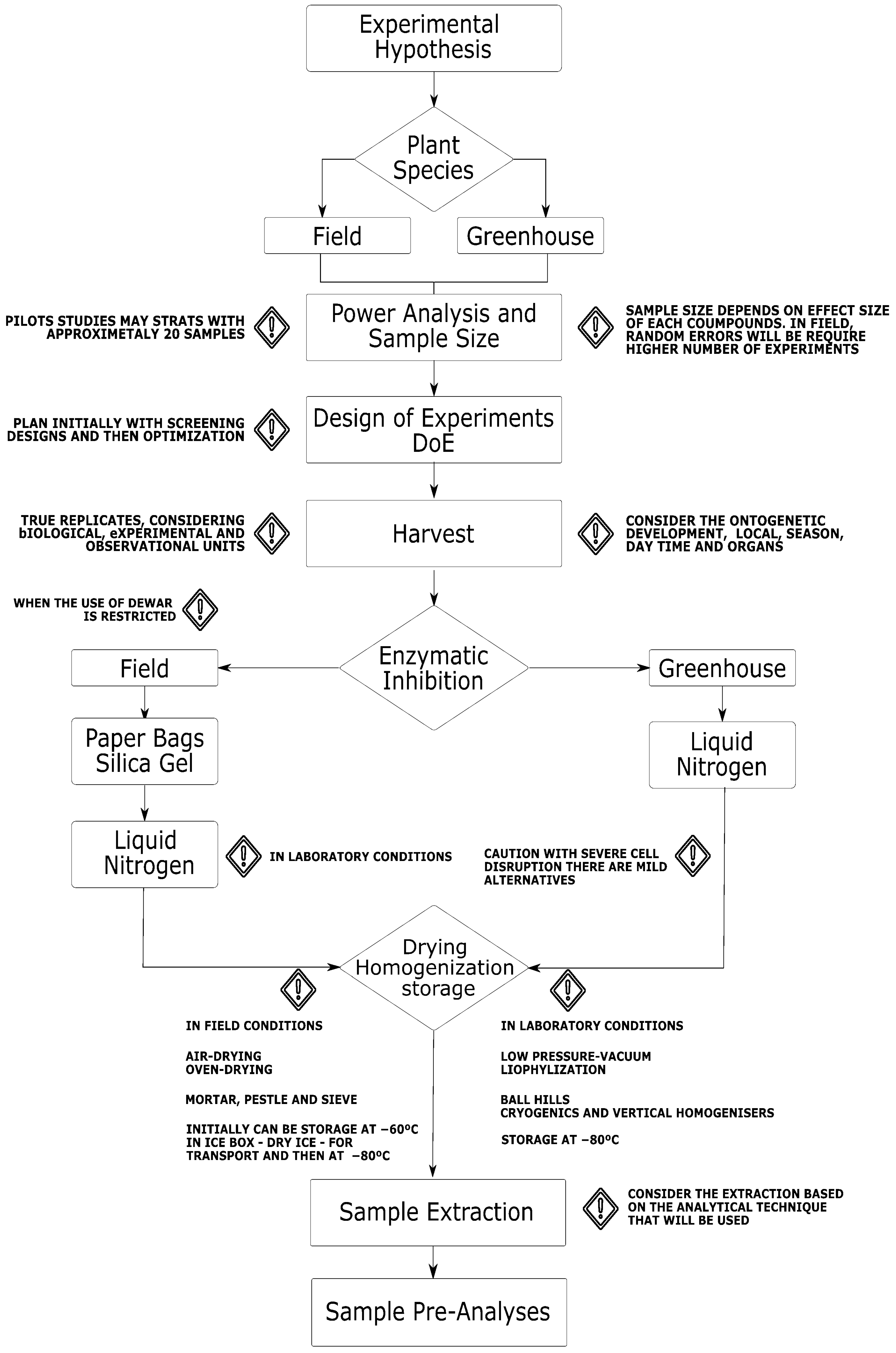
:1. Introduction
2. Experimental Hypothesis and Statistical Power Analysis
3. Design of Experiments
| DOE Approach | Analytical Technique | Optimization | Plant | Ref |
|---|---|---|---|---|
| Sample Preparation | ||||
| Box–Behnken design | GC-MS | Extraction and derivatization of polar compounds | Apple fruit | [57] |
| Central composite design | LC-UV | Extraction (solvent, time, and temperature) | Saffron | [63] |
| D-optimal | UPLC-MS | Accelerated solvent extractions | Camellia sinensis | [64] |
| GC-MS | Extraction and derivatization of compounds | Arabidopsis thaliana | [65] | |
| HS-SPME-GC-MS | Type of fiber, extraction time, equilibration time, and temperature | Grape berry | [66] | |
| ICP-OES | Digestion method (wet, dry, microwave) | Dried fruits | [59] | |
| Full factorial and Box–Behnken designs | NMR | Extraction (solvent, time, power, and solvent/material ratio) | Apricots | [67] |
| Simplex centroid | LC-DAD | Mixture of solvents of extraction | Jatropha species | [68] |
| Taguchi-based designs | LC-MS | Extraction (gridding solvent/material ratio, and stirring) | Lichens | [60] |
| Method Development and Validation | ||||
| Box–Behnken Design | LC-MS | Funnel technology and ion-source parameters | Standards | [69] |
| Box–Behnken Design | LC-MS | Four parameters for LC and six for ESI-MS | Meconopsi species | [70] |
| Central composite and factorial design | LC-DAD | LC solvent, injection volume, temperature | Jatropha species | [68] |
| Plackett–Burman and Factorial Design | GC-MS | Temperature, ramp rate, split, column flow | Olive oils | [60] |
| Data processing | ||||
| Plackett–Burman and Central Composite Design | LC-MS | XCMS parameters | Standards | [71] |
| Modified full factorial | LC-MS | XCMS parameters | Poplar | [72] |
4. Plant Ontology, Harvesting, and Sampling
5. Enzymatic Inhibition
6. Drying, Homogenization, and Storage
7. Extraction Process
8. Pre-Processing to Acquisition
9. Minimum Reports and Best Practices in mQACC
10. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rakusanova, S.; Cajka, T. Tips and Tricks for LC–MS-Based Metabolomics and Lipidomics Analysis. TrAC Trends Anal. Chem. 2024, 180, 117940. [Google Scholar] [CrossRef]
- Jacyna, J.; Kordalewska, M.; Markuszewski, M.J. Design of Experiments in Metabolomics-Related Studies: An Overview. J. Pharm. Biomed. Anal. 2019, 164, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Pilon, A.C.; Carneiro, R.L.; Carnevale Neto, F.; Da, S.; Bolzani, V.; Castro-Gamboa, I. Interval Multivariate Curve Resolution in the Dereplication of HPLC-DAD Data from Jatropha Gossypifolia. Phytochem. Anal. 2013, 24, 401–406. [Google Scholar] [CrossRef]
- Köfeler, H.C.; Ahrends, R.; Baker, E.S.; Ekroos, K.; Han, X.; Hoffmann, N.; Holčapek, M.; Wenk, M.R.; Liebisch, G. Recommendations for Good Practice in MS-Based Lipidomics. J. Lipid Res. 2021, 62, 100138. [Google Scholar] [CrossRef] [PubMed]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from Sample Preparation to Data Analysis: A Primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Y.; Zheng, L. Disease Ionomics: Understanding the Role of Ions in Complex Disease. Int. J. Mol. Sci. 2020, 21, 8646. [Google Scholar] [CrossRef] [PubMed]
- Filipiak, M.; Filipiak, Z.M. Application of Ionomics and Ecological Stoichiometry in Conservation Biology: Nutrient Demand and Supply in a Changing Environment. Biol. Conserv. 2022, 272, 109622. [Google Scholar] [CrossRef]
- Lin, G.; Ma, L.; He, X.; Tang, J.; Wang, L. Gene Regulation and Ionome Homeostasis in Rice Plants in Response to Arsenite Stress: Potential Connection between Transcriptomics and Ionomics. BioMetals 2023, 36, 1157–1169. [Google Scholar] [CrossRef] [PubMed]
- Schrader, M. Origins, Technological Development, and Applications of Peptidomics; Humana Press: New York, NY, USA, 2018; pp. 3–39. [Google Scholar] [CrossRef]
- Schrader, M.; Fricker, L.D. Current Challenges and Future Directions in Peptidomics; Humana Press: New York, NY, USA, 2024; pp. 485–498. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Szczepski, K.; Al-Younis, I.; Lachowicz, J.I.; Jaremko, M. Fluxomics—New Metabolomics Approaches to Monitor Metabolic Pathways. Front. Pharmacol. 2022, 13, 805782. [Google Scholar] [CrossRef]
- Ali, A.; Davidson, S.; Fraenkel, E.; Gilmore, I.; Hankemeier, T.; Kirwan, J.A.; Lane, A.N.; Lanekoff, I.; Larion, M.; McCall, L.-I.; et al. Single Cell Metabolism: Current and Future Trends. Metabolomics 2022, 18, 77. [Google Scholar] [CrossRef]
- de Souza, L.P.; Borghi, M.; Fernie, A. Plant Single-Cell Metabolomics—Challenges and Perspectives. Int. J. Mol. Sci. 2020, 21, 8987. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Salek, R.M.; Neumann, S.; Schober, D.; Hummel, J.; Billiau, K.; Kopka, J.; Correa, E.; Reijmers, T.; Rosato, A.; Tenori, L.; et al. COordination of Standards in MetabOlomicS (COSMOS): Facilitating Integrated Metabolomics Data Access. Metabolomics 2015, 11, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Azam, K.; Vadivelu, I.; Burant, C.; Edison, A.; Fiehn, O.; Higashi, R.; Nair, K.S.; et al. Metabolomics Workbench: An International Repository for Metabolomics Data and Metadata, Metabolite Standards, Protocols, Tutorials and Training, and Analysis Tools. Nucleic Acids Res. 2016, 44, D463–D470. [Google Scholar] [CrossRef] [PubMed]
