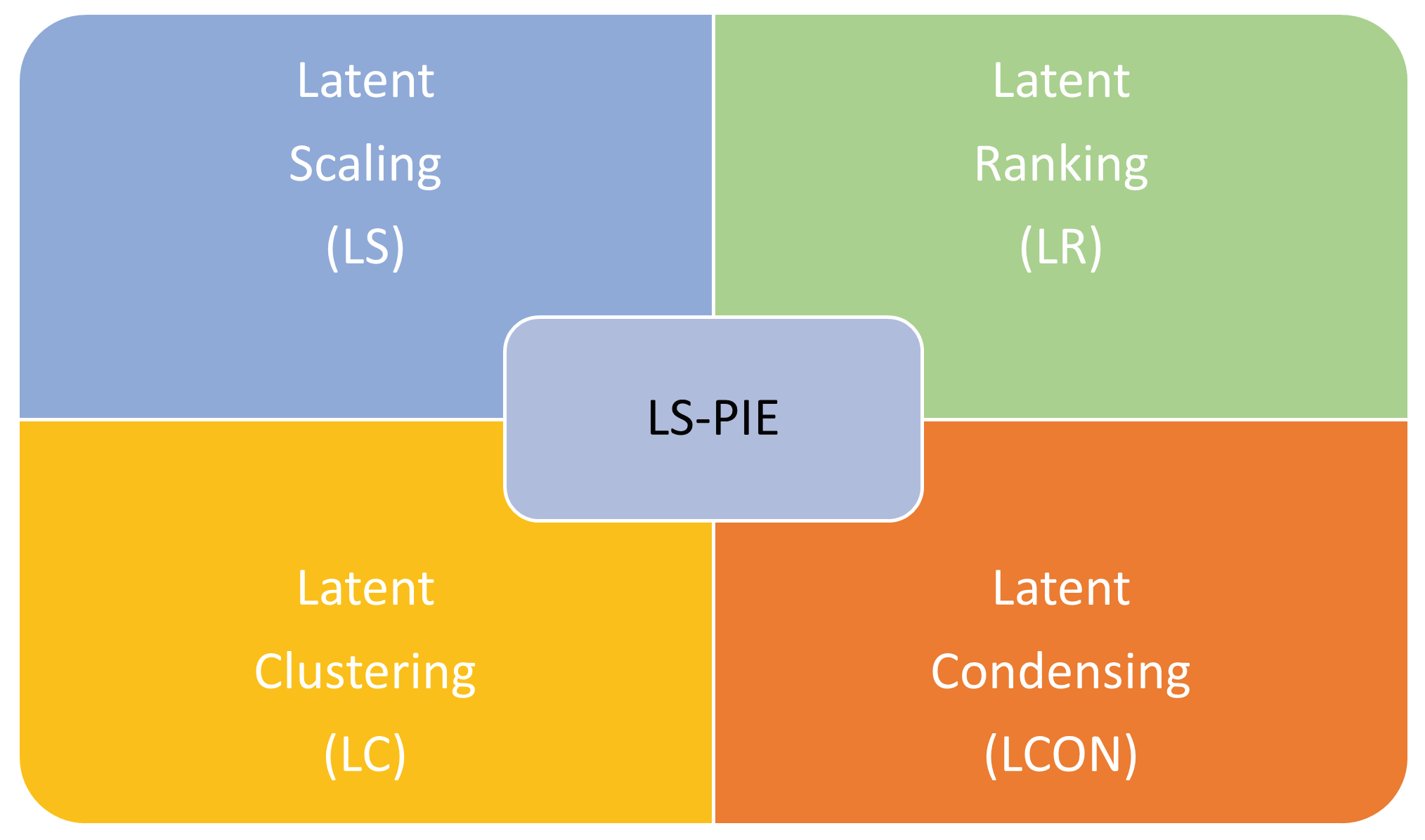
Latent Space Perspicacity and Interpretation Enhancement (LS-PIE) Framework
Abstract
1. Introduction
2. Required Background
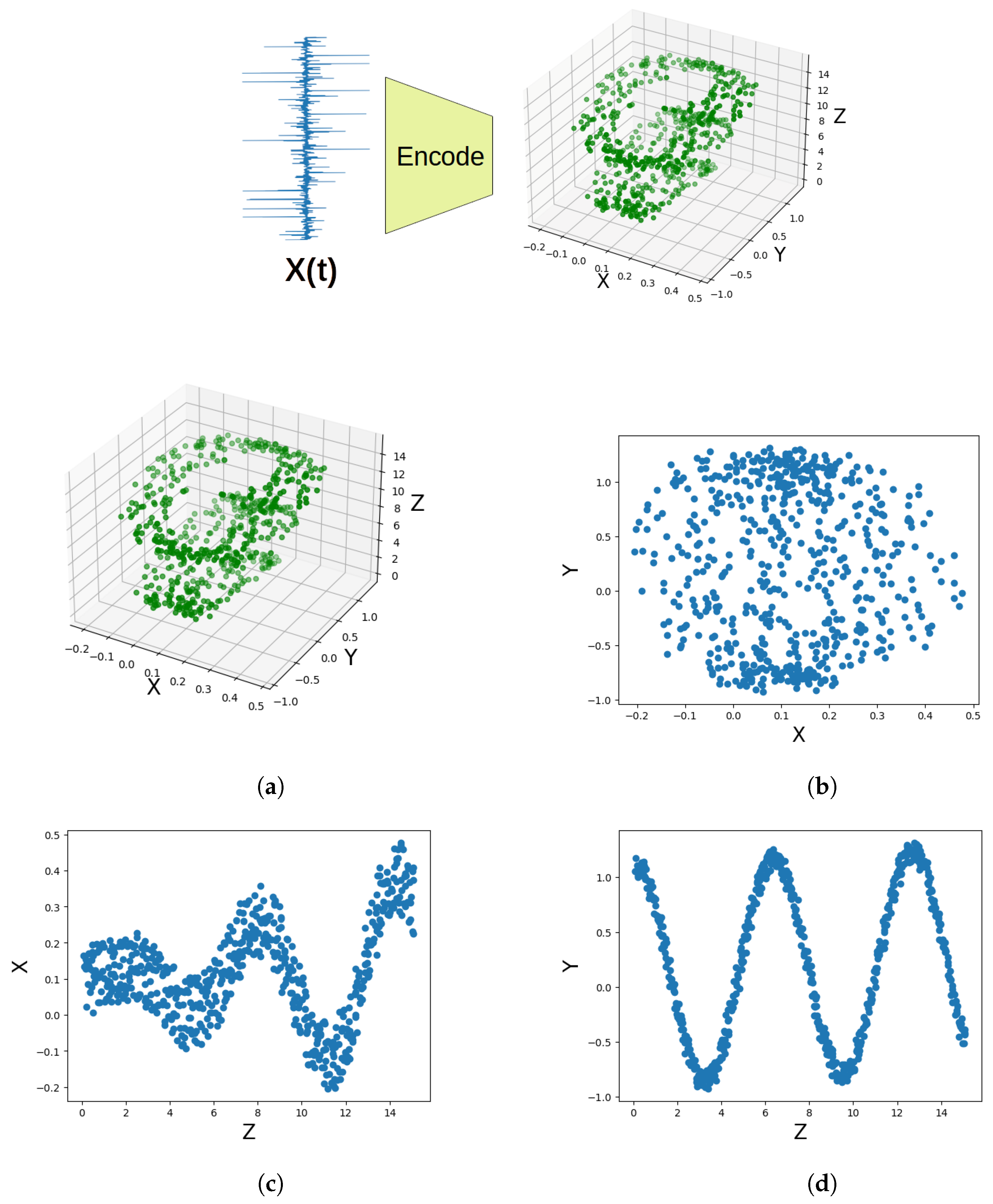
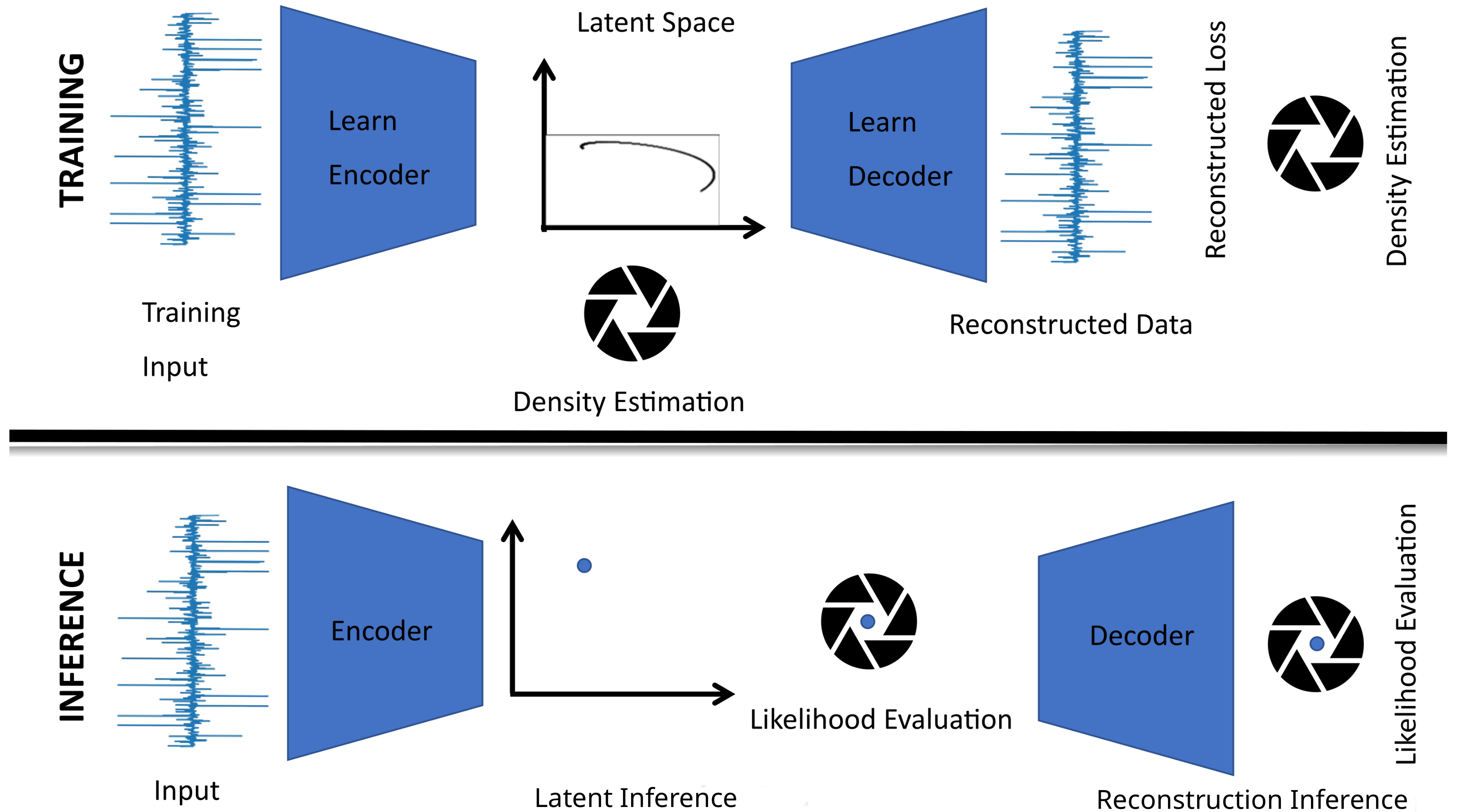
2.1. Latent Variable Models
- Data standardisation through mean centring or whitening of the data, .
- Compute the covariance matrix of the standardised time-series data .
- Find latent directions for the data by maximising or minimising an objective function plus regularisation terms subject to equality (equality constraints between latent directions are often enforced, e.g., orthogonality of the latent directions or some transformed representation of the latent directions. This can always be solved by direct optimisation, but solving the first-order necessary optimality condition, or a matrix decomposition may in some cases be computationally more efficient [15]) and inequality constraints [15]. PCA diagonalises the covariance matrix by finding its eigenvectors and eigenvalues, where eigenvectors represent the principal components and eigenvalues represent the variance each principal component explains.
- Select k latent directions from a maximum of . Eigenvalues and their associated eigenvectors are automatically sorted in descending order, from which k eigenvectors associated with the largest eigenvalues are usually selected. By choosing eigenvectors corresponding to the largest eigenvalues, the LVM prioritises reconstruction as they capture the most significant variation in the time-series data.
- The latent representation for a sample is obtained by projecting standardised time-series data onto k selected latent directions (a.k.a. loadings) to obtain a k-dimensional latent representation of the sample often referred to as a k-dimensional score.
- Reconstructing the sample from the latent representation merely requires the summation of each component of the k-dimensional score multiplied by their respective latent direction.
2.2. Data Sources and Channels
3. Materials and Methods
4. Related Work
5. Software Description
5.1. Latent Scaling (LS)
| Algorithm 1 Latent scaling (LS) |
|
5.2. Latent Ranking (LR)
| Algorithm 2 Latent ranking (LR) |
|
5.3. Latent Clustering (LC)
| Algorithm 3 Latent clustering (LC) |
|
5.4. Latent Condensing (LCON)
- Systematically reducing the latent dimensions solved by the algorithm and minimising or maximising a selected clustering index to find the optimal number of clusters.
