The Structural Aspects of Mutually Trans-X-Cu(I)-X (X = OL, NL, CL, PL, SL, SeL, Cl or Br) Complexes
Abstract
1. Introduction
2. Structural Aspects of Mutually Trans (X-Cu(I)-X) Complexes
2.1. Homo -X-Cu(I)-X (X = OL, NL) Type
2.2. Homo -C-Cu(I)-C- Type
2.3. Homo -X-Cu(I)-X- (X = PL, SL, SeL) Types
2.4. Homo -X-Cu(I)-X- (X = Cl, Br) Types
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BPh41− | tetraphenylborate |
| C2H3N | acetonitrile |
| C3H9SiS | trimethylsitanethotate |
| C3H9SiSe | trimethylsilaneselenotate |
| C4H12N | n-butylammonium |
| C4H8O | tetrahydrofuran |
| C5H6N2 | pyridine-4-amine |
| C5H8N4 | 3,5-dimethyl-1H-pyrazole |
| C6F18P | trifluoro(tris(pentafluoroethyl)phosphate |
| C6F18P1 | trifluoro[bis(pentafluoroethyl)phosphate |
| C6F5 | pentafluorophenyl |
| C6H14 | hexane |
| C6H5 | phenyl |
| C7H12N2 | 1,3,4,5-tetramethylimidazol-2-ylidene |
| C7H13N2 | (1,3,4,5-tetramethylimidazol-2-ylidene) |
| C8H5 | phenylethynyl |
| C9H11 | mesityl |
| C9H9 | (1-phenylprop-1-en-1-yl) |
| C10H12ClN4 | bis(pyridine-4-amine) |
| C10H14F4Si | ((2-dimethyl(phenyl)sibyl)1,1,2,2-tetrafluoro-ethyl) |
| C10H14NS | 3,3-dimethyl-3,4-dihydro-2H-1,3-benzothiazim-ium |
| C10H20BO4 | bis(2,2-dimethylpropane-1,3-diolate) |
| C10H9O2 | (2-phenylbut-2-enoato) |
| C12H24O6 | 18-crown-6 |
| C13H20N5S | 1-[bis(dimethylamino)methylidene]-2-(3-methyl-1,3-benzothiazol-2(3H)-ylidene) |
| C13H25N3 | (2,6-di-t-butyl-2,3,5,6-tetrahydro-1H-imidazo [1,2_a]-imidazole |
| C13H26N3 | 2,6-di-t-butyl-2,3,5,6-tetrahydro-1H-imidazol [1,2,a]imidazol-7-ium |
| C14H20N2O | (1-(2-hydroxyethyl-3-(2,4,6-trimethylphenyl)imidazolidine-2-ylidene) |
| C15H32N3 | 1,3,5-tri-t-butyl-2,3,4,5-tetrahydro-1,3,5-triazin-1-ium |
| C16H36N1 | tetra-n-butylammonium |
| C18H15F3NSi | (3-(3-cyanophenyl)-3-(dimethyl(phenyl)sibyl)-1,1,1-trifluoropropan-2-yl) |
| C20H24N2 | (1,3-bis(2,6-dimethylphenyl)hexahydro-pyrimidin-2-ylidne) |
| C20H27Si | (4-((t-1-butyl(diphenyl)sibyl)oxy)-1,1,1-trifluorobutan-2-yl) |
| C21H24N2Se | [1,3-bis(2,4,6-trimethylphenyl)-1.3-dihydro-2H-imidazole-2-selone] |
| C21H28N2 | (1,3-bis(2,4,6-trimethylphenyl)imidazolidine-2-ylidene) |
| C21H46N10Cl | chloro-(2,2′,2″-((nitrilo)truthane-2,1-diyl)tris(1,1,3,3-tetramethylquanidine) |
| C22H28N2 | (1,3-dimesityltetrahydropyrimidin-2(1H)-ylidene) |
| C22H28N6 | 2,2′-hydrazinediylildenebis(1,3-diethyl-2,3-dihydro-1H-benzimidazole) |
| C22H29N2 | (1,3-dimesitylhexahydropyrimidin-2-yl) |
| C22H29N2 | (1,3-dimesityltetrahydropyrimidin-2(1H)-ylidene) |
| C22H33N | (2-(2,6-di-isopropylyphenyl)-1,4,5-trimethyl-2-azabicyclo(2,2,2)octan-3-ylidne) |
| C23H25N3 | (2,6-di-t-butyl—2,3,5,6-tetrahydro-1H-ionidazo(1,2-a)imidazole |