- Lippa, K.A.; Aristizabal-Henao, J.J.; Beger, R.D.; Bowden, J.A.; Broeckling, C.; Beecher, C.; Clay Davis, W.; Dunn, W.B.; Flores, R.; Goodacre, R.; et al. Reference Materials for MS-Based Untargeted Metabolomics and Lipidomics: A Review by the Metabolomics Quality Assurance and Quality Control Consortium (mQACC). Metabolomics 2022, 18, 24. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Sánchez, B.; Priego-Capote, F.; de Castro, M.L. Metabolomics Analysis I. Selection of Biological Samples and Practical Aspects Preceding Sample Preparation. TrAC Trends Anal. Chem. 2010, 29, 111–119. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-Based Metabolomic Analysis of Plants. Nat. Protoc. 2010, 5, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Sánchez, B.; Priego-Capote, F.; de Castro, M.L. Metabolomics Analysis II. Preparation of Biological Samples Prior to Detection. TrAC Trends Anal. Chem. 2010, 29, 120–127. [Google Scholar] [CrossRef]
- Rodrigues, A.M.; Ribeiro-Barros, A.I.; António, C. Experimental Design and Sample Preparation in Forest Tree Metabolomics. Metabolites 2019, 9, 285. [Google Scholar] [CrossRef]
- Blaise, B.J.; Correia, G.; Tin, A.; Young, J.H.; Vergnaud, A.-C.; Lewis, M.; Pearce, J.T.M.; Elliott, P.; Nicholson, J.K.; Holmes, E.; et al. Power Analysis and Sample Size Determination in Metabolic Phenotyping. Anal. Chem. 2016, 88, 5179–5188. [Google Scholar] [CrossRef] [PubMed]
- Biais, B.; Bernillon, S.; Deborde, C.; Cabasson, C.; Rolin, D.; Tadmor, Y.; Burger, J.; Schaffer, A.A.; Moing, A. Precautions for Harvest, Sampling, Storage, and Transport of Crop Plant Metabolomics Samples; Humana Press: New York, NY, USA, 2012; pp. 51–63. [Google Scholar] [CrossRef]
- William Allwood, J.; Winder, C.L.; Dunn, W.B.; Goodacre, R. Considerations in Sample Preparation, Collection, and Extraction Approaches Applied in Microbial, Plant, and Mammalian Metabolic Profiling. In Methodologies for Metabolomics; Cambridge University Press: Cambridge, UK, 2013; pp. 79–118. [Google Scholar] [CrossRef]
- Lin, C.Y.; Wu, H.; Tjeerdema, R.S.; Viant, M.R. Evaluation of Metabolite Extraction Strategies from Tissue Samples Using NMR Metabolomics. Metabolomics 2007, 3, 55–67. [Google Scholar] [CrossRef]
- Römisch-Margl, W.; Prehn, C.; Bogumil, R.; Röhring, C.; Suhre, K.; Adamski, J. Procedure for Tissue Sample Preparation and Metabolite Extraction for High-Throughput Targeted Metabolomics. Metabolomics 2012, 8, 133–142. [Google Scholar] [CrossRef]
- Segers, K.; Declerck, S.; Mangelings, D.; Heyden, Y.V.; Eeckhaut, A.V. Analytical Techniques for Metabolomic Studies: A Review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef] [PubMed]
- Manickam, S.; Rajagopalan, V.R.; Kambale, R.; Rajasekaran, R.; Kanagarajan, S.; Muthurajan, R. Plant Metabolomics: Current Initiatives and Future Prospects. Curr. Issues Mol. Biol. 2023, 45, 8894–8906. [Google Scholar] [CrossRef] [PubMed]
- Vinay, C.M.; Udayamanoharan, S.K.; Prabhu Basrur, N.; Paul, B.; Rai, P.S. Current Analytical Technologies and Bioinformatic Resources for Plant Metabolomics Data. Plant Biotechnol. Rep. 2021, 15, 561–572. [Google Scholar] [CrossRef]
- Salt, D.E.; Baxter, I.; Lahner, B. Ionomics and the Study of the Plant Ionome. Annu. Rev. Plant Biol. 2008, 59, 709–733. [Google Scholar] [CrossRef]
- Yin, H.; Yan, Y.; Hu, W.; Liu, G.; Zeng, H.; Wei, Y.; Shi, H. Genome-Wide Association Studies Reveal Genetic Basis of Ionomic Variation in Cassava. Plant J. 2022, 112, 1212–1223. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.K.; Kumar, A. Fluxomics and Metabolic Flux Analysis. In Multi-Omics Analysis of the Human Microbiome: From Technology to Clinical Applications; Mani, I., Singh, V., Eds.; Springer Nature: Singapore, 2024; pp. 171–180. [Google Scholar] [CrossRef]
- Klein, S.; Heinzle, E. Isotope Labeling Experiments in Metabolomics and Fluxomics. WIREs Syst. Biol. Med. 2012, 4, 261–272. [Google Scholar] [CrossRef]
- Tian, B.; Chen, M.; Liu, L.; Rui, B.; Deng, Z.; Zhang, Z.; Shen, T. 13C Metabolic Flux Analysis: Classification and Characterization from the Perspective of Mathematical Modeling and Application in Physiological Research of Neural Cell. Front. Mol. Neurosci. 2022, 15, 883466. [Google Scholar] [CrossRef] [PubMed]
- de Falco, B.; Giannino, F.; Carteni, F.; Mazzoleni, S.; Kim, D.-H. Metabolic Flux Analysis: A Comprehensive Review on Sample Preparation, Analytical Techniques, Data Analysis, Computational Modelling, and Main Application Areas. RSC Adv. 2022, 12, 25528–25548. [Google Scholar] [CrossRef] [PubMed]
- Pandian, K.; Matsui, M.; Hankemeier, T.; Ali, A.; Okubo-Kurihara, E. Advances in Single-Cell Metabolomics to Unravel Cellular Heterogeneity in Plant Biology. Plant Physiol. 2023, 193, 949–965. [Google Scholar] [CrossRef]
- Cole, B.; Bergmann, D.; Blaby-Haas, C.E.; Blaby, I.K.; Bouchard, K.E.; Brady, S.M.; Ciobanu, D.; Coleman-Derr, D.; Leiboff, S.; Mortimer, J.C.; et al. Plant Single-Cell Solutions for Energy and the Environment. Commun. Biol. 2021, 4, 962. [Google Scholar] [CrossRef]
- Lazic, S.E.; Clarke-Williams, C.J.; Munafò, M.R. What Exactly Is ‘N’ in Cell Culture and Animal Experiments? PLoS Biol. 2018, 16, e2005282. [Google Scholar] [CrossRef] [PubMed]
