Abstract
This paper reports the synthesis and structural characterization of a novel azide-bridged polymeric manganese (III) Schiff base complex, using 2-((allylimino)methyl)-6-ethoxyphenol as a ligand. The crystal structure of the synthesized compound, elucidated by single-crystal X-ray diffraction analysis, indicates that it crystallizes in the monoclinic space group P21/c. The complex is found to display an octahedral geometry in which the central manganese Mn(III) coordinates with two bidentate donor Schiff base ligands via oxygen and nitrogen atoms. In addition, the metallic centers are linked together to form a one-dimensional chain bridged by end-to-end azide ligands. To offer a more thorough characterization of the synthesized compound, the study incorporates experimental data from FT-IR, UV-Vis, and cyclic voltammetry, alongside computational results from Hirshfeld surface analysis and DFT calculations conducted for both the ligand and complex. The computational analyses provided valuable insights into the intrachain and interchain interactions within the crystal structure, clarified the conformational characteristics of the isolated ligand molecule, and aided in the interpretation of the experimental IR spectra. Furthermore, an assessment of the compound’s drug-like properties was conducted using activity spectra for substances (PASS) predictions, revealing potential pharmacological activities.
1. Introduction
Schiff bases constitute an extensive group of versatile compounds characterized by the presence of a carbon–nitrogen double bond, denoted by the general structure R1R2C = NR3, where R1, R2, and R3 are typically composed of alkyl or aryl groups (R3 ≠ H) [1,2,3]. These compounds are known for their capacity to form ligands that interact effectively with most transition metals through their azomethine nitrogen atoms [4,5]. Schiff bases and their metal complexes have considerable potential as pharmaceutical agents, displaying a diverse spectrum of biological activities. These include anti-inflammatory, antitumor, and antimicrobial effects [5,6,7,8,9,10].
Some Schiff base complexes with allylamine derivatives as ligands have been reported in the literature [11,12,13,14,15]. For instance, Behnam et al. synthesized and characterized complexes of Zn(II), Ni(II), Co(III), Cu(II), and V(IV) with 5-bromo-2-((allylimino)methyl)phenol [12,13]. Identical studies were conducted by Kazemi et al. for complexes of Ni(II), Cu(II), Pd(II), and V(IV) with 2-hydroxy-1-allyliminomethyl-naphthalen [14]. In another study by Salehi et al., complexes of Zn(II), Mn(III), and Co(III) with the same Schiff base ligand were synthesized and investigated using a variety of theoretical (DFT) and experimental approaches [15]. Besides the synthesis and structural characterization, these novel compounds were also evaluated for their potential biological applications and molecular docking properties.
The objective of our current research is to synthesize and conduct a thorough characterization of a manganese (III) complex utilizing a Schiff base ligand derived from 3-ethoxy-hydroxybenzaldehyde and allylamine [2-((allylimino)methyl)-6-ethoxyphenol, Scheme 1]. The selection of this Schiff base ligand was mainly guided by its structural features, which offer the potential for different coordination types, such as chelation through the phenolate oxygen and imine nitrogen atoms, thereby facilitating the formation of stable and well-defined metal–ligand complexes. In addition, this choice of ligand builds on previous studies performed by some of the authors in analogous coordination systems [12,13,15]. This continuity of research thus enables the exploration of new ligand–metal combinations and their associated structural and biological properties. As we delve deeper into our investigation, this complex was found to represent an interesting example of an Mn(III)-based coordination polymer where the metallic centers are connected through an end-to-end μ-(1,3) azide linkage [16,17,18,19]. The incorporation of the allylamine-derived Schiff base ligand introduces distinct structural features and electronic properties, which may significantly influence the overall behavior of the complex. This ligand design has not been extensively explored in the context of Mn(III) coordination polymers [15], making our study a valuable addition to the existing reports on the structure of such systems [16,17,18,19,20,21,22]. Metal–organic coordination polymers have attracted significant interest in the scientific community due to their potential applications in diverse fields, including magnetism, catalysis, and sensing [23,24,25,26,27].
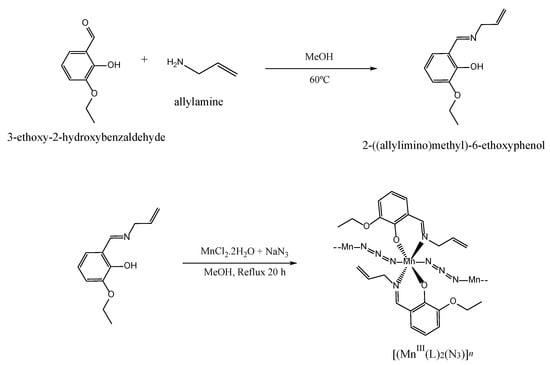
Scheme 1.
Synthetic route for the preparation of the polymeric Schiff base complex [MnIII(L)2(N3)]n (see Section 3.2 for details). The Schiff base ligand HL corresponds to 2-((allylimino)methyl)-6-ethoxyphenol.
In summary, the main structural features of the synthesized polymeric Mn(III) Schiff base complex have been thoroughly investigated by analyzing the most relevant geometrical parameters around the metallic center provided by X-ray diffraction data. In addition, Hirshfeld surfaces were utilized to study intermolecular interactions, providing valuable insights into the compound’s molecular arrangement in the crystal. Further investigations involving infrared and UV-Vis spectroscopy, cyclic voltammetry, and theoretical computations at the DFT level in both the ligand and the complex were also performed. These combined approaches provided a comprehensive insight into the properties and behavior of the compound. Furthermore, the synthetized complex was subjected to activity spectra for substances (PASS) analysis, which allowed the evaluation of its potential for combating pathogenic viruses.
2. Results and Discussion
2.1. Synthesis and Spectroscopic Characterization of the SCHIFF Base and Complex
The synthetic route for preparing the polymeric Schiff base complex [MnIII(L)2(N3)]n is outlined in Scheme 1. The bidentate Schiff base ligand (HL) was synthesized by reacting allylamine with 3-ethoxy-2-hydroxybenzaldehyde (1:1 molar ratio) in a methanol solution. Subsequently, the mixture was treated with MnCl2 and NaN3 methanolic solutions. For a detailed description of the experimental procedure used for synthesizing both the ligand and complex, please refer to Section 3.2.