| C23H30N2 | (1,3-bis(2,4,6-trimethylphenyl)1,3-diazepan-2-ylidene) |
| C24H20B | tetraphenylborate |
| C24H20P1+ | tetraphenylphosphonium |
| C24H28N2O2 | (1,3-dimesityl-5,5-dimethyl-4,6-diolotetrahydropyrimidin-2(1H)ylidene) |
| C24H36N6S2 | (N″,N′″-(ethane-1,2-diylbis(sulfanidiyl)2-phenylene))bis(N,N,N′,N′-tetramethyl-quanidine)) |
| C25H18N2 | (1-isoquinolin-1-yl)-N-phenylnaphtalen-2-amine) |
| C25H22P | benzyl(triphenyl)phosphonium |
| C26H38BF3N2O3Si | (4-((t-butyl(diphenyl)sibyl)oxy)-1,1,1-trifluoro-3-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)butan-2-yl) |
| C26H40N6S4Cl | (N-N″,N′″-(disulfanediylbis((ethane-2,1-diyl)sulfanediyl-2,1-phenylene))bis(N,N,N′,N′-tetramethylquanidine)-(u-chloro) |
| C27H33N2 | (1,3-bis(2,6-bis(propan-2-yl))phenyl]-2,3-dihydro-1H-imidazol-2-ylidene) |
| C27H36N2 | (1,3-bis(2,6-di-isopropylphenyl)-2,3-dihydro-1H-imidazol-2-ylidene) |
| C27H36N2 | (1,3-bis(2,6-disopropylphenyl)-imidazol-2-ylidene) |
| C27H36N2 | 1,3-bis[2,6-bis(propan-2-yl)phenyl]imidazole-2-ylidene) |
| C27H36N2Se | [1,3-bis(2,6-diisopropylphenyl)-1,3-dihydro-2H-imidazole-2-selone] |
| C28H35N | (1-(2,6-diisopropylphenyl)-3,3-dithyl-5)5-dimethylpyriolidin-2-ylidene) |
| C30H42N4 | (2-(N,N′-bis(2,6-diisopropylphenyl)carbamimidoyl)-1,3-dimethyl-1H-imidazol-3-iumato) |
| C33H40N2 | 1,3-bis[2,6-di-isopropylphenyl]-2-phenyl-1H-imidazol-3-ium-4-yl] |
| C33H60N9P | (N,N′,N″-tris(1,3-disopropyl-4,5-dimethyl-1,3-dihydro-2H-imidazol-2-ylidene)phosphorous triamide) |
| C36H45N3P2Si2 | (2,5-bis{[P,P-diphenyl-N-(trimethylsilyl)phosphorimidoyl)methyl)-1H-pyrrole) |
| C38H28N8S8 | 2-(4,5-bis((2-(methoxycarbonyl)phenyl)sulfanyl)-1,3-dithiol-2-ylidene)-4,5-bis((2-methoxycarbonyl)phenyl)sulfanyl-1,3-dithiol-1-ium |
| C42H36O8S8 | 2-(4,5-bis((4-ethoxycarbonyl)phenyl)sulfanyl)-1,3-dithiol-2-ylidene)-4,5-bis((4-(ethoxycarbonyl)phenyl)sulfanyl)-1,3-dithiol-1-ium |
| C44H62N2O8 | (4,6-dimethyl-1,3-bis(1,5,7-tripropyl-3-azabicyclo(3.3.1)none-2,4-dione-7-carboxylato)benzene) |
| C48H90N12Si2 | 2,2′,2″,2′″-(3,6-bis([tris(propan-2-yl)silyl]ethynyl)benzene-1,2,4,5 tetrayl)tetraphis (1,1,3,3-tetramethylquanidine) |
| Me | methyl |
| Met42 | tetraethylammonium |
| P≡CO | phosphinedyne |
| pz | pyrazole |
References
- Hattaway, B.J. Copper. Coord. Chem. Rev. 1981, 35, 211–252. [Google Scholar] [CrossRef]
- Hattaway, B.J. Copper. Coord. Chem. Rev. 1982, 41, 423–487. [Google Scholar] [CrossRef]
- Hattaway, B.J. Copper. Coord. Chem. Rev. 1983, 52, 87–169. [Google Scholar] [CrossRef]
- O’Brien, P. Copper. Coord. Chem. Rev. 1984, 58, 169–244. [Google Scholar] [CrossRef]
- Sigel, H. (Ed.) Metal Ions in Biological Systems: Properties of Copper Volume 12, 1st ed.; Marcel Dekker Inc: New York, NY, USA, 1981; 400p. [Google Scholar]