| Algorithm 4 Latent condensing (LCON) for Hankelised time-series data |
|
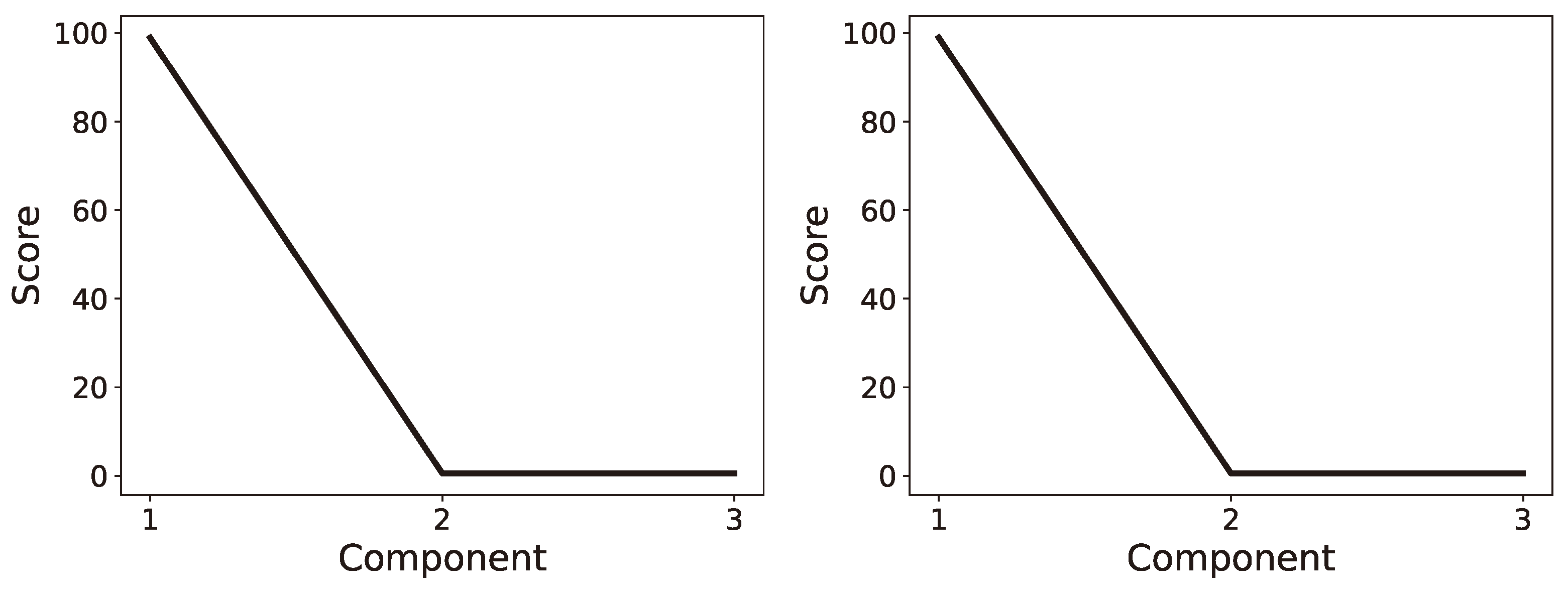
6. Numerical Investigation
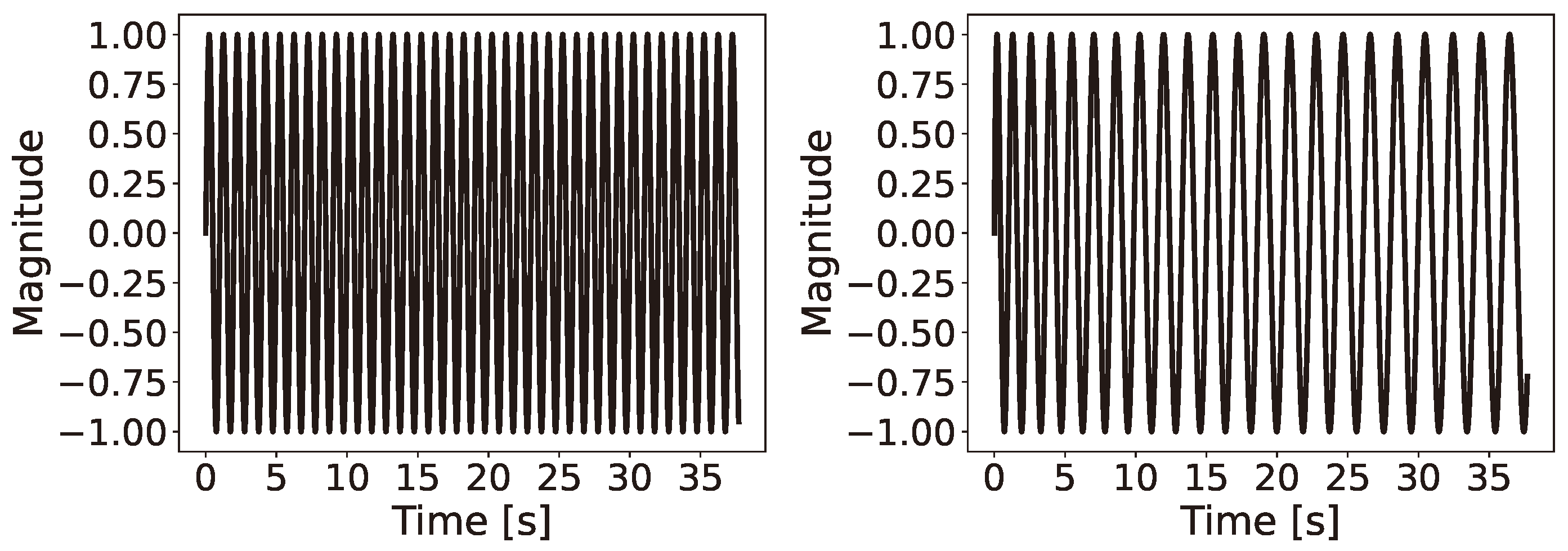
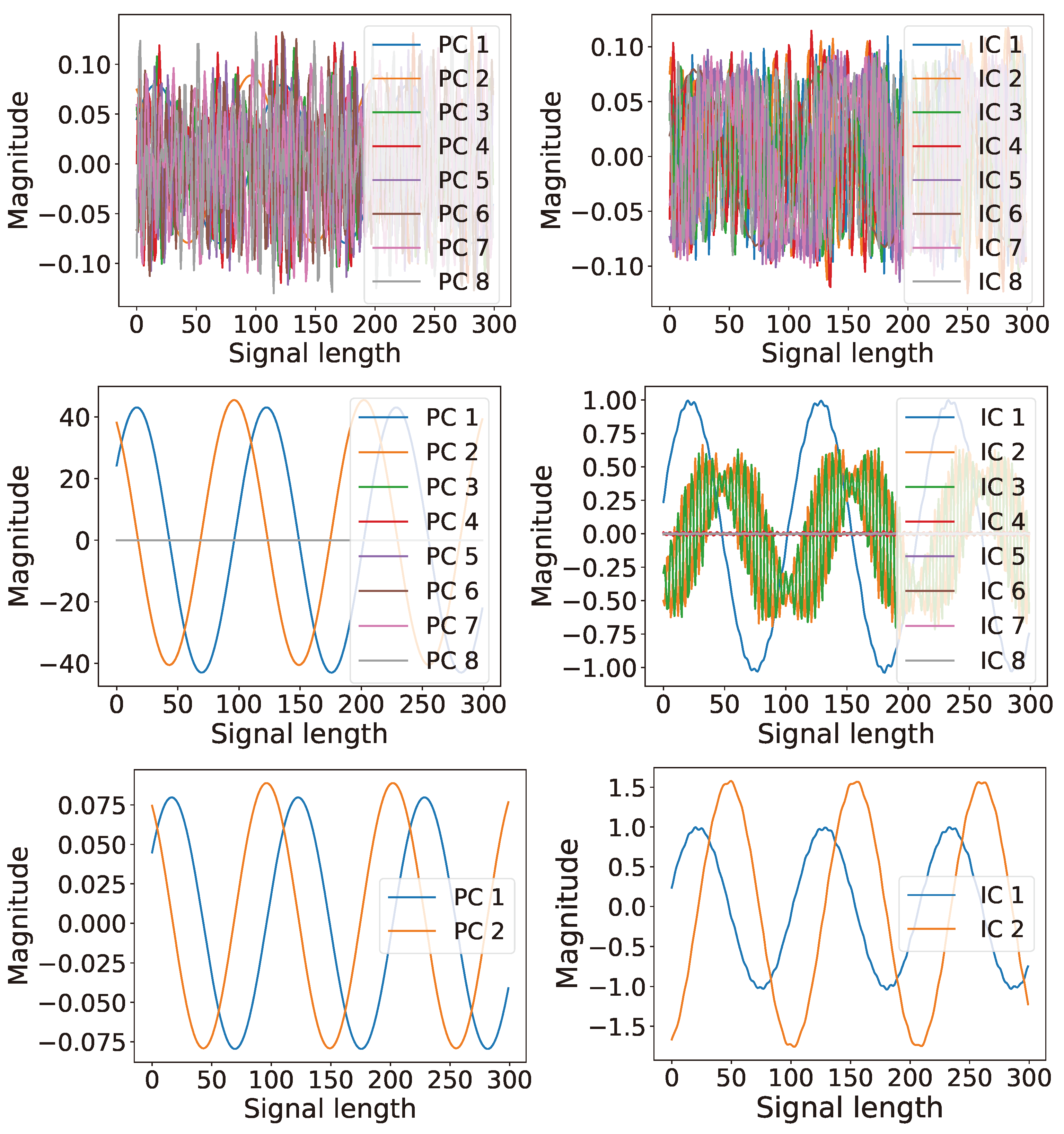
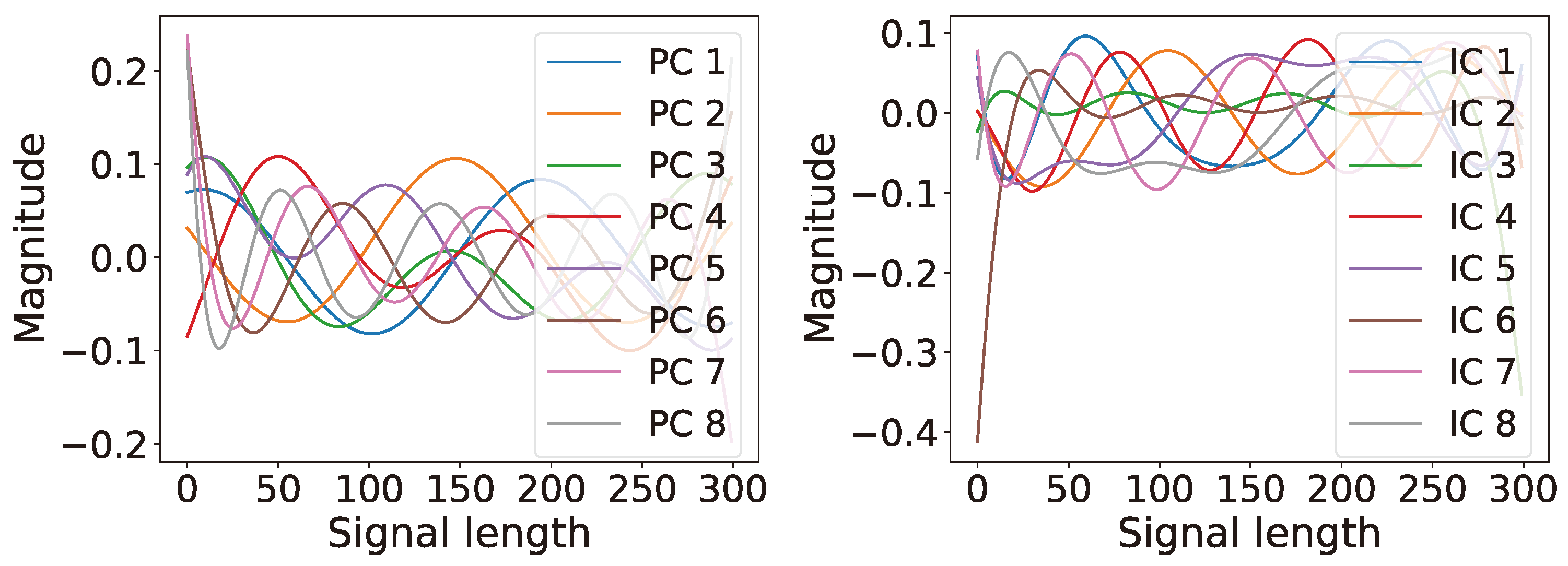
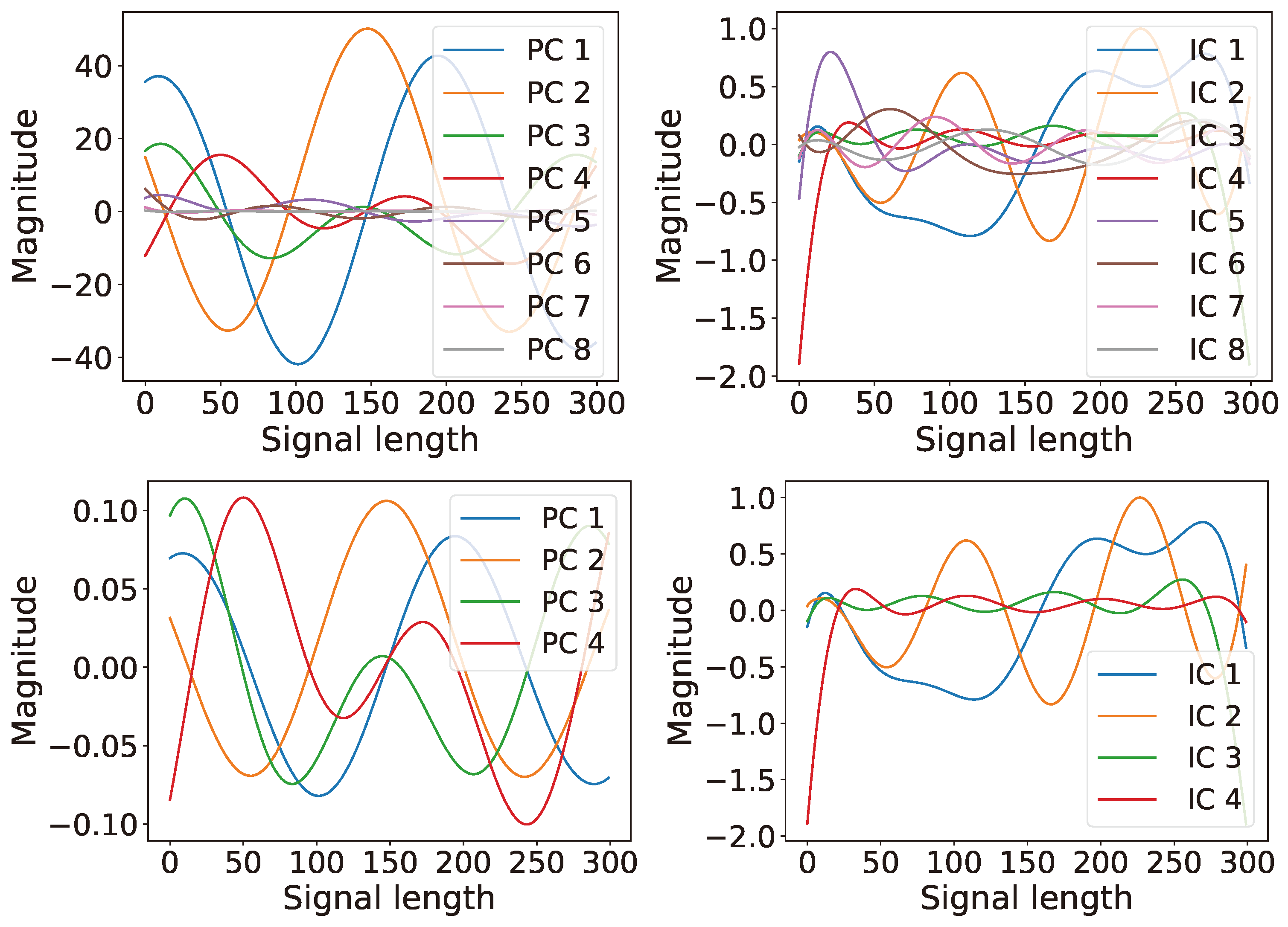
6.1. Single Channel: Latent Ranking (LR), Latent Scaling (LS), and Latent Condensing (LCON)
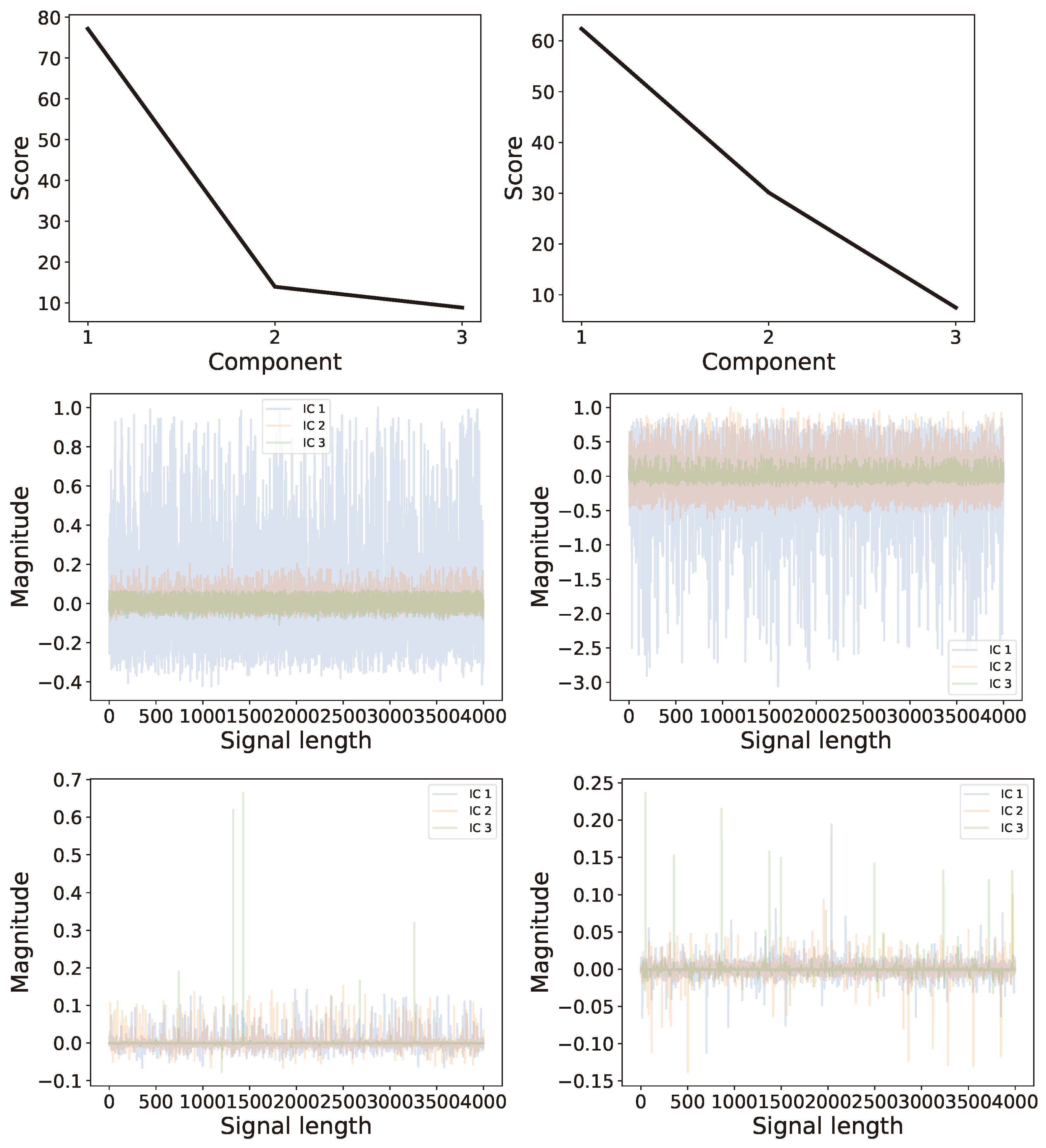
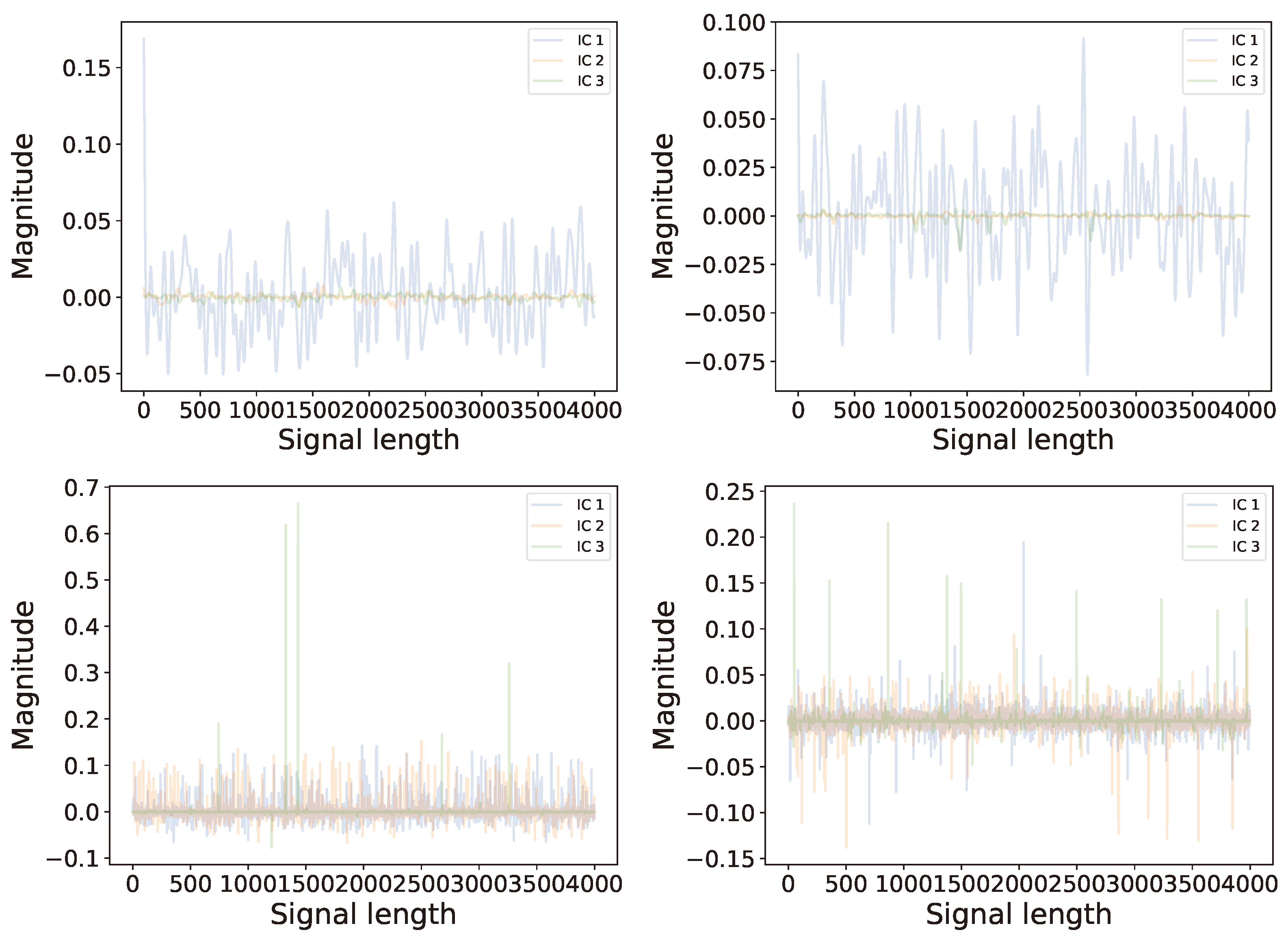
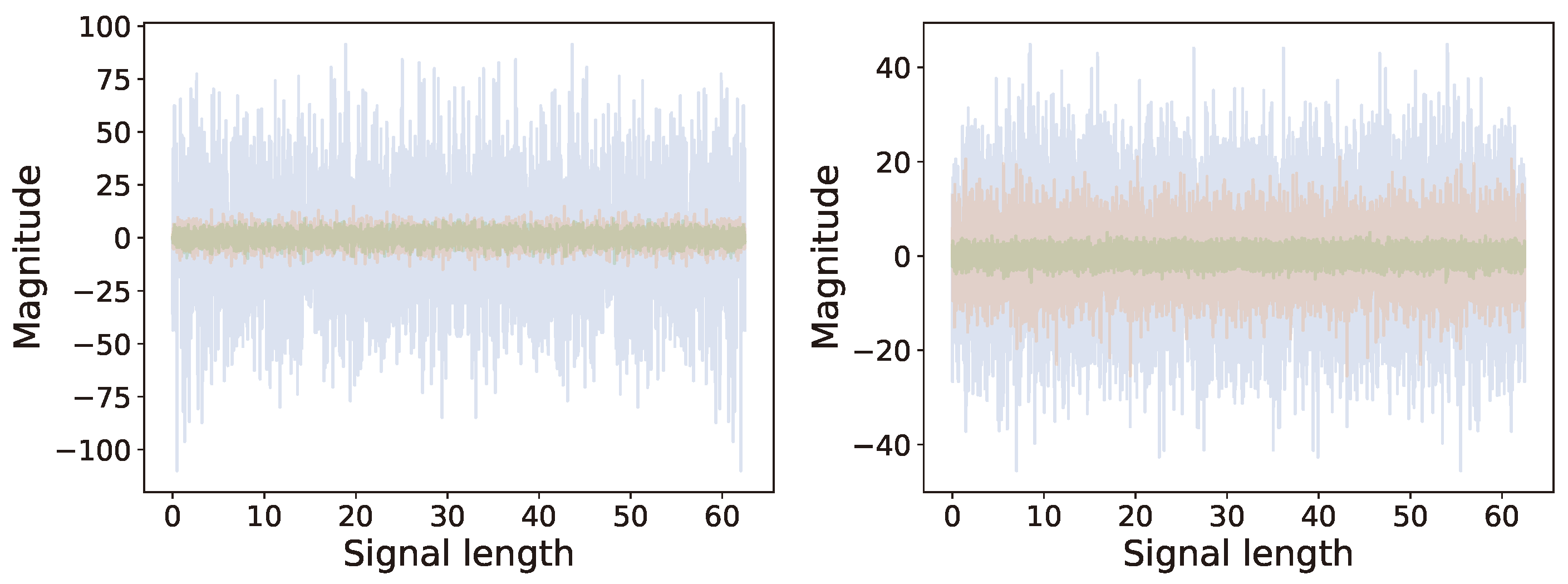
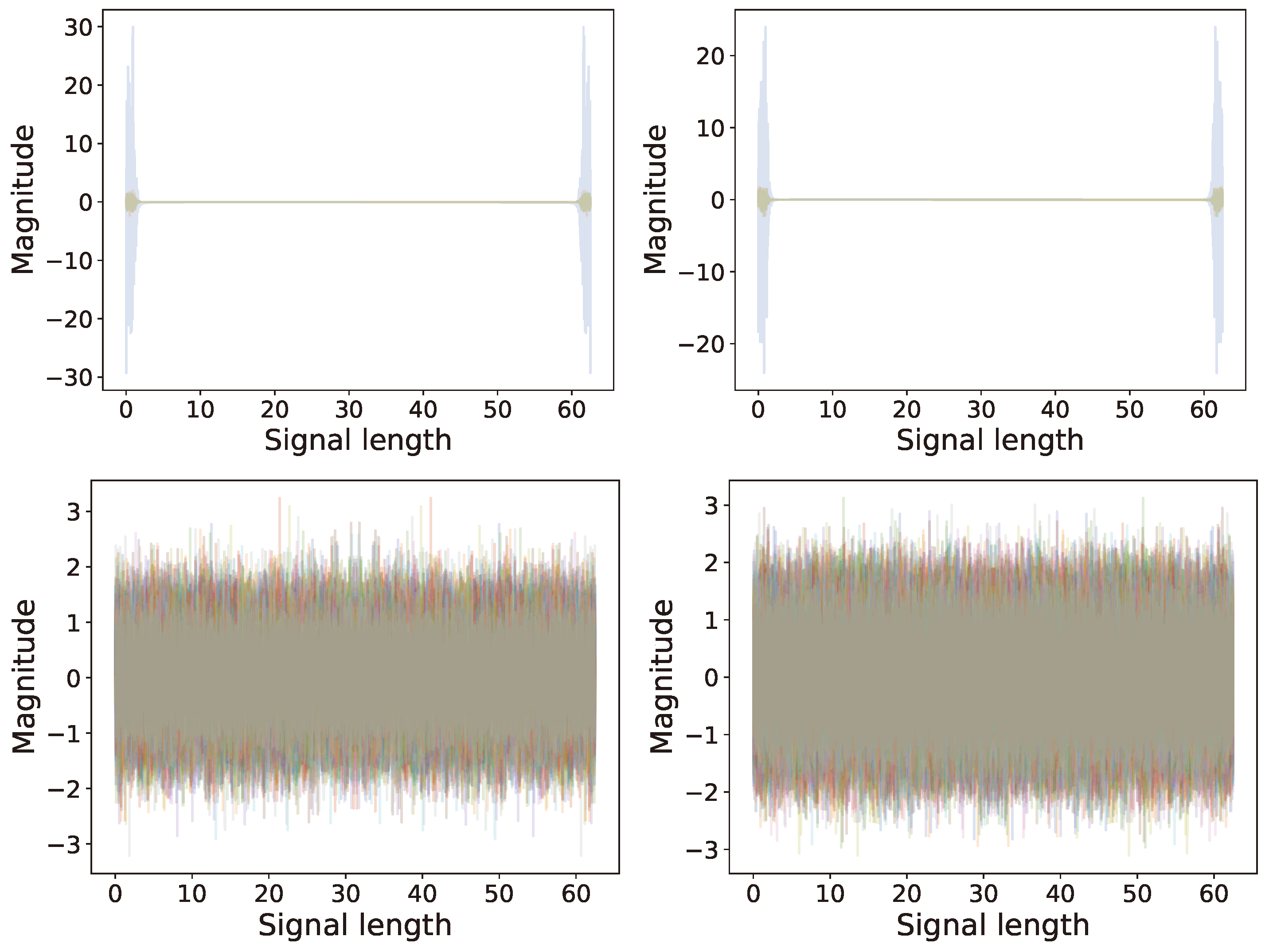
6.2. Multi-Channel Real World Data
6.2.1. Background
6.2.2. Analysis
6.2.3. Comparison of Results
6.3. Discussion
6.4. Discussion of Use Cases
7. Impact
8. Conclusions
- LS-PIE is applicable to source- and reconstruction-focused LVMs;
- It returns interpretable components;
- (a)
- Ranking and scaling help to organize latent spaces;
- (b)
- Clustering and condensing help solve for optimal representations;
- The approach is applicable to both artificial and real-world data;
- (a)
- In both cases, it improves the latent interpretability;
- (b)
- The real-world data showcase their applicability as pre-processing metrics;
- The algorithm is designed to be fully customisable to a users requirements.
9. Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| LS-PIE | Latent Space Perspicacity and Interpretation Enhancement |
| ICA | Independent Component Analysis |
| PCA | Principal Component Analysis |
| CCA | Canonical correlation analysis |
| SVD | Singular Value Decomposition |
| FA | Factor Analysis |
| LR | Latent Ranking |
| LS | Latent Scaling |
| LC | Latent Clustering |
| LCON | Latent Condensing |
| LVM | Latent Variable model |
| LDLVM | Lightweight Deep Latent Variable Model |
| AE | Autoencoder |
| VAE | Variational Autoencoder |
| SSA | Singular Spectrum Analysis |
| BSS | Blind Source Separation |
| PARAFAC | Parallel Factor Analysis |
References
- Booyse, W.; Wilke, D.N.; Heyns, S. Deep digital twins for detection, diagnostics and prognostics. Mech. Syst. Signal Process. 2020, 140, 106612. [Google Scholar] [CrossRef]
- Wilke, D.N. Digital Twins for Physical Asset Lifecycle Management. In Digital Twins: Basics and Applications; Lv, Z., Fersman, E., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 13–26. [Google Scholar] [CrossRef]
- Wilke, D.N. Lecture Notes in Optimum Design for Information Extraction from Data: Mastering Unsupervised Learning; Department of Mechanical and Aeronautical Engineering, Univerrsity of Pretoria: Pretoria, South Africa, 2021. [Google Scholar]