The IR spectra of the Schiff base ligand and complex are provided in Figure S1a,b, and the identified bands are characterized in Table S1. The vibrational calculations performed at the B3LYP/6-311++G(d,p) level on the fully optimized geometries of both species were used to help in the assignment of the most important experimental bands (the computed vibrational data are provided in Tables S2 and S3). In terms of what the IR spectrum of HL is concerned with, it is worth noting the presence of a strong feature at 1659/1647 cm−1, which is undoubtedly ascribed to the C=N stretching vibration of the azomethine group [13,28,29]. This band is theoretically predicted at 1652.8 (I = 198.3 km mol−1). The observation of this band, together with the absence of bands due to the antisymmetric and symmetric NH2 stretching vibrations, which have been observed in the IR spectra of allylamine within the 3390–3260 cm−1 range [30] (refer to Figure S1 for more details), unequivocally confirms the condensation of all primary amino groups during the synthesis of the ligand. The other prominent bands observed in the fingerprint region of the HL spectrum are located at 1464, 1254, and 739 cm−1. In the spectrum calculated for the isolated molecule (Table S2), these bands are predicted at 1463.4 (I = 246.1 km mol−1), 1252.7 (I = 437.2 km mol−1), and 728.8 cm−1 (I = 53.2 km mol−1). The first is assigned to the aromatic C–C and C1–O1 stretching vibrations, the second to the C2–O2 stretching vibration, and the third to ring C–H out-of-plane deformations (atom numbering is presented in Figure 1). In the high-frequency region of the IR spectrum, the broad band centered at around 3440 cm−1 is most likely assigned to the stretching vibration of the OH groups involved in intermolecular H-bonds. It is important to note that the spectrum calculated for the most stable conformer of HL predicts a very intense absorption at 3038 cm−1 (I = 547.8 km mol−1), which is ascribed to the stretching vibration of the OH group engaged in an intramolecular hydrogen bond with the azomethine nitrogen atom (OH⋯N=C) (see Section 2.5 for details). However, no prominent experimental band is evident in the vicinity of this spectral position, confirming the absence of such a hydrogen bond in the synthetized Schiff base ligand. The weak band observed at 2982 cm−1 (as shown in Figure S1) is likely associated with CH stretching vibrations (see Table S2).
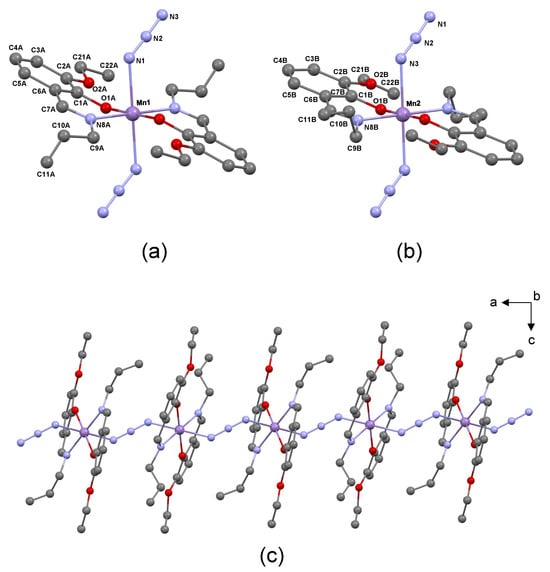
Figure 1.
(a,b) Coordination environment around the Mn1 and Mn2 cations in the Mn(III) complex. (c) Segment of the one-dimensional zigzag chain along the a-axis connecting individual monomeric MnIII(L2) units through the azide ligand.
Regarding the IR spectrum of the complex, the band corresponding to the C=N stretching vibration is shifted to lower frequencies as compared to the free ligand, being observed at 1614 cm−1 (calculated at 1662.0 cm−1, I = 607.7 km mol−1). This experimentally observed red shift is most likely attributed to the electron donation from the nitrogen to the empty d-orbitals of the metallic center [31]. The strongest absorption band observed at 2046 cm−1 (calculated at 2172 cm−1, I = 1878 km mol−1), which is not present in the spectrum of the ligand, is assigned to the antisymmetric stretching vibration of the azide group (N3), in consonance with the IR spectroscopic data obtained for other azide-bridged polymeric Mn(III) Schiff base complexes [15,32]. In the lower-frequency region of the IR spectrum, it is noteworthy to observe the bands at 624 and 495 cm−1, which also do not have a corresponding presence in the IR spectrum of the ligand. These bands can be attributed to the antisymmetric stretching vibrations of the Mn–O1 and Mn–N8 bonds. In the computed IR spectrum, the corresponding absorptions are predicted at 582.8 cm−1 (I = 66.4 km mol−1) and 461.7 cm−1 (I = 66.1 km mol−1). An intense absorption corresponding to the Mn–N1 (or N3) stretching vibration is predicted to occur at 411.9 cm−1 (I = 159.7 km mol−1). The non-observation of this band in the experimental spectrum may be attributed to the fact that it is most likely situated below 400 cm−1, placing it outside the detection range of the used IR spectrometer.
The UV-vis absorption spectrum of the Mn(III) complex recorded in a DMF solution is displayed in Figure S2. This spectrum follows the general pattern of that previously reported for other similar Mn(III) complexes [12,13,15]. In the UV region, the absorption bands at 235 and 274 nm correspond to π-π* transitions involving molecular orbitals located on phenolic chromophores, while that at 384 nm is most likely associated with intra-ligand (C=N) transitions. In the visible region, the complex exhibits a characteristic broad and low-intensity band at 624 nm, indicative of d → d transitions occurring within its complex.
2.2. Electrochemical Studies
The cyclic voltammogram of the synthesized complex in a DMF solution, recorded between −2.3 and 0.2 V, is shown in Figure S3. In this voltammogram, the cathodic peaks at −0.13 and −1.77 V correspond, to the Mn(III) → Mn(II) and Mn(II) → Mn(I) reductions. No anodic peaks were observed during the reverse scan, thus revealing the irreversibility of these redox processes. This behavior is in line with that previously observed for other structurally similar manganese (III) Schiff base complexes [15,33]. The observed irreversibility can be attributed to the inherent instability of the reduced manganese species in the given experimental conditions. Additionally, the electronic properties of the Schiff base ligand could further influence redox behavior, potentially affecting the ease with which these reduced forms revert to their higher oxidation state. This insight underscores the importance of both the ligand environment and the stability of the metal centers in determining the overall redox characteristics of the complex.
2.3. Crystal Structure
The X-ray diffraction analysis of the synthesized Mn(III) complex reveals that it crystallizes in the monoclinic system with the space group P21/c. The asymmetric unit of the crystal comprises two Mn(III) cations, identified as Mn1 and Mn2, along with two Schiff base ligands and one azide ion (N3−). Both cations are positioned at distinct centers of inversion within the space group P21/c, located at coordinates (½, ½, ½) and (0, ½, ½). As shown in Figure 1, the coordination environment around each manganese cation involves two deprotonated Schiff base ligands (Ls), where each ligand is connected to the metallic center through a nitrogen (N8) and oxygen (O1) atom, thus acting as a bidentate NO-donor (Figure 1a,b). In the complex, these two pairs of atoms are arranged in the equatorial plane of an octahedral structure, enveloping the manganese cation. The azide ion (N3−) serves as a bridging ligand, linking the two metallic centers (see Figure 1c for details). The most relevant geometric characteristics of the Mn(III) complex, obtained from the X-ray data, are provided in Table 1.

Table 1.
Selected parameters extracted from the X-ray data bond lengths (Å) and bond angles (deg.) around the metallic center (Mn1 or Mn2) of the studied polymeric Mn(III) complex.