- Sigel, H. Metal Ions in Biological Systems: Copper Proteins Volume 13, 1st ed.; Sigel, H., Sigel, A., Eds.; Marcel Dekker Inc: New York, NY, USA, 1982; 400p. [Google Scholar]
- Lontie, R. (Ed.) Copper Proteins and Copper Enzymes, Volume 1, 1st ed.; CRC Press: Boca Raton, FL, USA, 1984; 233p. [Google Scholar]
- Holloway, C.E.; Melník, M. Copper(I) Compounds: Classification and analysis of Crystallographic and structural Data. Rev. Inorg. Chem. 1995, 15, 147–386. [Google Scholar] [CrossRef]
- Le Cloux, D.D.; Lippard, S.J. Synthesis and Characterization of a Novel Class of Dicopper(I) Bis(carboxylate)-Bridged Complexes. Inorg. Chem. 1997, 36, 4035–4046. [Google Scholar] [CrossRef]
- Collins, L.R.; Riddlestone, I.M.; Mahon, M.F.; Whittlesey, M.K. A Comparison of the Stability and Reactivity of Diamido- and Diaminocarbene Copper Alkoxide and Hydride Complexes. Chem. Eur. J. 2015, 21, 14075–14084. [Google Scholar] [CrossRef] [PubMed]
- Luk’yanov, M.; Slyvka, Y.; Kinzhybalo, V.; Mys´kiv, M. Synthesis, crystal structure and Hirshfeld surface analysis of the [Cu2(3,5-dimethyl-1H-pyrazole)4(ClO4)2] complex. Chem. Met. Alloys 2016, 9, 27–33. [Google Scholar] [CrossRef]
- Marquez, A.; Avila, E.; Urbaneja, C.; Alvarez, E.; Palma, P.; Campora, J. Copper(I) Complexes of Zwitterionic Imidazolium-2-Amidinates, a Promising Class of Electroneutral, Amidinate-Type Ligands. Inorg. Chem. 2015, 54, 11007–11017. [Google Scholar] [CrossRef]
- Ramirez-Lopez, P.; Ros, A.; Romero-Arenas, A.; Iglesias-Siguenza, J.; Fernandez, R.; Lassaletta, J.M. Synthesis of IAN-type N,N-Ligands via Dynamic Kinetic Asymmetric Buchwald-Hartwig Amination. J. Am. Chem. Soc. 2016, 138, 12053–12056. [Google Scholar] [CrossRef]
- Jha, V.K.; Das, S.; Subramaniyan, V.; Guchhait, T.; Dakua, K.K.; Mishra, S.; Mani, G. Synthesis, structural characterization, and bonding analysis of two-coordinate copper(i) and silver(i) complexes of pyrrole-based bis(phosphinimine): New metal–pyrrole ring π-interactions. Dalton Trans. 2021, 50, 8036–8044. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Cui, X.Y.; Tan, S.M.; Jiang, H.; Ren, J.; Lee, N.; Lee, R.; Tan, C.H. Guanidine-Copper Complex Catalyzed Allylic Borylation for the Enantioconvergent Synthesis of Tertiary Cyclic Allylboronates. Angew. Chem. 2019, 58, 2382–2386. [Google Scholar] [CrossRef]
- Skelton, B.W. (Crystallography Centre and Department of Chemistry, University of Western Australia, Nedlands, WA 6907, Australia); Healy, P.C. (Faculty of Science and Technology, Griffith University, Nathan, Brisbane, QLD 4111, Australia). CSD Communication (2018) CBEWHOD, 2022. (Private communication).