- Dudzik, D.; Barbas-Bernardos, C.; García, A.; Barbas, C. Quality Assurance Procedures for Mass Spectrometry Untargeted Metabolomics. a Review. J. Pharm. Biomed. Anal. 2018, 147, 149–173. [Google Scholar] [CrossRef] [PubMed]
- Godzien, J.; Alonso-Herranz, V.; Barbas, C.; Armitage, E.G. Controlling the Quality of Metabolomics Data: New Strategies to Get the Best out of the QC Sample. Metabolomics 2015, 11, 518–528. [Google Scholar] [CrossRef]
- Mosley, J.D.; Schock, T.B.; Beecher, C.W.; Dunn, W.B.; Kuligowski, J.; Lewis, M.R.; Theodoridis, G.; Ulmer Holland, C.Z.; Vuckovic, D.; Wilson, I.D.; et al. Establishing a Framework for Best Practices for Quality Assurance and Quality Control in Untargeted Metabolomics. Metabolomics 2024, 20, 20. [Google Scholar] [CrossRef]
- Nyamundanda, G.; Gormley, I.; Fan, Y.; Gallagher, W.M.; Brennan, L. MetSizeR: Selecting the Optimal Sample Size for Metabolomic Studies Using an Analysis Based Approach. BMC Bioinform. 2013, 14, 338. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Lu, Y.; Zhou, G.; Hui, F.; Xu, L.; Viau, C.; Spigelman, A.F.; MacDonald, P.E.; Wishart, D.S.; Li, S.; et al. MetaboAnalyst 6.0: Towards a Unified Platform for Metabolomics Data Processing, Analysis and Interpretation. Nucleic Acids Res. 2024, 52, W398–W406. [Google Scholar] [CrossRef]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Ulmer, C.Z.; Ragland, J.M.; Koelmel, J.P.; Heckert, A.; Jones, C.M.; Garrett, T.J.; Yost, R.A.; Bowden, J.A. LipidQC: Method Validation Tool for Visual Comparison to SRM 1950 Using NIST Interlaboratory Comparison Exercise Lipid Consensus Mean Estimate Values. Anal. Chem. 2017, 89, 13069–13073. [Google Scholar] [CrossRef]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A Lipidome Atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef]
- Can Eylem, C.; Nemutlu, E.; Dogan, A.; Acik, V.; Matyar, S.; Gezercan, Y.; Altintas, S.; Okten, A.I.; Basci Akduman, N.E. Optimized High-Throughput Protocols for Comprehensive Metabolomic and Lipidomic Profiling of Brain Sample. Talanta 2025, 282, 126953. [Google Scholar] [CrossRef] [PubMed]
- Bednarski, T.K.; Rahim, M.; Young, J.D. In Vivo 2H/13C Flux Analysis in Metabolism Research. Curr. Opin. Biotechnol. 2021, 71, 1–8. [Google Scholar] [CrossRef]
- Ahn, E.; Kumar, P.; Mukha, D.; Tzur, A.; Shlomi, T. Temporal Fluxomics Reveals Oscillations in TCA Cycle Flux throughout the Mammalian Cell Cycle. Mol. Syst. Biol. 2017, 13, 953. [Google Scholar] [CrossRef] [PubMed]
- Hellinger, R.; Sigurdsson, A.; Wu, W.; Romanova, E.V.; Li, L.; Sweedler, J.V.; Süssmuth, R.D.; Gruber, C.W. Peptidomics. Nat. Rev. Methods Primers 2023, 3, 25. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, W.; Picciani, M.; The, M.; Wilhelm, M. Deep Learning-Assisted Analysis of Immunopeptidomics Data. In Peptidomics: Methods and Strategies; Schrader, M., Fricker, L.D., Eds.; Springer: New York, NY, USA, 2024; pp. 457–483. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Haro, C.; Montes-Borrego, M.; De La Fuente, L.; Navas-Cortés, J.A.; Landa, B.B. Metabolomic, Ionomic and Microbial Characterization of Olive Xylem Sap Reveals Differences According to Plant Age and Genotype. Agronomy 2021, 11, 1179. [Google Scholar] [CrossRef]
- Bruns, R.; Scarminio, I.; de Barros Neto, B. Statistical Design—Chemimetrics, 1st ed.; Bruns, R., Scarmínio, I., de Barros Neto, B., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Leardi, R. Experimental Design in Chemistry: A Tutorial. Anal. Chim. Acta 2009, 652, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G.; Jansen, J.; Lopes, J.; Marini, F.; Pomerantsev, A.; Rodionova, O.; Roger, J.M.; Walczak, B.; Tauler, R. Chemometrics in Analytical Chemistry—Part I: History, Experimental Design and Data Analysis Tools. Anal. Bioanal. Chem. 2017, 409, 5891–5899. [Google Scholar] [CrossRef] [PubMed]
- Bekele, E.A.; Annaratone, C.E.P.; Hertog, M.L.; Nicolai, B.M.; Geeraerd, A.H. Multi-Response Optimization of the Extraction and Derivatization Protocol of Selected Polar Metabolites from Apple Fruit Tissue for GC–MS Analysis. Anal. Chim. Acta 2014, 824, 42–56. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, M.; Sun, B. Novel Approach for Extraction of Grape Skin Antioxidants by Accelerated Solvent Extraction: Box–Behnken Design Optimization. J. Food Sci. Technol. 2019, 56, 4879–4890. [Google Scholar] [CrossRef] [PubMed]
- Altundag, H.; Tuzen, M. Comparison of Dry, Wet and Microwave Digestion Methods for the Multi Element Determination in Some Dried Fruit Samples by ICP-OES. Food Chem. Toxicol. 2011, 49, 2800–2807. [Google Scholar] [CrossRef] [PubMed]
- Parrot, D.; Peresse, T.; Hitti, E.; Carrie, D.; Grube, M.; Tomasi, S. Qualitative and Spatial Metabolite Profiling of Lichens by a LC-MS Approach Combined with Optimised Extraction. Phytochem. Anal. 2015, 26, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Boggia, R.; Borgogni, C.; Hysenaj, V.; Leardi, R.; Zunin, P. Direct GC–(EI)MS Determination of Fatty Acid Alkyl Esters in Olive Oils. Talanta 2014, 119, 60–67. [Google Scholar] [CrossRef]
- Merz, M.; Fischer, L.; Sia, J.; Born, P.; Weiss, R.; Grieb, N.; Rade, M.; Boldt, A.; Fricke, S.; Franz, P.; et al. Single Cell Multi-Omic Dissection of Response and Resistance to Chimeric Antigen Receptor T Cells Against BCMA in Relapsed Multiple Myeloma. Blood 2023, 142 (Suppl. S1), 453. [Google Scholar] [CrossRef]