Analyzing the provided geometrical parameters obtained from the X-ray data, several observations can be made. Firstly, the axial Mn–N1 (or N3) bonds are approximately 0.237–0.213 Å longer than the equatorial Mn–N8 and Mn–O1 bonds, indicating a deviation from an ideal octahedral geometry. This observed deviation is consistent with the patterns identified in previously reported Mn(III) Schiff base complexes that share similar structural features with our complex [15,17,18,19,34], and is typical in high-spin Mn (III) octahedral complexes that undergo Jahn–Teller distortions [35,36]. Secondly, within the equatorial plane, the Mn–O1 bonds are shorter than the Mn–N8 bonds, which is expected due to the higher electron density around the oxygen atom. This geometrical feature is consistent with the IR spectroscopic data presented in Section 2.1, where the Mn–O1 stretching vibration was observed at a higher frequency than the Mn–N8 stretching vibration. Lastly, a comparison between the two metal centers, Mn1 and Mn2, reveals variations in bond lengths. In Mn2, both the axial Mn–N1(or N3) and equatorial Mn–O1 bonds are shorter by approximately 0.015 and 0.019 Å, respectively, compared to Mn1. Conversely, the equatorial Mn–N8 bond in Mn1 is only 0.009 Å shorter than in Mn2. These differences suggest that, globally, the arrangement of ligands around the Mn2 center is more compact than that around the Mn1 center. Examining the bond angles around the central metal, it can be observed that the O1–Mn–N8, O–Mn–O, and O1–Mn–N1 (or N3) and N8–Mn–N1 (or N3) angles fall within the range of 88–93°, while the O1–Mn–O1, N8–Mn–N8, and N1–Mn–N1 (or N3–Mn–N3) angles are equal to 180°.
The crystal structure of the complex reveals the presence of a coordination polymer that extends along the [100] direction, as depicted in Figure 1c. This supramolecular structure is obtained by linking individual monomeric MnIII(L2) units through the azide ligand, involving an end-to-end μ-(1,3) bridging mode [15,18,34,37]. The bridging arrangement gives rise to a chain-like structure, exhibiting a one-dimensional zigzag pattern. Within this chain, the Mn1 and Mn2 metal centers are arranged in an alternating manner. The distance between adjacent Mn1 and Mn2 centers within the chain is 5.763 Å, while the shortest distance between Mn atoms in different chains is 10.291 Å. These data agree with the those reported for other azide-bridged polymeric Mn(III) Schiff base complexes [17,19,34].
Apart from the coordination bonds, the crystal structure of the compound is characterized by the formation of short C–H⋯N, C–H⋯O, and C–H⋯C intermolecular interactions. These interactions are observed within the polymeric chains, as well as between different chains. Although these contacts are associated with the formation of weak stabilizing interactions, they play a significant role in the arrangement of molecules in the crystal lattice. To better comprehend these intermolecular interactions and their influence on the crystal structure, an analysis of Hirshfeld surfaces is presented below.
2.4. Hirshfeld Surface Analysis
The Hirshfeld surfaces generated for the Schiff base complex are presented in Figure 2a. The larger red spots observed on these surfaces correspond to the axial Mn–N1/N3 coordination bonds, which have been already characterized in Section 2.3 and are therefore excluded from this analysis. The majority of the remaining red spots is attributed to the formation of weak stabilizing C–H⋯A interactions (A = N, O, or C) [38], which are denoted in Figure 2 by letters a–h. Red spots labeled as i, j are associated with the formation of C⋯C repulsive interactions. The geometrical parameters characterizing all these interactions are presented in Table 2, with the most relevant ones highlighted in Figure 3.
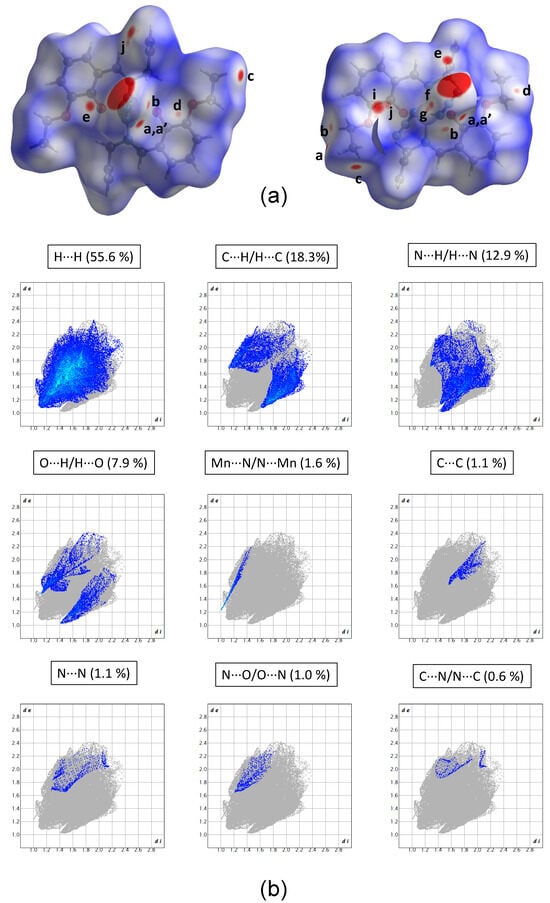
Figure 2.
(a) Three-dimensional Hirshfeld surfaces generated for the MnIII(L2)(N3)2− fragment. For the left surface, the metallic center is Mn1, while for the right surface, the metallic center is Mn2. The surfaces are mapped with the normalized contact distance (dnorm) and the relevant intermolecular contacts are denoted by small letters. (b) Fingerprint plots calculated for the Mn (III) complex, decomposed into different contacts. The di and de values correspond, to the closest internal and external distances (Å) from given points on the Hirshfeld surface contacts. The contributions (in %) of the different contacts to the total Hirshfeld surface area are included.

Table 2.
Geometrical characterization of the most relevant intermolecular contacts found in the crystalline structure of the polymeric Mn(III) complex a.
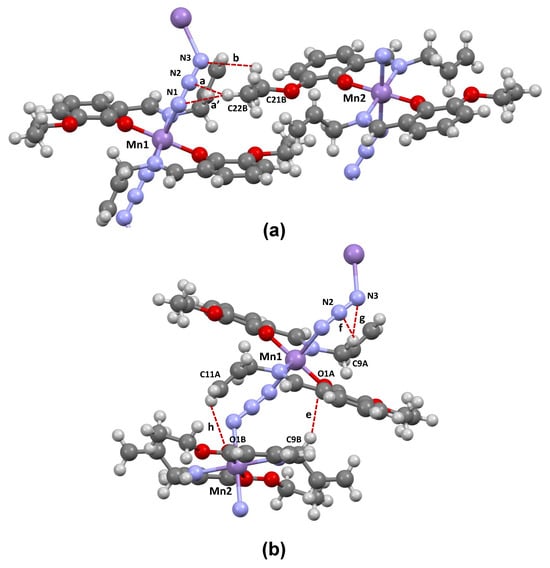
Figure 3.
Most relevant interchain (a) and intrachain (b) hydrogen bond-like interactions formed in the crystalline structure of the polymeric Schiff base complex [MnIII(L)2(N3)]n.