- Sorrell, T.N.; Jameson, D.L. Synthesis, structure, and reactivity of monomeric two-coordinate copper(I) complexes. J. Am. Chem. Soc. 1983, 105, 6013–6018. [Google Scholar] [CrossRef]
- Ho, N.K.T.; Reichmann, S.O.; Rottschafer, D.; Herbst-Irmer, R.; Ghadwal, R.S. Expanding the Scope of Cu(I) Catalyzed “Click Chemistry” with Abnormal NHCs: Three-Fold Click to Tris-Triazoles. Catalysts 2017, 7, 262. [Google Scholar] [CrossRef]
- Fochrenbach, S.A.; Kuhfuss, M.J.; Zaff, L.; Friedroch, A.; Ignalev, N.V.; Finze, M.; Radius, U. Tris (pentafluoroethyl) difluorophosphorane: A Versatile Fluoride Acceptor for Transition Metal Chemistry. Chem. A Eur. J. 2021, 27, 3504–3516. [Google Scholar] [CrossRef] [PubMed]
- Seifert, T.P.; Boukis, A.C.; Feuerstein, T.J.; Roesky, P.W. A straightforward synthetic route to symmetric bis(acetylide) metallates of the coinage metals. J. Organomet. Chem. 2018, 867, 92–97. [Google Scholar] [CrossRef]
- Shi, S.; Collins, L.R.; Mahon, M.F.; Djurovich, P.I.; Thompson, M.E.; Whittlesey, M.K. Synthesis and characterization of phosphorescent two-coordinate copper(i) complexes bearing diamidocarbene ligands. Dalton Trans. 2017, 46, 745–752. [Google Scholar] [CrossRef]
- Thiel, N.O.; Brechmann, L.T.; Teichert, J.F. Catalytic Hydrogenations with Cationic Heteroleptic Copper(I)/N-Heterocyclic Carbene Complexes. Synlett 2019, 30, 783–786. [Google Scholar] [CrossRef]
- Plotzitzka, J.; Kleeberg, C. [(NHC)CuI–ER3] Complexes (ER3 = SiMe2Ph, SiPh3, SnMe3): From Linear, Mononuclear Complexes to Polynuclear Complexes with Ultrashort CuI···CuI Distances. Inorg. Chem. 2016, 55, 4813–4823. [Google Scholar] [CrossRef]
- Hall, J.; Unson, D.; Brunel, P.; Collins, L.; Cybulski, M.; Mahon, M.; Whittlesey, M.K. Copper-NHC-Mediated Semi-Hydrogenation and Hydroboration of Alkynes: Enhanced Catalytic Activity Using Ring-Expanded Carbenes. Organometallics 2018, 37, 3102–3110. [Google Scholar] [CrossRef]
- Liu, L.L.; Ruiz, D.A.; Dahcheh, F.; Bertrand, G.; Suter, R.; Tondreau, A.M.; Grutzmacher, H. Isolation of Au-, Co-η1PCO and Cu-η2PCO complexes, conversion of an Ir–η1PCO complex into a dimetalladiphosphene, and an interaction-free PCO anion. Chem. Sci. 2016, 7, 2335–2341. [Google Scholar] [CrossRef] [PubMed]
- Paioti, P.H.S.; del Pozo, J.; Mikus, M.S.; Lee, J.; Koh, M.J.; Romiti, F.; Torker, S.; Hoveyda, A.H. Catalytic Enantioselective Boryl and Silyl Substitution with Trifluoromethyl Alkenes: Scope, Utility, and Mechanistic Nuances of Cu–F β-Elimination. J. Am. Chem. Soc. 2019, 141, 19917–19934. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, H.; Okashi, M.; Ogoshi, S. Fluorinated Vinylsilanes from the Copper-Catalyzed Defluorosilylation of Fluoroalkene Feedstocks. Angew. Chem. Int. Ed. Engl. 2018, 57, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Manar, K.K.; Chakrabortty, S.; Porwal, V.K.; Prakash, D.; Thakur, S.K.; Choudhury, A.R.; Singh, S. Two-Coordinate Cu(I) and Au(I) Complexes Supported by BICAAC and CAAC Ligands. ChemistrySelect 2020, 5, 9900–9907. [Google Scholar] [CrossRef]
- Ritter, F.; Mukherjee, D.; Spaniol, T.P.; Hoffmann, A.; Okuda, J. A Masked Cuprous Hydride as a Catalyst for Carbonyl Hydrosilylation in Aqueous Solutions. Angew. Chem. Int. Ed. 2019, 58, 1818–1822. [Google Scholar] [CrossRef]
- Drescher, W.; Borner, C.; Kleeberg, C. Stability and decomposition of copper(i) boryl complexes: [(IDipp)Cu–Bneop], [(IDipp*)Cu–Bneop] and copper clusters. New J. Chem. 2021, 45, 14957–14964. [Google Scholar] [CrossRef]
- Mehlmann, P.; Muck-Lichtenfeld, C.; Tan, T.Y.T.; Dielmann, F. Tris(imidazolin-2-ylidenamino)phosphine: A Crystalline Phosphorus(III) Superbase That Splits Carbon Dioxide. Chem. A Eur. J. 2017, 23, 5929–5933. [Google Scholar] [CrossRef]
- Guschlbauer, J.; Vollgraff, T.; Xie, X.; Weigend, F.; Sundermeyer, J. A Series of Homoleptic Linear Trimethylsilylchalcogenido Cuprates, Argentates and Aurates Cat[Me3SiE–M–ESiMe3] (M = Cu, Ag, Au; E = S, Se). Inorg. Chem. 2020, 59, 17565–17572. [Google Scholar] [CrossRef]
- Srinivas, K.; Prabusankar, G. Role of C, S, Se and P donor ligands in copper(i) mediated C–N and C–Si bond formation reactions. RSC Adv. 2018, 8, 32269–32282. [Google Scholar] [CrossRef]
- Florke, U. (Department Chemie, Fakultät für Naturwissenschaften, Universität Paderborn, Warburgerstraße 100, 33098 Paderborn, Germany); Nagel, C. (Institut für Anorganische Chemie, Julius-Maximilians-Universität Würzburg, Am Hubland, 97074 Würzburg, Germany); Henkel, G. CSD Communication (IVANKAR), 2017. (Private communication).