- Sarfarazi, M.; Jafari, S.M.; Rajabzadeh, G. Extraction Optimization of Saffron Nutraceuticals Through Response Surface Methodology. Food Anal. Methods 2015, 8, 2273–2285. [Google Scholar] [CrossRef]
- Kellogg, J.J.; Wallace, E.D.; Graf, T.N.; Oberlies, N.H.; Cech, N.B. Conventional and Accelerated-Solvent Extractions of Green Tea (Camellia sinensis) for Metabolomics-Based Chemometrics. J. Pharm. Biomed. Anal. 2017, 145, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, J.; Jonsson, P.; Nordström, A.; Sjöström, M.; Moritz, T. Design of Experiments: An Efficient Strategy to Identify Factors Influencing Extraction and Derivatization of Arabidopsis Thaliana Samples in Metabolomic Studies with Gas Chromatography/Mass Spectrometry. Anal. Biochem. 2004, 331, 283–295. [Google Scholar] [CrossRef]
- Fedrizzi, B.; Carlin, S.; Franceschi, P.; Vrhovsek, U.; Wehrens, R.; Viola, R.; Mattivi, F. D-Optimal Design of an Untargeted HS-SPME-GC-TOF Metabolite Profiling Method. Analyst 2012, 137, 3725. [Google Scholar] [CrossRef]
- Tsiaka, T.; Fotakis, C.; Lantzouraki, D.Z.; Tsiantas, K.; Siapi, E.; Sinanoglou, V.J.; Zoumpoulakis, P. Expanding the Role of Sub-Exploited DOE-High Energy Extraction and Metabolomic Profiling towards Agro-Byproduct Valorization: The Case of Carotenoid-Rich Apricot Pulp. Molecules 2020, 25, 2702. [Google Scholar] [CrossRef]
- Pilon, A.C.; Carnevale Neto, F.; Freire, R.T.; Cardoso, P.; Carneiro, R.L.; Da Silva Bolzani, V.; Castro-Gamboa, I. Partial Least Squares Model and Design of Experiments toward the Analysis of the Metabolome of Jatropha Gossypifolia Leaves: Extraction and Chromatographic Fingerprint Optimization. J. Sep. Sci. 2016, 39, 1023–1030. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Redestig, H.; Saito, K. Metabolomic Approaches toward Understanding Nitrogen Metabolism in Plants. J. Exp. Bot. 2011, 62, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Song, J.-Z.; Choi, F.F.-K.; Wu, H.-F.; Qiao, C.-F.; Ding, L.-S.; Gesang, S.-L.; Xu, H.-X. An Experimental Design Approach Using Response Surface Techniques to Obtain Optimal Liquid Chromatography and Mass Spectrometry Conditions to Determine the Alkaloids in Meconopsi Species. J. Chromatogr. A 2009, 1216, 7013–7023. [Google Scholar] [CrossRef]
- Zheng, H.; Clausen, M.R.; Dalsgaard, T.K.; Mortensen, G.; Bertram, H.C. Time-Saving Design of Experiment Protocol for Optimization of LC-MS Data Processing in Metabolomic Approaches. Anal. Chem. 2013, 85, 7109–7116. [Google Scholar] [CrossRef] [PubMed]
- Eliasson, M.; Rännar, S.; Madsen, R.; Donten, M.A.; Marsden-Edwards, E.; Moritz, T.; Shockcor, J.P.; Johansson, E.; Trygg, J. Strategy for Optimizing LC-MS Data Processing in Metabolomics: A Design of Experiments Approach. Anal. Chem. 2012, 84, 6869–6876. [Google Scholar] [CrossRef]
- Teahan, O.; Gamble, S.; Holmes, E.; Waxman, J.; Nicholson, J.K.; Bevan, C.; Keun, H.C. Impact of Analytical Bias in Metabonomic Studies of Human Blood Serum and Plasma. Anal. Chem. 2006, 78, 4307–4318. [Google Scholar] [CrossRef]
- Chen, S.; Pang, X.; Song, J.; Shi, L.; Yao, H.; Han, J.; Leon, C. A Renaissance in Herbal Medicine Identification: From Morphology to DNA. Biotechnol. Adv. 2014, 32, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Pilon, A.C.; Selegato, D.M.; Fernandes, R.P.; Bueno, P.C.P.; Pinho, D.R.; Neto, F.C.; Freire, R.T.; Castro-Gamboa, I.; Bolzani, V.S.; Lopes, N.P. Metabolômica de Plantas: Métodos e Desafios. Química Nova 2020, 43, 329–354. [Google Scholar] [CrossRef]
- Consortium, T.P.O. The Plant Ontology Consortium and Plant Ontologies. Comp. Funct. Genom. 2002, 3, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Walls, R.L.; Cooper, L.; Elser, J.; Gandolfo, M.A.; Mungall, C.J.; Smith, B.; Stevenson, D.W.; Jaiswal, P. The Plant Ontology Facilitates Comparisons of Plant Development Stages Across Species. Front. Plant Sci. 2019, 10, 631. [Google Scholar] [CrossRef] [PubMed]
- Urbanczyk-Wochniak, E.; Baxter, C.; Kolbe, A.; Kopka, J.; Sweetlove, L.J.; Fernie, A.R. Profiling of Diurnal Patterns of Metabolite and Transcript Abundance in Potato (Solanum tuberosum) Leaves. Planta 2005, 221, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Selegato, D.M.; Freire, R.T.; Pilon, A.C.; Biasetto, C.R.; de Oliveira, H.C.; de Abreu, L.M.; Araujo, A.R.; da Silva Bolzani, V.; Castro-Gamboa, I. Improvement of Bioactive Metabolite Production in Microbial Cultures—A Systems Approach by OSMAC and Deconvolution-Based 1HNMR Quantification. Magn. Reson. Chem. 2019, 57, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.; Villas-Boas, S.; Aggio, R. Analysis of Intracellular Metabolites from Microorganisms: Quenching and Extraction Protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef]
- Teng, Q.; Huang, W.; Collette, T.W.; Ekman, D.R.; Tan, C. A Direct Cell Quenching Method for Cell-Culture Based Metabolomics. Metabolomics 2009, 5, 199–208. [Google Scholar] [CrossRef]
- Mushtaq, M.Y.; Choi, Y.H.; Verpoorte, R.; Wilson, E.G. Extraction for Metabolomics: Access to The Metabolome. Phytochem. Anal. 2014, 25, 291–306. [Google Scholar] [CrossRef]
- Moreira, F.D.L.; Riul, T.B.; Moreira, M.D.L.; Pilon, A.C.; Dias-Baruffi, M.; Araújo, M.S.S.; Lopes, N.P.; de Oliveira, A.R.M. Leishmanicidal Effects of Piperlongumine (Piplartine) and Its Putative Metabolites. Planta Medica 2018, 84, 1141–1148. [Google Scholar] [CrossRef]