The short contacts labeled in the Hirshfeld surfaces as a, a’, and b correspond to the formation of C–H⋯N hydrogen-bond like interactions that occur between adjacent polymeric chains (interchain). A common characteristic of these interactions is that the carbon atoms serving as donors are part of the ethoxy fragment, while the nitrogen azide atoms function as acceptors. In the case of the first two interactions (a, a’), C22B (from the methyl group of the ethoxy fragment) acts as the donor atom, while the azide nitrogen atoms N1 or N2 act as acceptors. The third interaction (b) involves one of the hydrogens connected to C21B (from the methylene group of the ethoxy fragment) and the other azide N3 atom. The geometrical parameters presented in Table 2 indicate that interaction b is weaker when compared to interactions a and a’. This difference in strength is mainly attributed to the significantly smaller C–H⋯N angle observed in interaction b (114°), in contrast to the wider angles found in the other interactions (137–151°).
The remaining stabilizing interchain contacts (labeled as c and d) are characterized as very weak C–H⋯C interactions and are not shown in Figure 3. Interactions e-h take place within the same chain (intrachain). The positioning of the carbon atoms acting as donors is an important difference from the interchain interactions. Unlike the former atoms, the donor carbon atoms form part of the allyl fragment. Two of the intrachain interactions (e and h) are of the C–H⋯O type, where C9B or C11A serve as donors while O1A or O1B act as acceptors, respectively. Based on the geometrical parameters, interaction e (C9B–H⋯O1A) emerges as the strongest of all the ones present in the crystal structure of the complex. The other two intrachain interactions (f, g) are classified as C–H⋯N, involving C9A as a donor and the azide N2 or N3 atoms as acceptors. It is worth highlighting that the formation of the C11A–H⋯O1B interaction (h) has two significant structural consequences: (i) the conformation of the allyl fragment (–CH2–CH=CH2) in the Schiff base ligands connected to Mn2 differs from that connected to Mn1, as illustrated in Figure 1; and (ii) C11A approaches the C1B and C2B atoms of the phenyl ring of a Schiff base situated in adjacent intrachain complexes, resulting in the formation of C11A⋯C1B and C11A⋯C2B contacts, which are also characterized in Table 2.
The 2D-fingerprint plots, extracted from the Hirshfeld surfaces, divided into contributions stemming from key atomic interactions, are included in Figure 2b. Given the substantial number of hydrogen atoms within the molecular structure of the analyzed compound, it is not surprising to observe a predominance of the H⋯H contacts, representing 55.6% of the Hirshfeld surface. In line with our expectations from the discussion above, the subsequent noteworthy contributions arise from the H⋯C/C⋯H, H⋯N/N⋯H, and H⋯O/O⋯H contacts, accounting for approximately 18.3%, 12.9%, and 7.9% of the total surface area, respectively. On the other hand, less prominent contributions are attributed to Mn⋯N, C⋯C, N⋯N, N⋯O/O⋯N, and C⋯N/N⋯C contacts, which together represent only 5.4% of the surface.
2.5. Conformational Analysis of the Schiff Base Ligand
In addition to the structure of the complex, it is also instructive to investigate the structure of the Schiff base ligand (HL), with particular emphasis on exploring its conformational behavior. The potential energy surfaces around the C7=N8-C9-C10 (θ1) and N8-C9-C10=C11 (θ2) torsions (see Figure S4a,b) led to the identification of eight conformers. The B3LYP/6-311++G(d,p) fully optimized geometries of two representative conformers are shown in Figure 4, while the geometries of all eight identified conformers, along with their energetic characterizations, are presented in Figure S5 and Table S4, respectively. The conformers are labeled as HL1 or HL2, depending on whether the O1H group is directed toward the N8 or O2 atoms, respectively, followed by the labels “c” (cis, ~0°) or “sk±” (skew±, ~±120°), defining the orientation around the θ1 and θ2 dihedrals, which define the conformation of the allyl fragment. According to the calculations, the HL1 conformers are more stable than the HL2 ones. This difference in stability is attributed to the formation of an O1–H⋯N8 intramolecular hydrogen bond in HL1 [d(H⋯N) ~1.7Å; ∠(O–H⋯8) ~147–148°], which is much stronger than the O1–H⋯O2 intramolecular bond formed in the HL2 conformers [d (H⋯N) ~2.0Å; ∠(O–H⋯N) ~116–117°]. For a given orientation of the θ1 and θ2 torsions, the energy difference between these two sets of conformers is ~30 kJ mol−1. Among the HL1 conformers, the most stable one is characterized by sk− and sk+ (or sk+ and sk−) orientations around the θ1 and θ2 dihedrals (θ1 = ±121.7°; θ2 = ±128.5°) respectively (see Figure 4, left). It is worth noting that the arrangement of the allyl fragment in this conformer is not significantly different from its arrangement in the crystalline structure of the Schiff base ligand coordinated to the Mn2 cation (θ1 = ±74.1°; θ2 = ±126.5°). This is consistent with the fact that, in the crystalline structure of the complex, the allyl fragment of the ligand is not involved in intrachain or interchain intermolecular interactions allowing its arrangement to closely resemble that of the most stable form of the free molecule. On the other hand, the conformation of the allyl group in the Schiff base coordinated to the Mn1 cation (θ1 = ±8.9°; θ2 = ±122.9°) is very close to the fifth most stable conformer (θ1 = +18.3°; θ2 = −120.0°), and this conformational variation is attributed to the fact that the terminal CH2 group of the allyl fragment of the ligand is involved in the formation of an intrachain C–H⋯O interaction, as described in Section 2.4.
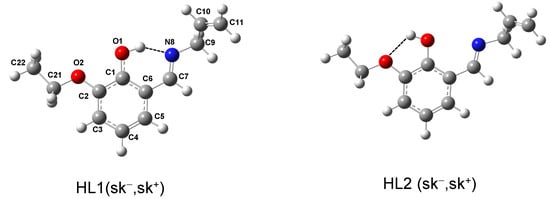
Figure 4.
Geometries of the most stable HL1- and HL2-type conformers of the Schiff base ligand (HL), fully optimized at the B3LYP/6-311++G(d,p) level of theory.
2.6. In Silico PASS Analysis of Biological Activity
In this study, we employed PASS (Prediction of Activity Spectra for Substances), a software tool designed to evaluate the broad biological potential of organic drugs (see Section 3.7 for details). PASS utilizes structural information from chemical compounds to predict multiple types of biological activities simultaneously. We harnessed the capabilities of PASS to generate virtual molecular biological activity profiles for our synthesized complex. The “to be active” probability estimates the likelihood that our compound belongs to the sub-class of active compounds, sharing structural similarities with the most common molecular structures found in a sub-set of “actives” from the PASS training dataset. Table 3 summarizes the results of the PASS analysis, which offers valuable insights into the potential pharmacological actions of our synthesized polymeric Mn(III) complex. This analysis is crucial for understanding the diverse biological activities that this compound may exhibit based on its structural characteristics.

Table 3.
Predicted biological activity spectrum of polymeric Mn(III) complex by PASS analysis.
As one can see from Table 3, the polymeric Mn(III) complex shows promising predictions for a wide spectrum of pharmacological functions. Notably, the “Pa” values (probability of being active) consistently surpass the “Pi” values (probability of being inactive) for various biological activities. Among the most notable predictions, the compound is found to exhibit substantial “Pa” values for antiviral activities against Rhinovirus (0.444), Influenza (0.350) and Picornavirus (0.352). This indicates a strong potential for the complex to exert specific effects in these areas (Pa > 0.3). Overall, the results of this study suggest that these Schiff base complex may be considered for further investigation as a possible therapeutic agent for antiviral properties.
