Effect of the Manufacturing Process on the Microbiota, Organoleptic Properties and Volatilome of Three Salmon-Based Products
Abstract
:1. Introduction
2. Material and Methods
2.1. Salmon Products, Raw Material Process and Storage
2.2. Sampling Dates and Type of Analyses
2.3. Classical Microbiological Analysis
2.4. Metabarcoding of 16S rRNA Gene
2.5. Sensory Analysis
2.6. Biochemical Analysis
2.6.1. Physicochemical Parameters
2.6.2. Biogenic Amines Measurement
2.6.3. Headspace–Solid-Phase Microextraction (HS-SPME) and Gas Chromatography/Mass Spectrometry (GC/MS) Analysis of the Volatilome
2.7. Multiblock Data Integration
3. Results
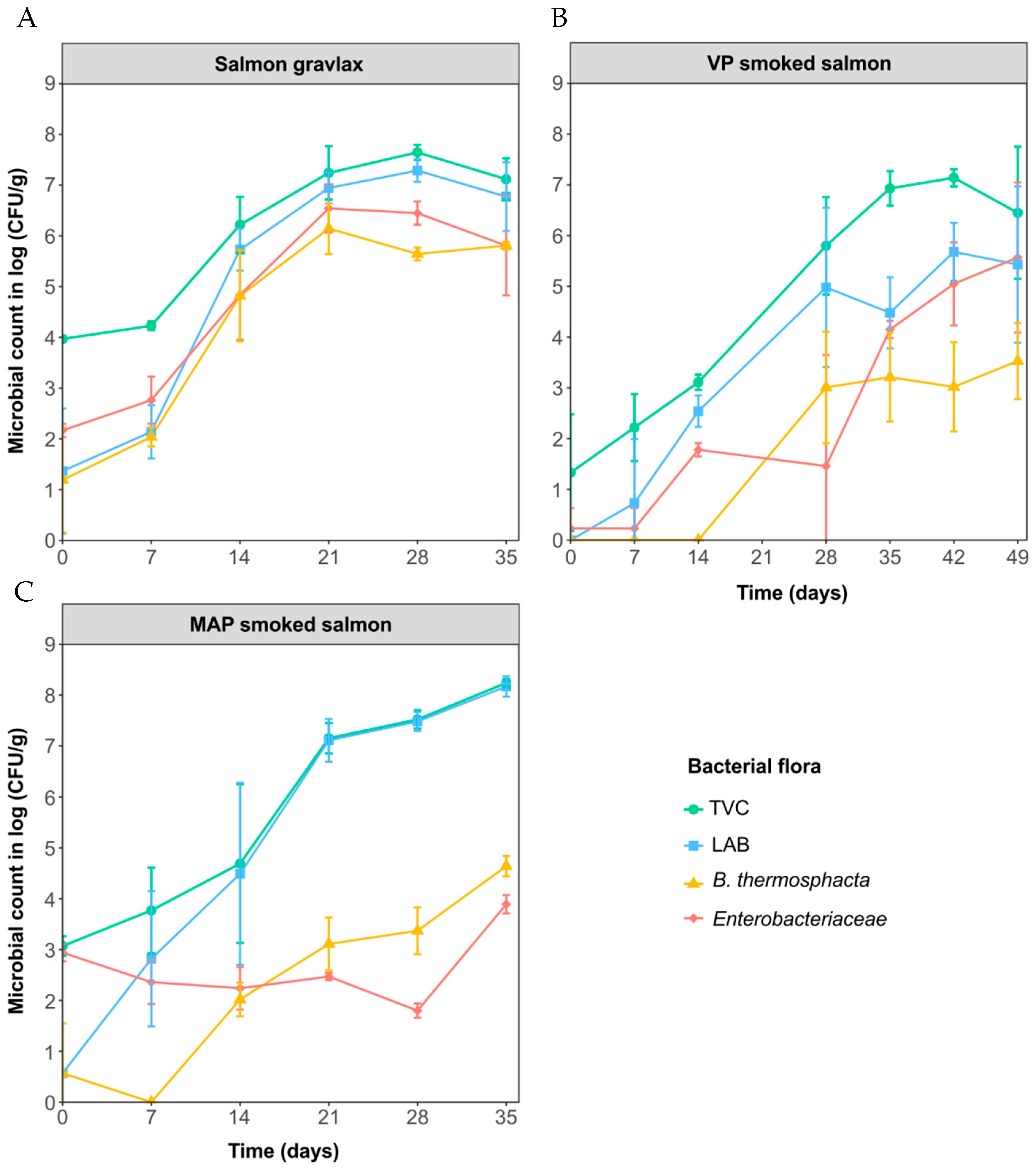
3.1. Microbial Analysis
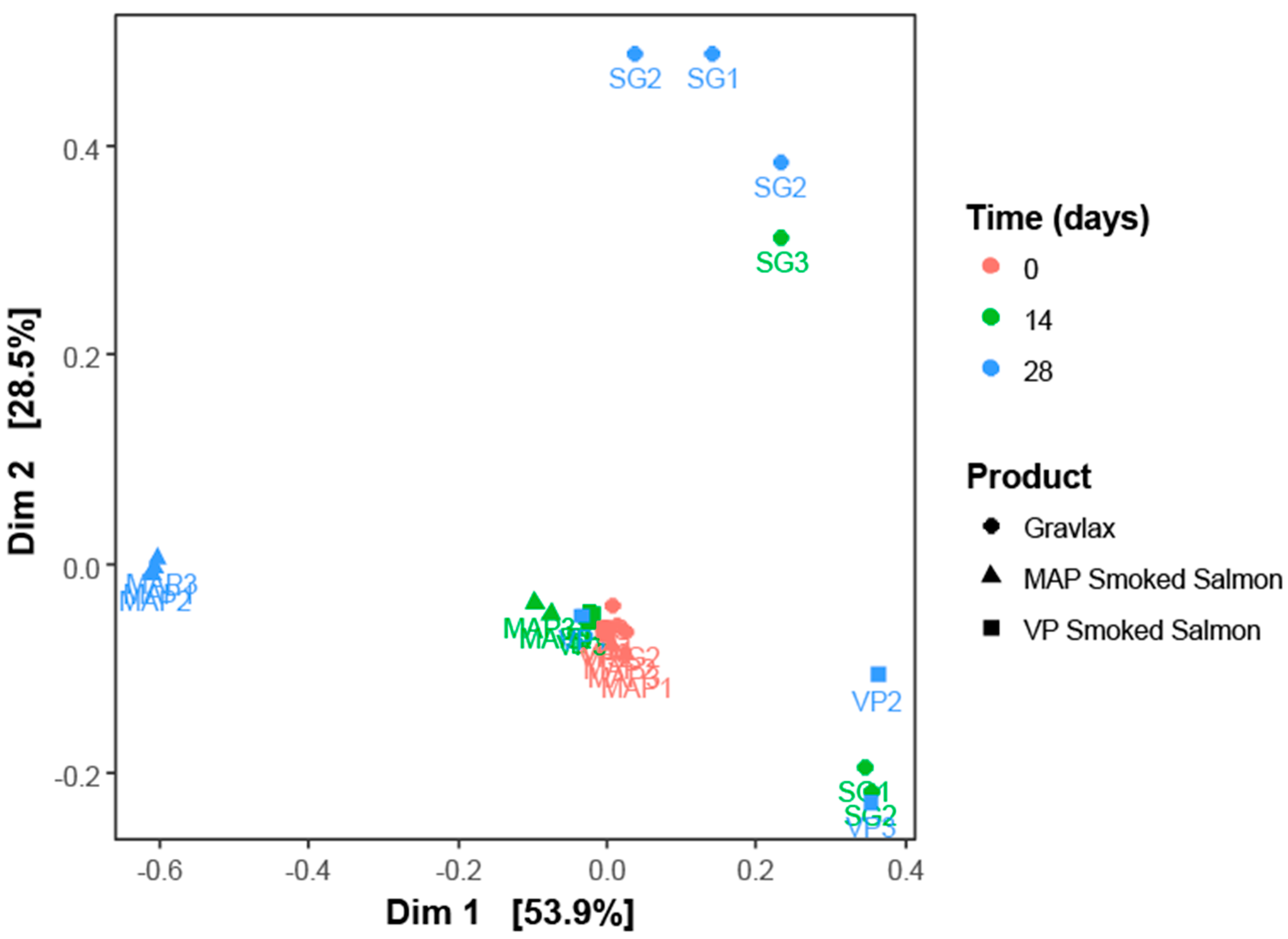
3.2. Ecosystem Monitoring through Metabarcoding
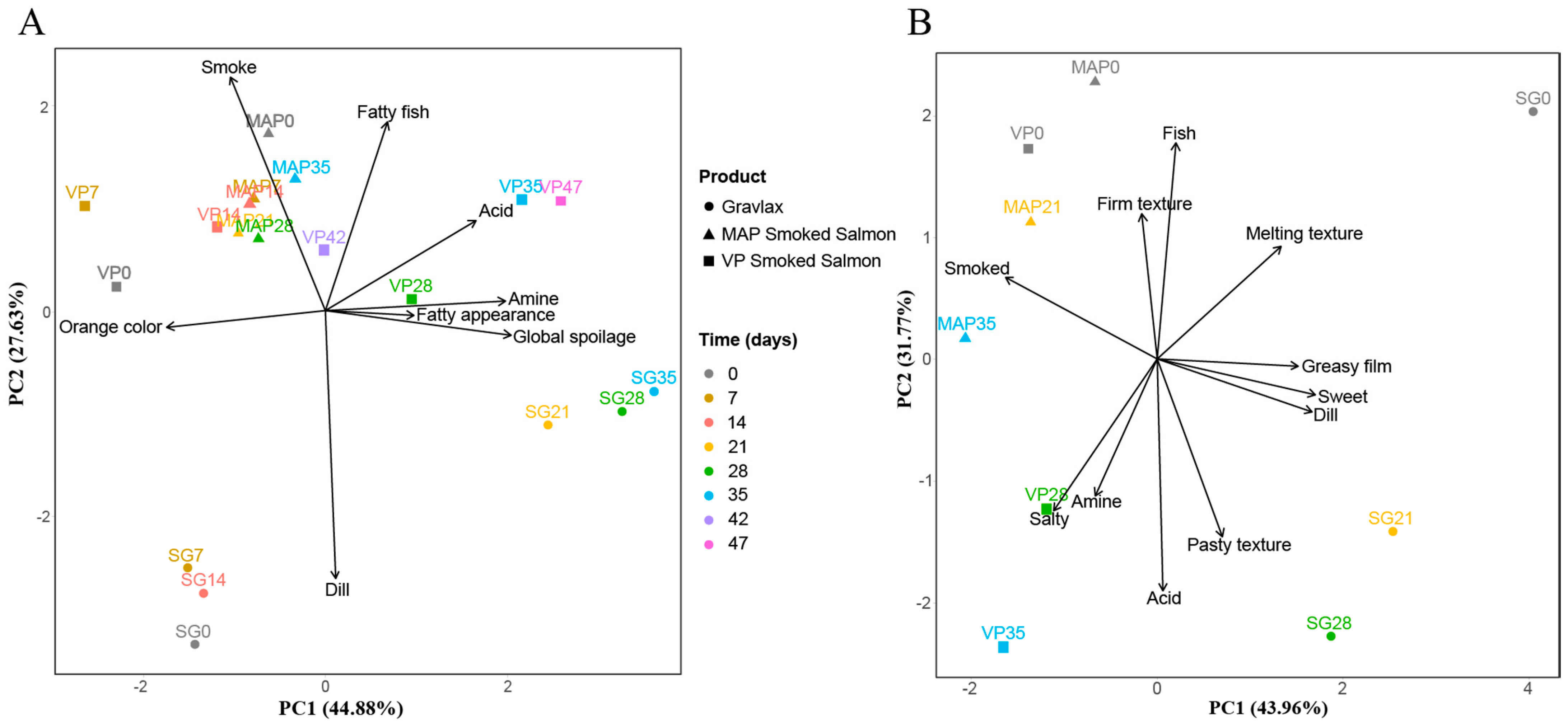