- Faijes, M.; Mars, A.E.; Smid, E.J. Comparison of Quenching and Extraction Methodologies for Metabolome Analysis of Lactobacillus Plantarum. Microb. Cell Factories 2007, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Sköld, K.; Svensson, M.; Norrman, M.; Sjögren, B.; Svenningsson, P.; Andrén, P.E. The Significance of Biochemical and Molecular Sample Integrity in Brain Proteomics and Peptidomics: Stathmin 2-20 and Peptides as Sample Quality Indicators. Proteomics 2007, 7, 4445–4456. [Google Scholar] [CrossRef]
- Hu, J.; Lu, J.; Zhang, X.; Wang, C.; Ren, K.; Chang, Q.; Ji, M.; Pan, W.; Ma, B.; Fan, W. Peptidomic Analysis on Synovial Tissue Reveals Galectin-1 Derived Peptide as a Potential Bioactive Molecule against Rheumatoid Arthritis. Cytokine 2020, 131, 155020. [Google Scholar] [CrossRef]
- Barashkova, A.S.; Rogozhin, E.A. Isolation of Antimicrobial Peptides from Different Plant Sources: Does a General Extraction Method Exist? Plant Methods 2020, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Tao, R.; Yang, B.; Zhang, H.; Chen, Y.Q.; Chen, H.; Chen, W. Optimization of the Quenching and Extraction Procedures for a Metabolomic Analysis of Lactobacillus plantarum. Anal. Biochem. 2018, 557, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wang, X.; Zhuang, Y.; Wang, G. A Systematic Evaluation of Quenching and Extraction Procedures for Quantitative Metabolome Profiling of HeLa Carcinoma Cell under 2D and 3D Cell Culture Conditions. Biotechnol. J. 2023, 18, 2200444. [Google Scholar] [CrossRef]
- Canelas, A.B.; Ten Pierick, A.; Ras, C.; Seifar, R.M.; Van Dam, J.C.; Van Gulik, W.M.; Heijnen, J.J. Quantitative Evaluation of Intracellular Metabolite Extraction Techniques for Yeast Metabolomics. Anal. Chem. 2009, 81, 7379–7389. [Google Scholar] [CrossRef]
- Jenkins, H.; Hardy, N.; Beckmann, M.; Draper, J.; Smith, A.R.; Taylor, J.; Fiehn, O.; Goodacre, R.; Bino, R.J.; Hall, R.; et al. A Proposed Framework for the Description of Plant Metabolomics Experiments and Their Results. Nat. Biotechnol. 2004, 22, 1601–1606. [Google Scholar] [CrossRef]
- Athersuch, T. Metabonomics. In Unraveling the Exposome: A Practical Review; Springer: Cham, Switzerland, 2018; Volume 455, pp. 141–181. [Google Scholar] [CrossRef]
- Harbourne, N.; Marete, E.; Jacquier, J.C.; O’Riordan, D. Effect of Drying Methods on the Phenolic Constituents of Meadowsweet (Filipendula ulmaria) and Willow (Salix alba). LWT-Food Sci. Technol. 2009, 42, 1468–1473. [Google Scholar] [CrossRef]
- Moonlight, P.W.; Banda-R, K.; Phillips, O.L.; Dexter, K.G.; Pennington, R.T.; Baker, T.R.; de Lima, H.C.; Fajardo, L.; González, M.R.; Linares-Palomino, R.; et al. Expanding Tropical Forest Monitoring into Dry Forests: The DRYFLOR Protocol for Permanent Plots. Plants People Planet 2021, 3, 295–300. [Google Scholar] [CrossRef]
- Wolfender, J.L.; Marti, G.; Thomas, A.; Bertrand, S. Current Approaches and Challenges for the Metabolite Profiling of Complex Natural Extracts. J. Chromatogr. A 2015, 1382, 136–164. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, Z.; Yang, N.; Zhou, K.; Hu, W.; Ou, S.; Liu, P. Widely Targeted UHPLC-MS/MS Metabolomic Analysis on the Chemical Variation in Blueberry-Filled Pastries During Processing. Front. Nutr. 2020, 7, 569172. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.T.R.; do Prado, R.M.; Porto, C.; dos Santos, G.T.; Huws, S.A.; Pilau, E.J. Exploring the Rumen Fluid Metabolome Using Liquid Chromatography-High-Resolution Mass Spectrometry and Molecular Networking. Sci. Rep. 2018, 8, 17971. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Witkamp, G.-J.; Verpoorte, R.; Choi, Y.H. Natural Deep Eutectic Solvents as a New Extraction Media for Phenolic Metabolites in Carthamus tinctorius L. Anal. Chem. 2013, 85, 6272–6278. [Google Scholar] [CrossRef]
- Fujii, T.; Matsuda, S.; Tejedor, M.L.; Esaki, T.; Sakane, I.; Mizuno, H.; Tsuyama, N.; Masujima, T. Direct Metabolomics for Plant Cells by Live Single-Cell Mass Spectrometry. Nat. Protoc. 2015, 10, 1445–1456. [Google Scholar] [CrossRef]
- Višnjevec, A.M.; Barp, L.; Lucci, P.; Moret, S. Pressurized Liquid Extraction for the Determination of Bioactive Compounds in Plants with Emphasis on Phenolics. TrAC Trends Anal. Chem. 2024, 173, 117620. [Google Scholar] [CrossRef]
- Gligor, O.; Mocan, A.; Moldovan, C.; Locatelli, M.; Crișan, G.; Ferreira, I.C. Enzyme-Assisted Extractions of Polyphenols—A Comprehensive Review. Trends Food Sci. Technol. 2019, 88, 302–315. [Google Scholar] [CrossRef]
- Wong, S.; Ngadi, N.; Inuwa, I.M.; Hassan, O. Recent Advances in Applications of Activated Carbon from Biowaste for Wastewater Treatment: A Short Review. J. Clean. Prod. 2018, 175, 361–375. [Google Scholar] [CrossRef]
- Kabir, A.; Olayanju, B. Chapter 5—Membrane-Based Extraction Techniques. In Green Analytical Chemistry; Locatelli, M., Kaya, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2025; pp. 119–144. [Google Scholar] [CrossRef]
- Van Eygen, G.; Van der Bruggen, B.; Buekenhoudt, A.; Luis Alconero, P. Efficient Membrane-Based Affinity Separations for Chemical Applications: A Review. Chem. Eng. Process.-Process Intensif. 2021, 169, 108613. [Google Scholar] [CrossRef]