3. Experimental and Computational Methodologies
3.1. Materials and Instrumentation
All reagents used in the experiments were obtained from Merck (Rahway, NJ, USA) and Sigma-Aldrich (St. Louis, MI, USA) and were used without further purification. IR spectra of the synthesized solid compounds were recorded on a SHIMADZU FT-IR spectrometer (4000–400 cm−1) in KBr pellets, while UV-Vis absorbance spectra in a dimethylformamide (DMF) solution were measured using a UV-1650 PC SHIMADZU spectrophotometer. A Metrohm 757 VA Computrace instrument was utilized to conduct electrochemical analyses in a DMF solution.
3.2. Synthesis of the Polymeric Schiff Base Complex [MnIII(L)2(N3)]n
The methodology employed for synthesizing the Mn(III) complex followed that described elsewhere for an analogous complex [15]. Initially, for ligand (HL) preparation, 3-ethoxy-2-hydroxybenzaldehyde (0.185 g) was dissolved in a methanolic solution containing allylamine (0.058 g in 10 mL of methanol), and the resulting mixture was stirred for 2 h at 60 °C. To prepare the complex, 0.5 mmol of MnCl2.2H2O was dissolved in 10 mL of methanol and added dropwise to the ligand solution. The color of the solution changed from yellow to red upon addition. Subsequently, 0.065 g of NaN3 was dissolved in 2 mL of methanol and added to the solution, leading to a color change to black. The resulting mixture was allowed to stir magnetically under reflux for 20 h. After a few days, dark-brown single crystals were obtained, subsequently filtered, washed with a small quantity of cold n-hexane, and dried under vacuum. The resulting product proved suitable for X-ray analysis. Yield: 63%. FT-IR data (KBr, cm−1): 2975–2923 (νC–H), 2046 [νas (N3)]; 1616 (νC=N); 624 (νMn–O); 495 [ν(Mn–N)]. UV–Vis: λmax (nm) (ε, M−1cm−1) (DMF): 235 (62500), 274 (37500), 384 (12500), 625 (3000). In the Supplementary Materials, we provide the IR and UV-Vis spectra (Figures S1 and S2, respectively).
3.3. X-ray Data Collection and Refinement Parameters
Diffraction data were collected by the ω-scan technique, at 100(1) K, on a Rigaku XCalibur four-circle diffractometer with an EOS CCD detector, equipped with a graphite-monochromatized MoKα radiation source (λ = 0.71073 Å). The data were corrected for Lorentz polarization as well as for absorption effects [39]. The structures were solved with SHELXT [40] and refined with the full-matrix least-squares procedure on F2 by SHELXL-2013 [13]. All non-hydrogen atoms were refined anisotropically, while hydrogen atoms were placed in idealized positions and refined as ‘riding model’ with isotropic displacement parameters set at 1.2 (1.5 for methyl groups) times Ueq of appropriate carrier atoms. Crystal data: C24H28MnN5O4, Mr = 505.45, monoclinic, P21/c, a = 11.5257(5) Å, b = 13.9571(6) Å, c = 15.1254(5) Å, β = 104.333(4)°, V = 2357.42(17) Å3, Z = 4, dx = 1.424 g cm −3, F(000) = 1056, μ = 0.601 mm−1, 18,193 reflection collected, 4736 symmetry independent (Rint = 4.02%), and 3458 with I > 2σ(I). Final R[I > 2σ(I)] = 0.0463, wR2[I > 2σ(I)] = 0.1104, R [all reflections] = 0.0728, wR2 [all reflections] = 0.1293, S = 1.064, and (Δρmax/Δρmin) = 0.60/−0.59 e·Å−3. The CIF file containing the supplementary crystallographic data for the synthesized compound was deposited at the Cambridge Crystallographic Data Centre, with reference CCDC 2172659.
3.4. Cyclic Voltammetry
Cyclic voltammetry experiments were carried out for the complex at 298 K in a 10−3 M DMF solution, utilizing 0.1 M of tetrabutylammonium hexafluorophosphate (TBAH) as the supporting electrolyte. The experimental setup involved a three-electrode system, which consisted of an Ag/AgCl reference electrode, a platinum auxiliary electrode, and a glassy carbon working electrode. The voltammograms were recorded with a scan rate of 100 mV/s. Anodic and cathodic potentials were measured with reference to the Ferrocenium (Fc+)/Ferrocene (Fc) redox couple (E0 = 0.45 V).
3.5. Hirshfeld Surface Analysis
The CrystalExplorer software package (version 17.5) was used to perform a Hirshfeld surface (HS) analysis of the crystal structure of the Mn(III) complex [41,42,43,44,45]. This analysis serves as a valuable graphical tool, allowing for a comprehensive examination of the intermolecular interactions and close contacts within the crystal packing. The CIF file, obtained from the X-ray data, was used as the input for the software. The Hirshfeld surfaces were generated with high resolution and were mapped with the normalized contact distance (dnorm), calculated from the following equation [44,46]:
In this equation, di and de are, respectively, the distances from the point of the surface to the nearest atoms inside and outside the surface, while rivdW and revdW are the van der Waals radii of the respective atoms. Intermolecular contacts shorter than the sum of the van der Waals radii (dnorm < 0) are depicted as red spots on the surface. Contacts around the van der Waals separation (dnorm = 0) are shown as white, while contacts longer than the van der Waals distance (dnorm > 0) are represented by blue regions. Furthermore, as part of the Hirshfeld surface analysis, fingerprint plots were also generated. These plots provide a two-dimensional representation of the relationship between di and de at each surface point, assisting in understanding the nature and relative contribution of these contacts to the total Hirshfeld surface area, thereby aiding in the characterization of the complex’s supramolecular architecture.
3.6. Theoretical Calculations
All theoretical calculations were performed with the Gaussian 16 software package (Revision A.03) [47]. The HL conformers were identified by calculating the relaxed potential energy surfaces around the C7=N8-C9-C10 (θ1) and N8-C9-C10=C11 (θ2) dihedrals. These calculations were performed with the B3LYP (DFT) functional [48,49,50] and the 6-311++G(d,p) basis set [51,52]. The located energy minima were then fully optimized followed by Harmonic vibrational calculations at the same B3LYP/6-311++G(d,p) level of theory. For the Mn(III) complex, an MnIII(L)2(N3)2− fragment extracted from the crystalline structure was fully optimized, followed by a vibrational calculation by using the same DFT functional combined with the def2-svp basis set [53]. These computations were conducted for both the high-spin (HS, quintet) and low-spin (LS, triplet) electron configurations. At this level of theory, the Gibbs energy difference at 298.15 K between the two configurations was computed to be 43.5 kJ mol−1 in favor of the HS state. Consequently, the vibrational data employed in the interpretation of the experimental IR spectrum of the Mn(III) complex were extracted for the fully optimized HS MnIII(L)2(N3)2− fragment. From the atomic spin densities, a magnetic moment of 3.9 μB was estimated for this configuration, which is consistent with the experimentally determined magnetic moments reported for HS Mn(III) complexes [54], including azide-bridged manganese(III) complexes [16,20,21]. For all optimized structures of the ligand and complex, no imaginary frequencies were observed in the vibrational calculations, confirming that they are all minima on the respective potential energy surfaces. The B3LYP/6-311++G(d,p) wavenumbers computed for the ligand were multiplied by 0.980 for the region below 2000 cm−1, while for the vibrations above 2000 cm−1, which are typically more anharmonic, a scale factor of 0.950 was used [55,56]. Concerning the wavenumbers calculated for the complex using the B3LYP/def2-SVP model chemistry, a single scaling factor of 0.967 was applied [57].