- Florke, U. (Department Chemie, Fakultät für Naturwissenschaften, Universität Paderborn, Warburgerstraße 100, 33098 Paderborn, Germany); Bernard, M. (Institut Charles Sadron 6, rue Boussingault, F-67083 Strasbourg Cedex, France); Henkel, G. (Institut für Anorganische Chemie, Westfälische Wilhelms-Universität Münster, 4400 Münster, Germany). CSD Communication, ZAJBUK, 2017. (Private communication).
- Werr, M.; Kaifer, E.; Wadepohl, H.; Himmel, H.J. Tuneable Redox Chemistry and Electrochromism of Persistent Symmetric and Asymmetric Azine Radical Cations. Chem. A Eur. J. 2019, 25, 12981–12990. [Google Scholar] [CrossRef]
- Cui, X.Y.; Ge, Y.; Tan, S.M.; Jiang, H.; Tan, D.; Lu, Y.; Lee, R.; Tan, C.H. (Guanidine)copper Complex-Catalyzed Enantioselective Dynamic Kinetic Allylic Alkynylation under Biphasic Condition. J. Am. Chem. Soc. 2018, 140, 8448–8455. [Google Scholar] [CrossRef] [PubMed]
- Siegler, M.A. (Department of Chemistry, Johns Hopkins University, Baltimore, MD 21218, USA); Spek, A.L. (Bijvoet Center for Biomolecular Research, Crystal and Structural Chemistry, Utrecht University, Utrecht, The Netherlands); Sperotto, E. (Chemical Biology and Organic Chemistry, Utrecht University, Padualaan 8, 3584 CH, Utrecht, The Netherlands); Klein Gebbink, R.J.M. (Organic Chemistry and Catalysis, Debye Institute for NanoMaterials Science, Utrecht University, Universiteitsweg 99, 3584 CG, Utrecht, The Netherlands). CSD Communication BABRUV, 2015. (Private communication).
- Jones, P.G. (Fachbereich Chemie und Pharmazie, Technische Universität Braunschweig, Postfach 3329, 38023 Braunschweig, Germany); Daniliuc, C.G. (Organisch-Chemisches Institut, Universität Münster, Corrensstraße 40, Münster 48149, Germany); du Mont, W.W. (Fachbereich Chemie und Pharmazie, Technische Universität Braunschweig, Postfach 3329, 38023 Braunschweig, Germany). CSD Communication BUCJEF, 2015. (Private communication).
- Connor, B.A.; Smaha, R.W.; Li, J.; Gold-Parker, A.; Heyer, A.J.; Toney, M.F.; Lee, Y.S.; Karunadasa, H.I. Alloying a single and a double perovskite: A Cu+/2+ mixed-valence layered halide perovskite with strong optical absorption. Chem. Sci. 2021, 12, 8689–8697. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Kreis, F.; Popp, D.; Hubner, O.; Kaifer, R.; Himmel, H.J. 1,2,4,5-Tetrakis(tetramethylguanidino)-3,6-diethynyl-benzenes: Fluorescent Probes, Redox-Active Ligands and Strong Organic Electron Donors. Chemistry 2020, 26, 10336–10347. [Google Scholar] [CrossRef]
- Khatua, S.; Majumdar, A. Cleavage of carbon–nitrogen bond in 1,3,5-tri-tert-butyl-1,3,5-triazacyclohexane by copper(I) bromide. J. Mol. Struct. 2016, 1120, 267–273. [Google Scholar] [CrossRef]
- Ma, L.; Sun, J.; Li, X.; Zhang, S.; Qi, H.; Liu, L.; Shao, Y.; Shao, X. Copper ion salts of arylthiotetrathiafulvalenes: Synthesis, structure diversity and magnetic properties. Beilstein J. Org. Chem. 2015, 11, 850. [Google Scholar] [CrossRef]
- Chen, S.; Gao, J.; Chang, J.; Li, Y.; Huangfu, C.; Meng, H.; Wang, Y.; Xia, G.; Fang, L. Family of Highly Luminescent Pure Ionic Copper(I) Bromide Based Hybrid Materials. ACS Appl. Mat. Interfaces 2019, 11, 17513–17520. [Google Scholar] [CrossRef]
- Florke, U. (Department Chemie, Fakultät für Naturwissenschaften, Universität Paderborn, Warburgerstraße 100, 33098 Paderborn, Germany). CSD Communication OJIFIY, 2016. (Private communication).