- Khan, S.A.; Aslam, R.; Makroo, H.A. High Pressure Extraction and Its Application in the Extraction of Bio-Active Compounds: A Review. J. Food Process Eng. 2019, 42, e12896. [Google Scholar] [CrossRef]
- Funari, C.S.; Sutton, A.T.; Carneiro, R.L.; Fraige, K.; Cavalheiro, A.J.; da Silva Bolzani, V.; Hilder, E.F.; Arrua, R.D. Natural Deep Eutectic Solvents and Aqueous Solutions as an Alternative Extraction Media for Propolis. Food Res. Int. 2019, 125, 108559. [Google Scholar] [CrossRef] [PubMed]
- Borges, M.S.; Zanatta, A.C.; Souza, O.A.; Pelissari, J.H.; Camargo, J.G.S.; Carneiro, R.L.; Funari, C.S.; Bolzani, V.S.; Rinaldo, D. A Green and Sustainable Method for Monitoring the Chemical Composition of Soybean: An Alternative for Quality Control. Phytochem. Anal. 2021, 32, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global Metabolic Profiling of Animal and Human Tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Maltese, F.; Van Der Kooy, F.; Verpoorte, R. Solvent Derived Artifacts in Natural Products Chemistry. Nat. Prod. Commun. 2009, 4, 447–454. [Google Scholar] [CrossRef]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas Chromatography Mass Spectrometry-Based Metabolite Profiling in Plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Higgs, R.E.; Zahn, J.A.; Gygi, J.D.; Hilton, M.D. Rapid Method to Estimate the Presence of Secondary Metabolites in Microbial Extracts. Appl. Environ. Microbiol. 2001, 67, 371–376. [Google Scholar] [CrossRef]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted Large-Scale Plant Metabolomics Using Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; de Groot, J.; van Beek, T.A.; Vervoort, J.; de Vos, C.H.R. A Liquid Chromatography-Mass Spectrometry-Based Metabolome Database for Tomato. Plant Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef]
- Vorst, O.; de Vos, C.H.R.; Lommen, A.; Staps, R.V.; Visser, R.G.F.; Bino, R.J.; Hall, R.D. A Non-Directed Approach to the Differential Analysis of Multiple LC–MS-Derived Metabolic Profiles. Metabolomics 2005, 1, 169–180. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Stanley, G.H.S. A Simple Method for The Isolation and Purification of Total Lipides from Animal Tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid Extraction by Methyl-Tert-Butyl Ether for High-Throughput Lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Löfgren, L.; Ståhlman, M.; Forsberg, G.-B.; Saarinen, S.; Nilsson, R.; Hansson, G.I. The BUME Method: A Novel Automated Chloroform-Free 96-Well Total Lipid Extraction Method for Blood Plasma. J. Lipid Res. 2012, 53, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Löfgren, L.; Forsberg, G.-B.; Ståhlman, M. The BUME Method: A New Rapid and Simple Chloroform-Free Method for Total Lipid Extraction of Animal Tissue. Sci. Rep. 2016, 6, 27688. [Google Scholar] [CrossRef]
- Creydt, M.; Arndt, M.; Hudzik, D.; Fischer, M. Plant Metabolomics: Evaluation of Different Extraction Parameters for Nontargeted UPLC-ESI-QTOF-Mass Spectrometry at the Example of White Asparagus Officinalis. J. Agric. Food Chem. 2018, 66, 12876–12887. [Google Scholar] [CrossRef] [PubMed]
- Romsdahl, T.B.; Cocuron, J.-C.; Pearson, M.J.; Alonso, A.P.; Chapman, K.D. A Lipidomics Platform to Analyze the Fatty Acid Compositions of Non-Polar and Polar Lipid Molecular Species from Plant Tissues: Examples from Developing Seeds and Seedlings of Pennycress (Thlaspi arvense). Front. Plant Sci. 2022, 13, 1038161. [Google Scholar] [CrossRef]
- Hu, A.; Wei, F.; Huang, F.; Xie, Y.; Wu, B.; Lv, X.; Chen, H. Comprehensive and High-Coverage Lipidomic Analysis of Oilseeds Based on Ultrahigh-Performance Liquid Chromatography Coupled with Electrospray Ionization Quadrupole Time-of-Flight Mass Spectrometry. J. Agric. Food Chem. 2021, 69, 8964–8980. [Google Scholar] [CrossRef] [PubMed]
- Sedio, B.E.; Boya, P.C.A.; Rojas Echeverri, J.C. A Protocol for High-Throughput, Untargeted Forest Community Metabolomics Using Mass Spectrometry Molecular Networks. Appl. Plant Sci. 2018, 6, e1033. [Google Scholar] [CrossRef]
- Bijttebier, S.; Van der Auwera, A.; Foubert, K.; Voorspoels, S.; Pieters, L.; Apers, S. Bridging the Gap between Comprehensive Extraction Protocols in Plant Metabolomics Studies and Method Validation. Anal. Chim. Acta 2016, 935, 136–150. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Urdiola, R.; Borràs, E.; Valverde, F.; Matus, J.T.; Sabidó, E.; Riechmann, J.L. Peptidomics Methods Applied to the Study of Flower Development. In Flower Development: Methods and Protocols; Riechmann, J.L., Ferrándiz, C., Eds.; Springer: New York, NY, USA, 2023; pp. 509–536. [Google Scholar] [CrossRef]
- Van Dijck, A.; Hayakawa, E.; Landuyt, B.; Baggerman, G.; Van Dam, D.; Luyten, W.; Schoofs, L.; De Deyn, P.P. Comparison of Extraction Methods for Peptidomics Analysis of Mouse Brain Tissue. J. Neurosci. Methods 2011, 197, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Ernst, M.; Silva, D.B.; Silva, R.; Monge, M.; Semir, J.; Vêncio, R.Z.N.; Lopes, N.P. A Metabolomic Protocol for Plant Systematics by Matrix-Assisted Laser-Desorption/Ionization Time-of Flight Mass Spectrometry. Anal. Chim. Acta 2015, 859, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J.; Troncoso-Ponce, M.A.; Ratcliffe, R.G. 1H NMR Metabolite Fingerprinting and Metabolomic Analysis of Perchloric Acid Extracts from Plant Tissues. Nat. Protoc. 2008, 3, 1001–1012. [Google Scholar] [CrossRef]