- Wilke, D.N.; Heyns, P.S.; Schmidt, S. The Role of Untangled Latent Spaces in Unsupervised Learning Applied to Condition-Based Maintenance. In Modelling and Simulation of Complex Systems for Sustainable Energy Efficiency; Hammami, A., Heyns, P.S., Schmidt, S., Chaari, F., Abbes, M.S., Haddar, M., Eds.; Springer: Cham, Switzerland, 2022; pp. 38–49. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Altman, N. Points of Significance: Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef]
- Jaadi, Z.; Powers, J.; Pierre, S. Principal Component Analysis (PCA) Explained; Built In: Chicago, IL, USA, 2022. [Google Scholar]
- Golyandina, N.; Zhigljavsky, A. Singular Spectrum Analysis for Time Series; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar] [CrossRef]
- Wilke, D.N.; Schmidt, S.; Heyns, P.S. A Review of Singular Spectral Analysis to Extract Components from Gearbox Data. In International Workshop on Modelling and Simulation of Complex Systems for Sustainable Energy Efficiency; Applied Condition Monitoring; Springer: Cham, Switzerland, 2021; Volume 20, pp. 160–172. [Google Scholar]
- Tharwat, A. Independent component analysis: An introduction. Appl. Comput. Inform. 2018, 17, 222–249. [Google Scholar] [CrossRef]