3.7. Prediction of Probable Activity Spectra of Substances (PASS Online Software)
Prediction of Probable Activity Spectra of Substances (PASS, version 11) analysis is an online software (https://www.way2drug.com/passonline, accessed on 20 November 2023) [58,59] that uses computer-based algorithms to assess the biological and pharmacological potential of a compound. It provides insights into the compound’s interactions with various biological entities, enabling the evaluation of its utility in biological and pharmacological contexts. To perform this analysis, the software compares the 2D structure of a molecule to a vast training set consisting of over 46,000 drugs, drug candidates, and lead compounds. The molecular structure can be stored as a Mol file (*.mol) and directly uploaded to the PASS prediction website to forecast the biological activity spectrum. The molecule’s activity is predicted by comparing the new compound’s structure to that of a well-known biological active substrate stored in the database. The algorithm employs a Bayesian technique. The PASS tool provides a Pa:Pi ratio (active to inactive ratio) at prediction thresholds of Pa > 30%, Pa > 50%, and Pa > 70%. These thresholds indicate the predicted likelihood of a compound being active. According to leave-one-out cross-validation (LOO CV) estimates, the average prediction accuracy is approximately 95%. The accuracy of the PASS prediction relies on detailed information about the biological activity spectrum for each molecule in the PASS training set, making the biological activity estimate more accurate [59].
4. Conclusions
In this manuscript, we successfully synthesized a novel Mn(III) Schiff base complex. Its characterization was accomplished through FT-IR and UV–Vis spectroscopy. Furthermore, the molecular structure of the compound was elucidated using single-crystal X-ray diffraction analysis. The crystallographic results reveal a distinctive 1D-coordination polymer complex, where two nitrogen and oxygen atoms of the Schiff base ligand are coordinated to the central cation. The sodium azide was found to act as a bridging component, connecting the MnIII(L)2 complex units, originating a chain-like structure with a one-dimensional zigzag pattern. The Hirshfeld surface analysis revealed diverse C–H⋯A (A = N, O, or C) interactions both within the same chain or among chains. Interchain interactions prominently featured ethoxy carbon atoms (C21 and C22) as hydrogen donors and bridging azide nitrogen atoms (N1, N2, and N3) as acceptors. On the other hand, intrachain interactions were primarily driven by allyl carbon atoms (C9 and C11) as donors, involving azide nitrogens (N2 and N3) or phenoxyl oxygen (O1) as acceptors. This comprehensive analysis sheds light on the complex’s structural characteristics, providing valuable insights into potential implications for its behavior. Electrochemical studies revealed an irreversible reduction process for the Mn (III) complex, providing a greater understanding of its redox behavior. Besides the characterization of the complex, we also investigated the conformational features of the isolated HL Schiff base ligand, identifying its most stable conformation as one stabilized by an intramolecular O–H⋯N hydrogen bond. Finally, Activity Spectra for Substances (PASS) predictions indicated potential antiviral activity for the synthesized complex. These findings can inspire and guide future studies, particularly those aimed at gaining deeper insights into the structure and behavior of the complex in biologically relevant environments. Such research will be crucial for fully exploring its therapeutic potential.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/inorganics12090234/s1, Figure S1: IR spectra of the Schiff base ligand and the polymeric Mn (III) complex; Figure S2: UV-Vis spectrum of the complex; Figure S3: Cyclic voltammogram registered for a complex solution; Figure S4: Relaxed potential energy scans computed for the Schiff base ligand (HL) at the B3LYP/6-311++G(d,p) level; Figure S5: Geometries of the conformers of HL fully optimized at the B3LYP/6-311++G(d,p) level of theory. Tables: Table S1: Experimental wavenumbers and intensity of the bands observed in the IR spectra of HL and polymeric Mn(III) complex; Table S2: B3LYP/6-311++G(d,p) wavenumbers and IR intensities calculated for the HL ligand; Table S3: B3LYP/def2-SVP wavenumbers and IR intensities calculated for the MnIII(L)2(N3)2− fragment of the polymeric manganese (III) Schiff base fully optimized at the same level; Table S4: Geometric data computed for the conformers of HL fully optimized at the B3LYP/6-311++G(d,p) level of theory.
Author Contributions
A.T.: investigation and writing—original draft preparation. M.S.: validation, data curation, and project administration. A.J.L.J.: formal analysis, writing—review and editing, and visualization. M.K.: methodology. R.F.: validation. R.G.: resources. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Semnan University, Adam Mickiewicz University, and Payame Noor University. CQC-IMS is supported by FCT through projects UI0313B/QUI/2020, UI0313P/QUI/2020, and LA/P/0056/2020 (national funds).
Data Availability Statement
All original contributions presented in this study are included within the article. For further information, inquiries can be directed to the corresponding authors.