- Wu, X.; Li, N.; Li, H.; Tang, H. An Optimized Method for NMR-Based Plant Seed Metabolomic Analysis with Maximized Polar Metabolite Extraction Efficiency, Signal-to-Noise Ratio, and Chemical Shift Consistency. Analyst 2014, 139, 1769–1778. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Jones, R.R.; Mitchell, S.B.; Broeckling, C.D.; Heuberger, A.L.; Shafizadeh, T.; Watkins, S.; Prenni, J.E. Multielement Profiling of Diverse Food Samples. ACS Food Sci. Technol. 2023, 3, 459–464. [Google Scholar] [CrossRef]
- Nardin, R.; Tamasi, G.; Baglioni, M.; Bisozzi, F.; Consumi, M.; Costa, J.; Fattori, G.; Tozzi, C.; Riccaboni, A.; Rossi, C. Determination of Elemental Content in Vineyard Soil, Leaves, and Grapes of Sangiovese Grapes from the Chianti Region Using ICP-MS for Geographical Identification. ACS Food Sci. Technol. 2024, 4, 2585–2599. [Google Scholar] [CrossRef] [PubMed]
- Beyzi, E. Chemometric Assessment of the Chemical Profile of Tea Seed (Camellia sinensis) with Different Size Determined by GC and ICP/OES. Eur. Food Res. Technol. 2024, 250, 1229–1237. [Google Scholar] [CrossRef]
- Kruszewska, J.; Kur, A.; Kulpińska, D.; Grabowska-Jadach, I.; Matczuk, M.; Keppler, B.K.; Timerbaev, A.R.; Jarosz, M. An Improved Protocol for ICP-MS-Based Assessment of the Cellular Uptake of Metal-Based Nanoparticles. J. Pharm. Biomed. Anal. 2019, 174, 300–304. [Google Scholar] [CrossRef]
- Gonçalves, E.S.; Borges, R.M.; De Carvalho, L.V.B.; Alves, S.R.; André, L.C.; Moreira, J.C. Estratégias analíticas com cromatografia e espectrometria de massas para biomonitorização da exposição ao benzeno pela determinação do ácido S-fenilmercaptúrico urinário. Rev. Bras. Saúde Ocup. 2017, 42, e1s. [Google Scholar] [CrossRef]
- Nasiri, A.; Jahani, R.; Mokhtari, S.; Yazdanpanah, H.; Daraei, B.; Faizi, M.; Kobarfard, F. Overview, Consequences, and Strategies for Overcoming Matrix Effects in LC-MS Analysis: A Critical Review. Analyst 2021, 146, 6049–6063. [Google Scholar] [CrossRef]
- Balcells, C.; Foguet, C.; Tarragó-Celada, J.; de Atauri, P.; Marin, S.; Cascante, M. Tracing Metabolic Fluxes Using Mass Spectrometry: Stable Isotope-Resolved Metabolomics in Health and Disease. TrAC Trends Anal. Chem. 2019, 120, 115371. [Google Scholar] [CrossRef]
- Babele, P.K.; Young, J.D. Applications of Stable Isotope-Based Metabolomics and Fluxomics toward Synthetic Biology of Cyanobacteria. WIREs Syst. Biol. Med. 2020, 12, e1472. [Google Scholar] [CrossRef]
- Kašička, V. Peptide Mapping of Proteins by Capillary Electromigration Methods. J. Sep. Sci. 2022, 45, 4245–4279. [Google Scholar] [CrossRef] [PubMed]
- Foreman, R.E.; George, A.L.; Reimann, F.; Gribble, F.M.; Kay, R.G. Peptidomics: A Review of Clinical Applications and Methodologies. J. Proteome Res. 2021, 20, 3782–3797. [Google Scholar] [CrossRef] [PubMed]
- Rund, K.M.; Carpanedo, L.; Lauterbach, R.; Wermund, T.; West, A.L.; Wende, L.M.; Calder, P.C.; Schebb, N.H. LC-ESI-HRMS—Lipidomics of Phospholipids. Anal. Bioanal. Chem. 2024, 416, 925–944. [Google Scholar] [CrossRef]
- Liakh, I.; Sledzinski, T.; Kaska, L.; Mozolewska, P.; Mika, A. Sample Preparation Methods for Lipidomics Approaches Used in Studies of Obesity. Molecules 2020, 25, 5307. [Google Scholar] [CrossRef]
- ul Ain, Q.; Hocking, R.K.; Mahon, P.J.; Bhave, M.; Butardo, V. Chapter 3—Ionomics-Based Imaging, Localization and Quantification of Zinc and Other Micronutrients in Rice Grains for Biofortification Research. In Genetic Engineering and Genome Editing for Zinc Biofortification of Rice; Swamy, B.P.M., Macovei, A., Trijatmiko, K.R., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 31–43. [Google Scholar] [CrossRef]
- Dunn, W.B.; Kuligowski, J.; Lewis, M.; Mosley, J.D.; Schock, T.; Holland, C.U.; Zanetti, K.A.; Vuckovic, D. Metabolomics 2022 Workshop Report: State of QA/QC Best Practices in LC–MS-Based Untargeted Metabolomics, Informed through mQACC Community Engagement Initiatives. Metabolomics 2023, 19, 93. [Google Scholar] [CrossRef] [PubMed]
- Broeckling, C.D.; Beger, R.D.; Cheng, L.L.; Cumeras, R.; Cuthbertson, D.J.; Dasari, S.; Davis, W.C.; Dunn, W.B.; Evans, A.M.; Fernández-Ochoa, A.; et al. Current Practices in LC-MS Untargeted Metabolomics: A Scoping Review on the Use of Pooled Quality Control Samples. Anal. Chem. 2023, 95, 18645–18654. [Google Scholar] [CrossRef]
- Kirwan, J.A.; Gika, H.; Beger, R.D.; Bearden, D.; Dunn, W.B.; Goodacre, R.; Theodoridis, G.; Witting, M.; Yu, L.-R.; Wilson, I.D. Quality Assurance and Quality Control Reporting in Untargeted Metabolic Phenotyping: mQACC Recommendations for Analytical Quality Management. Metabolomics 2022, 18, 70. [Google Scholar] [CrossRef] [PubMed]
| Feature | Analytical Technique | |||||
|---|---|---|---|---|---|---|
| LC-MS | GC-MS | CE-MS | ICP-MS | MALDI-MS | NMR | |
| Amount of sample preparation | ++ | +++ | ++ | +++ | +++ | + |
| Volume for injection | ++ | ++ | + | +++ | ++ | +++ |
| Range of metabolites | RP: non-polar HILIC: polar | Volatile and thermostable | Polar | Elements | Polar and non-polar | Polar and non-polar |
| Sensitivity | +++ | +++ | + | +++ | +++ | + |
| Resolution | +++ | +++ | + | +++ | +++ | + |
| Quantification | IS needed | IS | IS | IS | IS | |
| Reproducibility | <NMR | <NMR | High | <NMR | High | |