- De Lathauwer, L.; De Moor, B.; Vandewalle, J. An introduction to independent component analysis. J. Chemom. 2000, 14, 123–149. [Google Scholar] [CrossRef]
- Hyvärinen, A.; Karhunen, J.; Oja, E. What is Independent Component Analysis? In Independent Component Analysis; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2001; Chapter 7; pp. 145–164. [Google Scholar] [CrossRef]
- Westad, F. Independent component analysis and regression applied on sensory data. J. Chemom. 2005, 19, 171–179. [Google Scholar] [CrossRef]
- Kohonen, T. An adaptive associative memory principle. IEEE Trans. Comput. 1974, 100, 444–445. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Snyman, J.A.; Wilke, D.N. Practical Mathematical Optimization, 2nd ed.; Springer Optimization and Its Applications; Springer: Cham, Switzerland, 2018; pp. XXVI+372. [Google Scholar] [CrossRef]
- Marino, J.; Chen, L.; He, J.; Mandt, S. Improving sequential latent variable models with autoregressive flows. Mach. Learn. 2022, 111, 1597–1620. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, C. Deep latent variable models for generating knockoffs. Stat 2019, 8, e260. [Google Scholar] [CrossRef]
- Candes, E.; Fan, Y.; Janson, L.; Lv, J. Panning for Gold: Model-X Knockoffs for High-dimensional Controlled Variable Selection. arXiv 2016, arXiv:1610.02351. [Google Scholar] [CrossRef]