3.3. Salmon Based Products Sensory Evolution
3.4. Chemical Analyses
3.5. Biogenic Amines Content
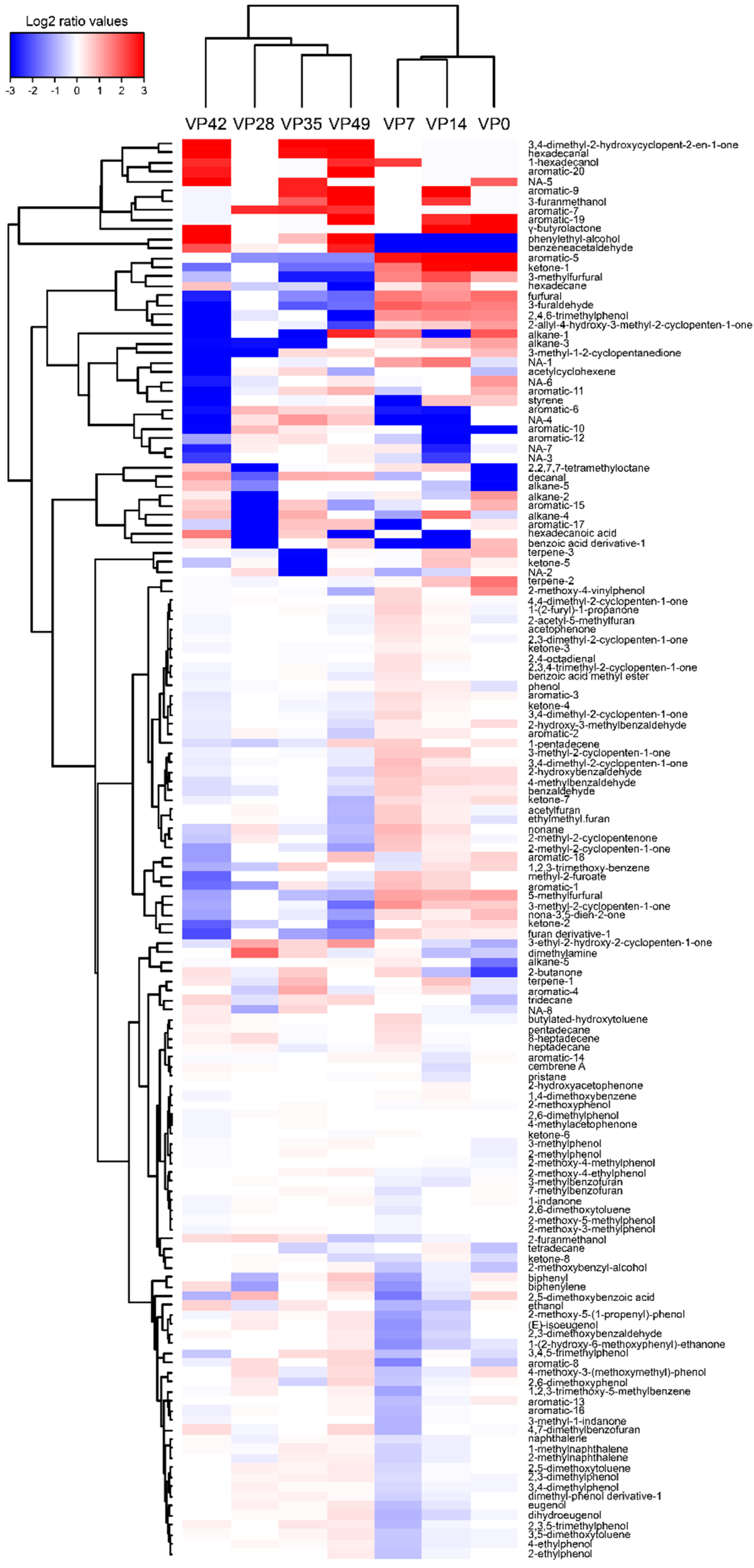
3.6. VOCs Profile
3.7. Integration of the Various Blocks of Data
4. Discussion
| Genus | Ecological Origin | References |
|---|---|---|
| Bacteria | ||
| Brachybacterium | Salt-fermented seafood, lake sediment, soil, seawater, animal feces | [85,86,87,88] |
| Duganella | Soil, plant roots, water | [89,90,91] |
| Terribacillus | Soil, salted lake sediment, plant material | [92,93,94] |
| Spelaicoccus | Soil | [95] |
| Brevibacterium | Soil, seawater, feces, compost, plant material, milk, cheese, poultry, CSS | [96,97,98,99,100] |
| Comamonas | Freshwater, plant, compost, soil, fish gut | [101,102,103,104,105] |