Acknowledgments
This work results from a collaborative effort between the scientific members of Semnan University (Aynaz Talebi and M. Salehi), University of Coimbra (Rui Fausto and A. J. Lopes Jesus), Payame Noor University (Reza Golbedaghi), and Institute Sciences Adam Mickiewicz University (Maciej Kubicki). The authors consider it their duty to thank Rui Fausto (University of Coimbra) for his guidance during the revision of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Schiff, H. Mittheilungen aus dem Universitätslaboratorium in Pisa: Eine neue Reihe organischer Basen. Justus Liebigs Ann. Chem. 1864, 131, 118–119. [Google Scholar] [CrossRef]
- Fabbrizzi, L. Beauty in Chemistry: Making Artistic Molecules with Schiff Bases. J. Org. Chem. 2020, 85, 12212–12226. [Google Scholar] [CrossRef] [PubMed]
- Raczuk, E.; Dmochowska, B.; Samaszko-Fiertek, J.; Madaj, J. Different Schiff Bases mdash—Structure, Importance and Classification. Molecules 2022, 27, 787. [Google Scholar] [CrossRef] [PubMed]
- Vigato, P.A.; Tamburini, S. The challenge of cyclic and acyclic schiff bases and related derivatives. Coord. Chem. Rev. 2004, 248, 1717–2128. [Google Scholar]
- Golbedaghi, R.; Tabanez, A.M.; Esmaeili, S.; Fausto, R. Biological Applications of Macrocyclic Schiff Base Ligands and Their Metal Complexes: A Survey of the Literature (2005–2019). Appl. Organomet. Chem. 2020, 34, e5884. [Google Scholar] [CrossRef]
- Soroceanu, A.; Bargan, A. Advanced and Biomedical Applications of Schiff-Base Ligands and Their Metal Complexes: A Review. Crystals 2022, 12, 1436. [Google Scholar] [CrossRef]
- Shaygan, S.; Pasdar, H.; Foroughifar, N.; Davallo, M.; Motiee, F. Cobalt (II) Complexes with Schiff Base Ligands Derived from Terephthalaldehyde and ortho-Substituted Anilines: Synthesis, Characterization and Antibacterial Activity. Appl. Sci. 2018, 8, 385. [Google Scholar] [CrossRef]
- Sinicropi, M.S.; Ceramella, J.; Iacopetta, D.; Catalano, A.; Mariconda, A.; Rosano, C.; Saturnino, C.; El-Kashef, H.; Longo, P. Metal Complexes with Schiff Bases: Data Collection and Recent Studies on Biological Activities. Int. J. Mol. Sci. 2022, 23, 14840. [Google Scholar] [CrossRef]
- Naeimi, H.; Sadat Nazifi, Z.; Matin Amininezhad, S.; Amouheidari, M. Synthesis, characterization and in vitro antimicrobial activity of some new Schiff bases and their complexes. J. Antibiot. 2013, 66, 687–689. [Google Scholar] [CrossRef]
- Iacopetta, D.; Ceramella, J.; Catalano, A.; Mariconda, A.; Giuzio, F.; Saturnino, C.; Longo, P.; Sinicropi, M.S. Metal Complexes with Schiff Bases as Antimicrobials and Catalysts. Inorganics 2023, 11, 320. [Google Scholar] [CrossRef]
- Amiri Rudbari, H.; Khorshidifard, M.; Askari, B.; Habibi, N.; Bruno, G. New asymmetric Schiff base ligand derived from allylamine and 2,3-dihydroxybenzaldehyde and its molybdenum(VI) complex: Synthesis, characterization, crystal structures, computational studies and antibacterial activity together with synergistic effect against Pseudomonas aeroginosa PTTC 1570. Polyhedron 2015, 100, 180–191. [Google Scholar]
- Deilami, A.B.; Salehi, M.; Amiri, A.; Arab, A. New copper(II) and vanadium(IV) complexes based on allylamine-derived Schiff base ligand; synthesis, crystal structure, electrochemical properties and DFT calculations. J. Mol. Struct. 2019, 1181, 190–196. [Google Scholar] [CrossRef]
- Deilami, A.B.; Salehi, M.; Arab, A.; Amiri, A. Synthesis, crystal structure, electrochemical properties and DFT calculations of three new Zn(II), Ni(II) and Co(III) complexes based on 5-bromo-2-((allylimino)methyl)phenol Schiff-based ligand. Inorg. Chim. Acta 2018, 476, 93–100. [Google Scholar] [CrossRef]
- Kazemi, Z.; Rudbari, H.A.; Sahihi, M.; Mirkhani, V.; Moghadam, M.; Tangestaninejad, S.; Mohammadpoor-Baltork, I.; Gharaghani, S. Synthesis, characterization and biological application of four novel metal-Schiff base complexes derived from allylamine and their interactions with human serum albumin: Experimental, molecular docking and ONIOM computational study. J. Photochem. Photobiol. B 2016, 162, 448–462. [Google Scholar] [CrossRef] [PubMed]
- Ramezanipoor, S.; Parvarinezhad, S.; Salehi, M.; Grześkiewicz, A.M.; Kubicki, M. Crystal structures, electrochemical properties and theoretical studies of three New Zn(II), Mn(III) and Co(III) Schiff base complexes derived from 2-hydroxy-1-allyliminomethyl-naphthalen. J. Mol. Struct. 2022, 1257, 132541. [Google Scholar] [CrossRef]
- Miyasaka, H.; Saitoh, A.; Abe, S. Magnetic assemblies based on Mn(III) salen analogues. Coord. Chem. Rev. 2007, 251, 2622–2664. [Google Scholar] [CrossRef]
- Chakrabarty, P.P.; Saha, S.; Schollmeyer, D.; Boudalis, A.K.; Jana, A.D.; Luneau, D. Azide-bridged manganese(III) one-dimensional chain: Synthesis, structure, and magnetic study. J. Coord. Chem. 2013, 66, 9–17. [Google Scholar] [CrossRef]
- Ko, H.H.; Lim, J.H.; Kim, H.C.; Hong, C.S. Coexistence of Spin Canting and Metamagnetism in a One-Dimensional Mn(III) Complex Bridged by a Single End-to-End Azide. Inorg. Chem. 2006, 45, 8847–8849. [Google Scholar] [CrossRef]
- Panja, A.; Shaikh, N.; Vojtíšek, P.; Gao, S.; Banerjee, P. Synthesis, crystal structures and magnetic properties of 1D polymeric [MnIII(salen)N3] and [MnIII(salen)Ag(CN)2] complexes. New J. Chem. 2002, 26, 1025–1028. [Google Scholar] [CrossRef]
- Li, W.; Li, Z.; Li, L.; Liao, D.; Jiang, Z. A one-dimensional azido-bridged manganese(III) complex with bidentate Schiff base: Crystal structure and magnetic properties. J. Solid State Chem. 2007, 180, 2973–2977. [Google Scholar] [CrossRef]
- Qian, K.; Xu, Y.; Wang, Z.; Yang, J. Synthesis, crystal structure, and magnetic properties of an azido-bridged Mn(II) complex [C3H5NH3][Mn(N3)3]. Z. Für Naturforschung B 2017, 72, 409–413. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, W.R.; Ryu, D.W.; Lee, J.W.; Yoon, S.W.; Suh, B.J.; Kim, H.C.; Hong, C.S. End-to-End Azide-Bridged Manganese(III) Chain Compounds: Field-Induced Magnetic Phase Transitions and Variation of TC to 38 K depending on the Side Groups of the Schiff Bases. Inorg. Chem. 2011, 50, 10777–10785. [Google Scholar] [CrossRef]
- Hong, C.S.; Koo, J.-e.; Son, S.-K.; Lee, Y.S.; Kim, Y.-S.; Do, Y. Unusual Ferromagnetic Couplings in Single End-to-End Azide-Bridged Cobalt(II) and Nickel(II) Chain Systems. Chem. Eur. J. 2001, 7, 4243–4252. [Google Scholar] [CrossRef] [PubMed]
- Masoomi, M.Y.; Morsali, A. Morphological study and potential applications of nano metal–organic coordination polymers. RSC Adv. 2013, 3, 19191–19218. [Google Scholar] [CrossRef]