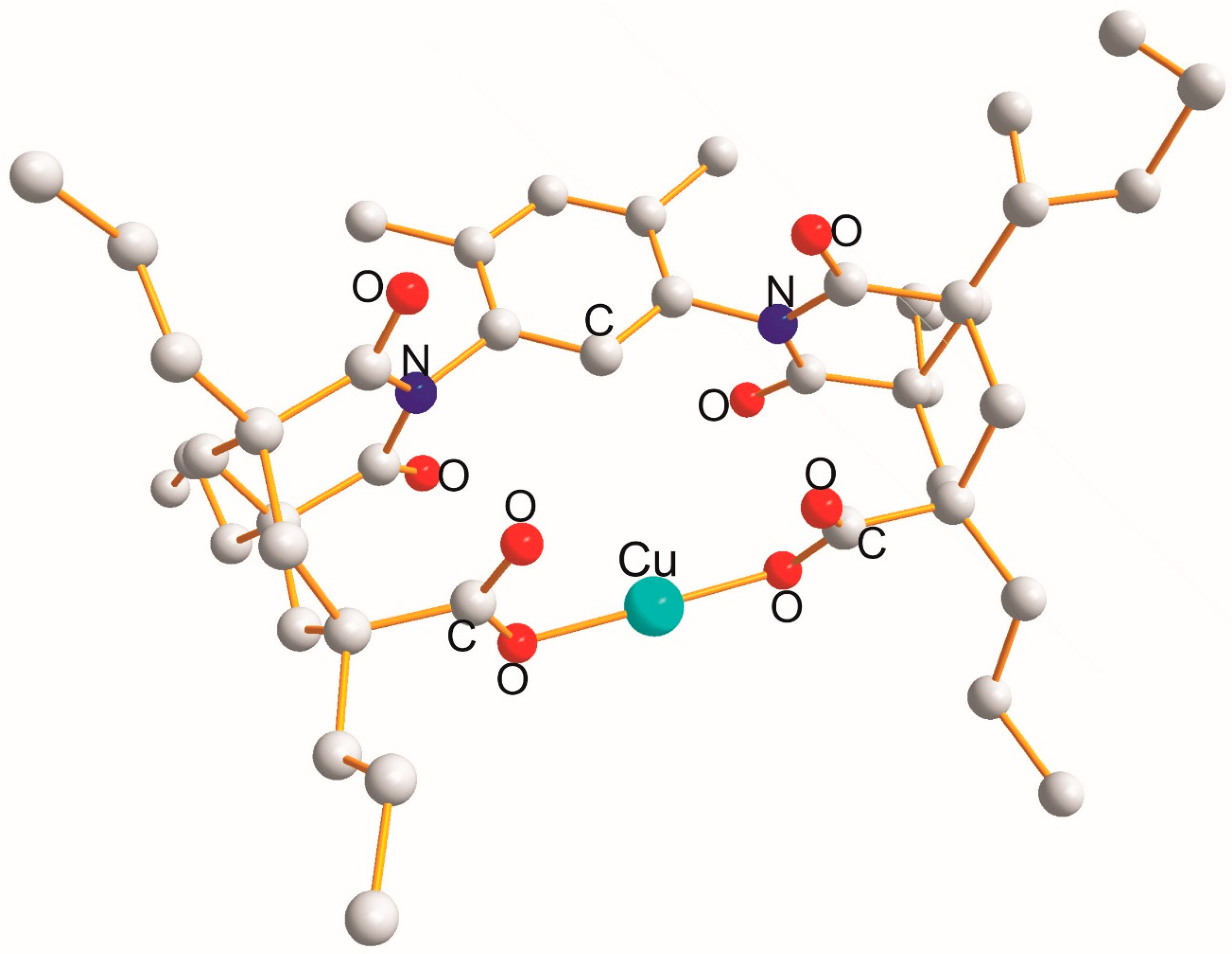

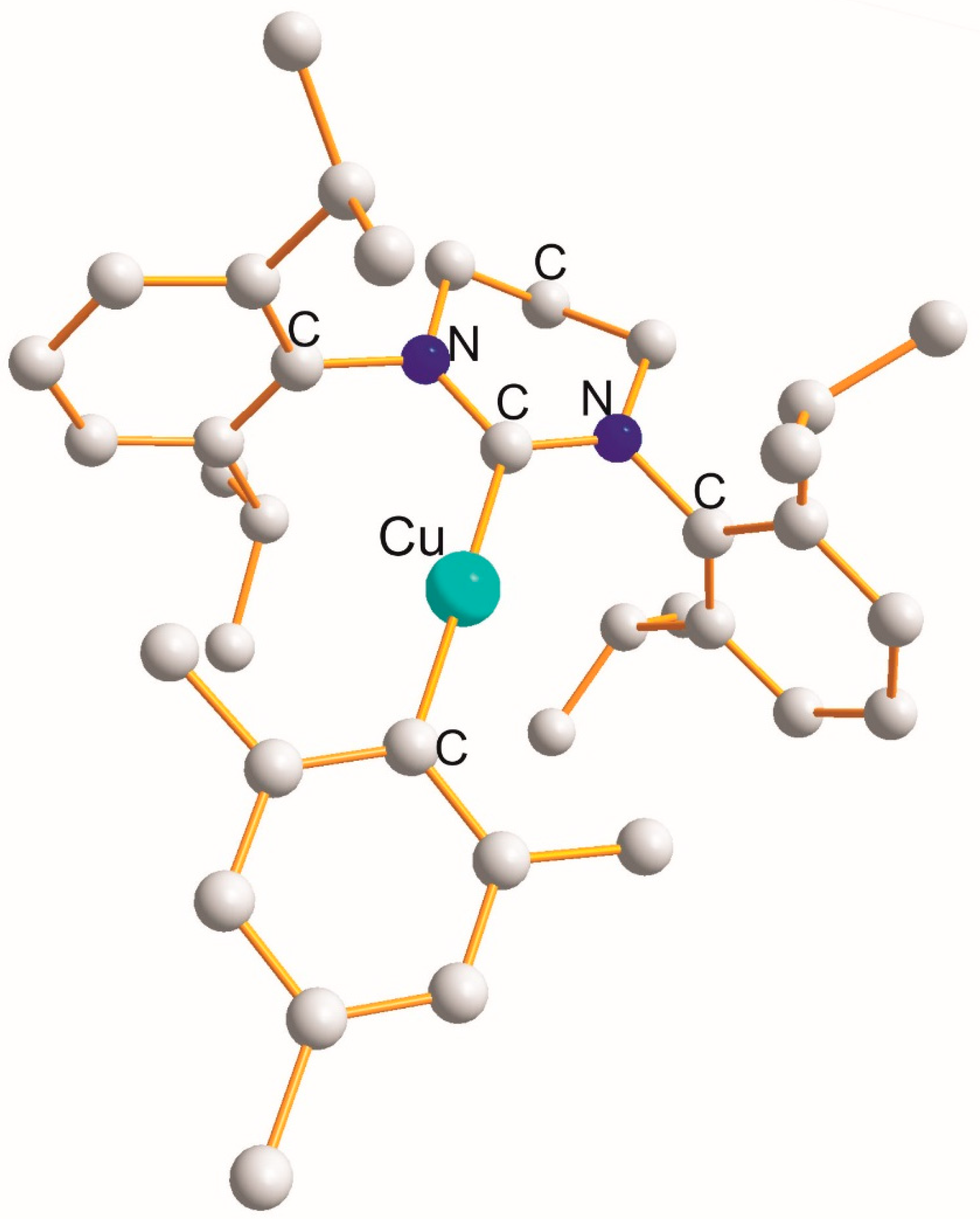
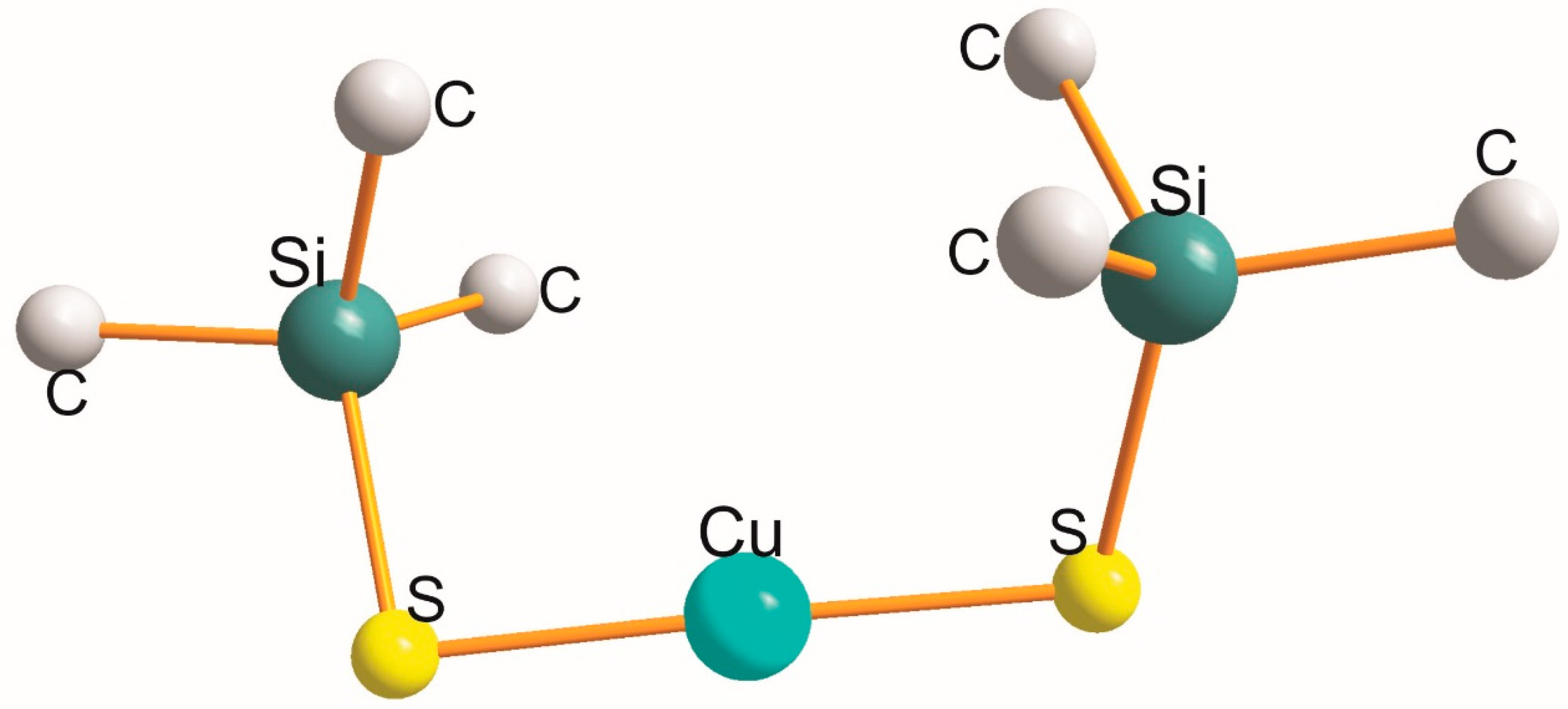
| X-Cu(I)-X | Cu-X [Å] | X (cov. r.) [Å] | X-Cu-X [°] |
|---|---|---|---|
| O-Cu(I)-O | 1.849 | O (0.73) | 176.5 |
| N-Cu(I)-N | 1.886 | N (0.75) | 174.5 |
| C-Cu(I)-C | 1.900 | C (0.77) | 174.0 |
| Cl-Cu(I)-Cl | 2.104 | Cl (0.99) | 175.9 |
| S-Cu(I)-S | 2.137 | S (1.02) | 178.6 |
| P-Cu(I)-P | 2.236 | P (1.06) | 172.6 |
| Br-Cu(I)-Br | 2.244 | Br (1.14) | 178.4 |