- Chandra, R.; Jain, M.; Maharana, M.; Krivitsky, P.N. Revisiting Bayesian Autoencoders With MCMC. IEEE Access 2022, 10, 40482–40495. [Google Scholar] [CrossRef]
- Song, G.; Wang, S.; Huang, Q.; Tian, Q. Harmonized Multimodal Learning with Gaussian Process Latent Variable Models. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 43, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Jiang, X.; Zhang, B.; Yuan, J.; Ge, Z. Latent variable models in the era of industrial big data: Extension and beyond. Annu. Rev. Control 2022, 54, 167–199. [Google Scholar] [CrossRef]
- Golyandina, N.; Dudnik, P.; Shlemov, A. Intelligent Identification of Trend Components in Singular Spectrum Analysis. Algorithms 2023, 16, 353. [Google Scholar] [CrossRef]
- Wadekar, S.; Mahalkari, A.; Ali, A.; Gupta, A. Abstract Proceedings of International Conference on 2022 IEEE International Conference on Current Development in Engineering and Technology (CCET): 23rd–24th December 2022; IEEE: Piscataway, NJ, USA, 2022. [Google Scholar] [CrossRef]
- Karhunen, J.; Honkela, A.; Raiko, T.; Harva, M.; Ilin, A.; Tornio, M.; Valpola, H. Bayesian Learning of Latent Variable Models. Available online: https://users.ics.aalto.fi/juha/biennial2007-bayes.pdf (accessed on 31 May 2024).