| Identification of metabolites | Difficult (few or no standard libraries) | Easy (large spectral libraries) | Difficult (databases with few data) | Easy | Difficult (few or no standard libraries) | Easy |
| Drying Method | Advantages | Limitations | Ref. |
|---|---|---|---|
| Oven Drying | Efficient and cost-effective; suitable for large sample volumes. | Prolonged exposure to heat may degrade heat-sensitive metabolites; risk of inconsistent drying if temperature fluctuates. | [19,75,79,93] |
| Air Drying | Low-cost; no specialized equipment needed. | Longer drying times; potential for microbial contamination in humid conditions. | [19,21,79,94] |
| Vacuum Drying | Minimizes oxidation and thermal degradation; effective for heat-sensitive compounds. | Equipment cost; potential loss of volatile compounds. | [19,24,79,85] |
| Freeze-Drying | Preserves heat-sensitive metabolites; produces highly reproducible results. | Expensive; loss of volatiles; thermal shock may affect metabolite adherence. | [23,24,79,95] |
| Drying with Silica Gel | Portable; suitable for fieldwork. | Limited capacity; inconsistent drying efficiency compared to lab-based methods. | [19,21,94] |
| Characteristics | Analytical Method | Ref. |
|---|---|---|
| Volatile | - Steam distillation- Microwave-assisted extraction- Cold plasma-assisted extraction- Liquid pressurization extraction (LPE)- Hyperbaric extraction | [18,85,99,109] |
| Non-Volatile | - Solvent extraction (maceration)- Supercritical fluid extraction (e.g., CO2)- Membrane extraction- Ultrasound-assisted extraction | [18,85,98,106,107] |
| Polar | - Methanol–water mixtures (50–90% methanol)- Methanol, water, and chloroform (2:2:1)- Buffers for NMR to control pH | [85,87,108,109,110] |
| Non-Polar | - Chloroform-free methods (e.g., MTBE-based protocols)- Lipid extraction protocols (e.g., Folch, Bligh and Dyer, Matyash, BUME) [108,109,110]- Supercritical CO2 | [115,116,117,118,119] |
| Analytical Method | Characteristics | Ref. |
|---|---|---|
| GC-ToF-MS | Established protocol with a large database for compound identification. Difficult to detect secondary metabolites. | [110] |
| GC-MS and LC-MS | Focused on pre- and post-treatment data. | [13] |
| LC-QToF-MS | Established protocol for analysis of semi-polar metabolites, mainly secondary metabolites. | [112] |
| UHPLC-MS | Protocol for analysis of large data sets using molecular networking (identifying tool). | [123] |
| LC-MS | Optimization of extraction steps evaluating efficiency, repeatability, and ionization efficiency | [124,125,126] |
| MALDI-ToF MS | Protocol for MALDI-TOF-MS with multivariate analysis and taxonomic approach. | [127] |
| NMR | Use perchloric acid as extraction solvent. Used mainly for primary metabolism of plants. | [128] |
| NMR | Relatively simple protocol including different groups of primary/secondary metabolites. | [19] |
| NMR | Quality for extraction of tissues for NMR. Also applied for quantitative metabolomics. | [129] |
| LC-MS | Used for live single-cell metabolomics. | [87] |
| ICP-OES and ICP-MS | Used for elemental analysis of plant material. | [130,131,132] |
| ICP-MS | Analysis of metals in nanoparticles | [133] |
| Quality Assurance and Control | Minimum Reports | Best Practices |
|---|---|---|
| QC Sample types | Specify the types of QC samples (e.g., pooled samples). | Use multiple QC types (e.g., intra- and inter-batch) and ensure consistency. |
| Blanks and System Suitability | Identify blank types used to detect contamination. | Differentiate between true blanks and process blanks; provide details of composition, suppliers, and preparation methods. |
| Internal Standards | Report concentration and source of internal standards used. | Include internal standard normalization details, signal tracking over time, and exact time of addition in analysis. |
| Preparation and Storage | Report preparation and storage conditions, including freeze–thaw cycles. | Include detailed aliquoting information and monitor freeze–thaw stability with evidence of sample integrity. |
| Data Normalization and Scaling | Provide basic normalization details, e.g., total area or selected features. | Report all normalization, scaling, and transformation steps applied, with justification for each. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, W.B.; Hispagnol, G.F.; dos Santos Nunes, E.V.; Castro-Gamboa, I.; Pilon, A.C. Plant Sample Preparation for Metabolomics, Lipidomics, Ionomics, Fluxomics, and Peptidomics. Separations 2025, 12, 21. https://doi.org/10.3390/separations12020021
da Silva WB, Hispagnol GF, dos Santos Nunes EV, Castro-Gamboa I, Pilon AC. Plant Sample Preparation for Metabolomics, Lipidomics, Ionomics, Fluxomics, and Peptidomics. Separations. 2025; 12(2):21. https://doi.org/10.3390/separations12020021
Chicago/Turabian Styleda Silva, Walace Breno, Gabriel Felipe Hispagnol, Emanuel Victor dos Santos Nunes, Ian Castro-Gamboa, and Alan Cesar Pilon. 2025. "Plant Sample Preparation for Metabolomics, Lipidomics, Ionomics, Fluxomics, and Peptidomics" Separations 12, no. 2: 21. https://doi.org/10.3390/separations12020021
APA Styleda Silva, W. B., Hispagnol, G. F., dos Santos Nunes, E. V., Castro-Gamboa, I., & Pilon, A. C. (2025). Plant Sample Preparation for Metabolomics, Lipidomics, Ionomics, Fluxomics, and Peptidomics. Separations, 12(2), 21. https://doi.org/10.3390/separations12020021