- Islamoglu, T.; Goswami, S.; Li, Z.; Howarth, A.J.; Farha, O.K.; Hupp, J.T. Postsynthetic Tuning of Metal–Organic Frameworks for Targeted Applications. Acc. Chem. Res. 2017, 50, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Aung, T.; Guo, N.; Weichselbaum, R.; Lin, W. Nanoscale Metal–Organic Frameworks for Therapeutic, Imaging, and Sensing Applications. Adv. Mater. 2018, 30, 1707634. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-C.J.; Kitagawa, S. Metal–Organic Frameworks (MOFs). Chem. Soc. Rev. 2014, 43, 5415–5418. [Google Scholar] [CrossRef]
- Keypour, H.; Salehzadeh, S.; Pritchard, R.G.; Parish, R.V. Synthesis and Crystal Structure Determination of Some Asymmetrical and Symmetrical CR-Type Macrocyclic Schiff Base Complexes, with a Single Pendant Coordinating 2-Aminoethyl Arm. Inorg. Chem. 2000, 39, 5787–5790. [Google Scholar] [CrossRef]
- Suydam, F.H. The C=N Stretching Frequency in Azomethines. Anal. Chem. 1963, 35, 193–195. [Google Scholar] [CrossRef]
- Verma, A.L.; Venkateswarlu, P. Vibrational spectra and rotational isomerism of allyl amine. J. Mol. Spectrosc. 1971, 39, 227–241. [Google Scholar] [CrossRef]
- İspir, E.; Kurtoğlu, M.; Purtaş, F.; Serin, S. Synthesis and Antimicrobial Activity of New Schiff Bases Having the –SiOR Group (R = CH3 or CH2CH3), and their Transition Metal Complexes. Transit. Met. Chem. 2005, 30, 1042–1047. [Google Scholar] [CrossRef]
- Darensbourg, D.J.; Frantz, E.B. Manganese(III) Schiff Base Complexes: Chemistry Relevant to the Copolymerization of Epoxides and Carbon Dioxide. Inorg. Chem. 2007, 46, 5967–5978. [Google Scholar] [CrossRef] [PubMed]
- Sepehrfar, S.; Salehi, M.; Parvarinezhad, S.; Grześkiewicz, A.M.; Kubicki, M. New Cu(II), Mn(II) and Mn(III) Schiff base complexes cause noncovalent interactions: X-ray crystallography survey, Hirshfeld surface analysis and molecular simulation investigation against SARS-CoV-2. J. Mol. Struct. 2023, 1278, 134857. [Google Scholar] [CrossRef]
- Yuan, M.; Zhao, F.; Zhang, W.; Wang, Z.-M.; Gao, S. Azide-Bridged One-Dimensional MnIII Polymers: Effects of Side Group of Schiff Base Ligands on Structure and Magnetism. Inorg. Chem. 2007, 46, 11235–11242. [Google Scholar] [CrossRef]
- Freitag, R.; Conradie, J. Understanding the Jahn–Teller Effect in Octahedral Transition-Metal Complexes: A Molecular Orbital View of the Mn(β-diketonato)3 Complex. J. Chem. Educ. 2013, 90, 1692–1696. [Google Scholar] [CrossRef]
- Bikas, R.; Shahmoradi, E.; Reinoso, S.; Emami, M.; Lezama, L.; Sanchiz, J.; Noshiranzadeh, N. The effect of the orientation of the Jahn–Teller distortion on the magnetic interactions of trinuclear mixed-valence Mn(II)/Mn(III) complexes. Dalton Trans. 2019, 48, 13799–13812. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Lee, J.W.; Ryu, D.W.; Yoon, S.W.; Suh, B.J.; Kim, H.C.; Hong, C.S. One-Dimensional End-To-End Azide-Bridged MnIII Complexes Incorporating Alkali Metal Ions: Slow Magnetic Relaxations and Metamagnetism. Chem. Eur. J. 2011, 17, 3028–3034. [Google Scholar] [CrossRef] [PubMed]
- Desiraju, G.; Steiner, T. The Weak Hydrogen Bond in Structural Chemistry and Biology; Oxford Univiversity Press: Oxford, UK, 2006. [Google Scholar]
- Rigaku Oxford Diffraction. CrysAlisPro, Version. 1.171.36.20; Rigaku Oxford Diffraction: Yarnton, UK, 2015. [Google Scholar]
- Altomare, A.; Cascarano, G.; Giacovazzo, C.; Guagliardi, A. Completion and refinement of crystal structures with SIR92. J. Appl. Crystallogr. 1993, 26, 343–350. [Google Scholar] [CrossRef]
- Hirshfeld, F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta 1977, 44, 129–138. [Google Scholar] [CrossRef]
- Spackman, M.A.; Byrom, P.G. A novel definition of a molecule in a crystal. Chem. Phys. Lett. 1997, 267, 215–220. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Spackman, M.A.; Mitchell, A.S. Novel tools for visualizing and exploring intermolecular interactions in molecular crystals. Acta Crystallogr. Sect. B Struct. Sci. 2004, 60, 627–668. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. A.03; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Lee, C.T.; Yang, W.T.; Parr, R.G. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron-Density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Becke, A.D. Density-Functional Exchange-Energy Approximation with Correct Asymptotic-Behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate Spin-Dependent Electron Liquid Correlation Energies for Local Spin Density Calculations: A Critical Analysis. Can. J. Phys. 1980, 58, 1200–1211. [Google Scholar] [CrossRef]
- Krishnan, R.; Binkley, J.S.; Seeger, R.; Pople, J.A. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980, 72, 650–654. [Google Scholar] [CrossRef]
- Francl, M.M.; Pietro, W.J.; Hehre, W.J.; Binkley, J.S.; Gordon, M.S.; DeFrees, D.J.; Pople, J.A. Self-consistent molecular orbital methods. XXIII. A polarization-type basis set for second-row elements. J. Chem. Phys. 1982, 77, 3654–3665. [Google Scholar] [CrossRef]
- Schäfer, A.; Horn, H.; Ahlrichs, R. Fully optimized contracted Gaussian basis sets for atoms Li to Kr. J. Chem. Phys. 1992, 97, 2571–2577. [Google Scholar] [CrossRef]
- Boucher, L.J.; Day, V.W. Manganese Schiff base complexes. 5. Synthesis and spectroscopy of some anion complexes of N,N′-ethylenebis(acetylacetone iminato)manganese(III). Inorg. Chem. 1977, 16, 1360–1367. [Google Scholar] [CrossRef]
- Ferreira, G.A.; Nunes, C.M.; Lopes Jesus, A.J.; Fausto, R. The meta and para OH Substitution Effect on C-Phenyl-Nitrilimine Bond-Shift Isomers. Eur. J. Org. Chem. 2023, 26, e202300310. [Google Scholar] [CrossRef]
- Lopes Jesus, A.J.; de Lucena Júnior, J.R.; Fausto, R.; Reva, I. Infrared Spectra and Phototransformations of meta-Fluorophenol Isolated in Argon and Nitrogen Matrices. Molecules 2022, 27, 8248. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, M.K.; Brauer, B.; Martin, J.M.L. Frequency and Zero-Point Vibrational Energy Scale Factors for Double-Hybrid Density Functionals (and Other Selected Methods): Can Anharmonic Force Fields Be Avoided? J. Phys. Chem. A 2015, 119, 1701–1714. [Google Scholar] [CrossRef] [PubMed]
- Filimonov, D.A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Druzhilovskii, D.S.; Pogodin, P.V.; Poroikov, V.V. Prediction of the Biological Activity Spectra of Organic Compounds Using the Pass Online Web Resource. Chem. Heterocycl. Compd. 2014, 50, 444–457. [Google Scholar] [CrossRef]
- Parasuraman, S. Prediction of activity spectra for substances. J. Pharmacol. Pharmacother. 2011, 2, 52–53. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).