| Se-Cu(I)-Se | 2.260 | Se (1.17) | 180.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melník, M.; Mikušová, V.; Mikuš, P. The Structural Aspects of Mutually Trans-X-Cu(I)-X (X = OL, NL, CL, PL, SL, SeL, Cl or Br) Complexes. Inorganics 2024, 12, 245. https://doi.org/10.3390/inorganics12090245
Melník M, Mikušová V, Mikuš P. The Structural Aspects of Mutually Trans-X-Cu(I)-X (X = OL, NL, CL, PL, SL, SeL, Cl or Br) Complexes. Inorganics. 2024; 12(9):245. https://doi.org/10.3390/inorganics12090245
Chicago/Turabian StyleMelník, Milan, Veronika Mikušová, and Peter Mikuš. 2024. "The Structural Aspects of Mutually Trans-X-Cu(I)-X (X = OL, NL, CL, PL, SL, SeL, Cl or Br) Complexes" Inorganics 12, no. 9: 245. https://doi.org/10.3390/inorganics12090245
APA StyleMelník, M., Mikušová, V., & Mikuš, P. (2024). The Structural Aspects of Mutually Trans-X-Cu(I)-X (X = OL, NL, CL, PL, SL, SeL, Cl or Br) Complexes. Inorganics, 12(9), 245. https://doi.org/10.3390/inorganics12090245