- Tucker, L. Some mathematical notes on three-mode factor analysis. Psychometrika 1966, 31, 279–311. [Google Scholar] [CrossRef]
- Andersson, C.A.; Bro, R. Improving the speed of multi-way algorithms: Part I. Tucker3. Chemom. Intell. Lab. Syst. 1998, 42, 93–103. [Google Scholar] [CrossRef]
- Gallo, M. Tucker3 Model for Compositional Data. Commun. Stat.-Theory Methods 2015, 44, 4441–4453. [Google Scholar] [CrossRef]
- Murphy, K.R.; Stedmon, C.A.; Graeber, D.; Bro, R. Fluorescence spectroscopy and multi-way techniques. PARAFAC. Anal. Methods 2013, 5, 6557–6566. [Google Scholar] [CrossRef]
- Bro, R. Parafac. Tutorial and Applications. Chemom. Intell. Lab. Syst. 1997, 38, 149–191. [Google Scholar] [CrossRef]
- Bach, F.R.; Jordan, M.I. Kernel independent component analysis. J. Mach. Learn. Res. 2002, 3, 1–48. [Google Scholar] [CrossRef]
- Cavicchia, C.; Vichi, M.; Zaccaria, G. Hierarchical disjoint principal component analysis. AStA Adv. Stat. Anal. 2023, 107, 537–574. [Google Scholar] [CrossRef]
- d’Aspremont, A.; Ghaoui, L.E.; Jordan, M.I.; Lanckriet, G.R. A direct formulation for sparse PCA using semidefinite programming. SIAM Rev. 2007, 49, 434–448. [Google Scholar] [CrossRef]
- Bertsimas, D.; Cory-Wright, R.; Pauphilet, J. Solving Large-Scale Sparse PCA to Certifiable (Near) Optimality. arXiv 2020, arXiv:2005.05195. [Google Scholar]
- Lamboy, W.F. Disjoint Principal Component Analysis: A Statistical Method of Botanical Identification. Source Syst. Bot. 1990, 15, 3–12. [Google Scholar] [CrossRef]
- Hyvärinen, A. Independent component analysis: Recent advances. Philos. Trans. Math. Phys. Eng. Sci. 2013, 371, 20110534. [Google Scholar] [CrossRef] [PubMed]
- Broomhead, D.; King, G.P. Extracting qualitative dynamics from experimental data. Phys. D Nonlinear Phenom. 1986, 20, 217–236. [Google Scholar] [CrossRef]
- Setshedi, I.I.; Loveday, P.W.; Long, C.S.; Wilke, D.N. Estimation of rail properties using semi-analytical finite element models and guided wave ultrasound measurements. Ultrasonics 2019, 96, 240–252. [Google Scholar] [CrossRef]
- Bach, F.R.; Jordan, M.I. Finding Clusters in Independent Component Analysis. In Proceedings of the 4th International Symposium on Independent Component Analysis and Signal Separation, Nara, Japan, 1–4 April 2003. [Google Scholar]
- Widom, H.; Society, A.M. Hankel Matrices. Trans. Am. Math. Soc. 1966, 121, 1–35. [Google Scholar] [CrossRef]
- Yao, F.; Coquery, J.; Lê Cao, K.A. Independent Principal Component Analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinform. 2012, 13, 24. [Google Scholar] [CrossRef]
- Zhao, X.; Ye, B. Similarity of signal processing effect between Hankel matrix-based SVD and wavelet transform and its mechanism analysis. Mech. Syst. Signal Process. 2009, 23, 1062–1075. [Google Scholar] [CrossRef]
- Dokmanic, I.; Parhizkar, R.; Ranieri, J.; Vetterli, M. Euclidean Distance Matrices: Essential Theory, Algorithms and Applications. IEEE Signal Process. Mag. 2015, 32, 12–30. [Google Scholar] [CrossRef]
- Singh, A. K-means with Three different Distance Metrics. Int. J. Comput. Appl. 2013, 67, 8887. [Google Scholar] [CrossRef]
- Lahitani, A.R.; Permanasari, A.E.; Setiawan, N.A. Cosine similarity to determine similarity measure: Study case in online essay assessment. In Proceedings of the 2016 4th International Conference on Cyber and IT Service Management, Bandung, Indonesia, 26–27 April 2016; pp. 1–6. [Google Scholar] [CrossRef]
- Ghorbani, H. Mahalanobis Distance and its application for detecting Multivariate Outliers. Facta Univ. Ser. Math. Inform. 2019, 34, 583. [Google Scholar] [CrossRef]
- Zhang, T.; Ramakrishnan, R.; Livny, M. BIRCH: An efficient data clustering method for very large databases. In Proceedings of the 1996 ACM SIGMOD International Conference on Management of Data, SIGMOD ’96, Montreal, QC, Canada, 4–6 June 1996; ACM: New York, NY, USA, 1996; pp. 103–114. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining (KDD), Portland, OR, USA, 2–4 August 1996; AAAI Press: Washington, DC, USA, 1996; pp. 226–231. [Google Scholar]
- Schubert, E.; Sander, J.; Ester, M.; Kriegel, H.P.; Xu, X. DBSCAN revisited, revisited: Why and how you should (still) use DBSCAN. ACM Trans. Database Syst. (TODS) 2017, 42, 19. [Google Scholar] [CrossRef]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. ECG Heartbeat Classification: A Deep Transferable Representation. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevens, J.; Wilke, D.N.; Setshedi, I.I. Latent Space Perspicacity and Interpretation Enhancement (LS-PIE) Framework. Math. Comput. Appl. 2024, 29, 85. https://doi.org/10.3390/mca29050085
Stevens J, Wilke DN, Setshedi II. Latent Space Perspicacity and Interpretation Enhancement (LS-PIE) Framework. Mathematical and Computational Applications. 2024; 29(5):85. https://doi.org/10.3390/mca29050085
Chicago/Turabian StyleStevens, Jesse, Daniel N. Wilke, and Isaac I. Setshedi. 2024. "Latent Space Perspicacity and Interpretation Enhancement (LS-PIE) Framework" Mathematical and Computational Applications 29, no. 5: 85. https://doi.org/10.3390/mca29050085
APA StyleStevens, J., Wilke, D. N., & Setshedi, I. I. (2024). Latent Space Perspicacity and Interpretation Enhancement (LS-PIE) Framework. Mathematical and Computational Applications, 29(5), 85. https://doi.org/10.3390/mca29050085