| Sphingobacterium | Soil, permafrost, glacier, animal feces and gut, milk, compost, plant material, lichen, freshwater | [106,107,108,109,110,111,112] |
| Archaea | ||
| Halorubrum | Salt-fermented seafood, rock salt, salted lake sediment, solar saltern | [113,114,115,116] |
| Halohasta | Salted lake water, aquaculture water | [117] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FranceAgriMer Consommation Des Produits de La Pêche et de l’aquaculture 2017. Available online: http://www.franceagrimer.fr/fam/content/download/52763/508694/file/STA-MER-CONSO%202016-juil2017.pdf (accessed on 10 July 2018).
- EUFOMA. The EU Fish Market. 2020 Edition. Available online: https://www.eumofa.eu/fr/the-eu-fish-market-2020-edition-is-now-online (accessed on 25 August 2021).
- Leroi, F.; Joffraud, J.-J.; Chevalier, F.; Cardinal, M. Study of the Microbial Ecology of Cold-Smoked Salmon during Storage at 8 °C. Int. J. Food Microbiol. 1998, 39, 111–121. [Google Scholar] [CrossRef]
- Leroi, F.; Joffraud, J.J.; Chevalier, F.; Cardinal, M. Research of Quality Indices for Cold-Smoked Salmon Using a Stepwise Multiple Regression of Microbiological Counts and Physico-Chemical Parameters. J. Appl. Microbiol. 2001, 90, 578–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paludan-Müller, C.; Dalgaard, P.; Huss, H.H.; Gram, L. Evaluation of the Role of Carnobacterium Piscicola in Spoilage of Vacuum- and Modified-Atmosphere-Packed Cold-Smoked Salmon Stored at 5°C. Int. J. Food Microbiol. 1998, 39, 155–166. [Google Scholar] [CrossRef]
- Truelstrup Hansen, L.; Huss, H.H. Comparison of the Microflora Isolated from Spoiled Cold-Smoked Salmon from Three Smokehouses. Food Res. Int. 1998, 31, 703–711. [Google Scholar] [CrossRef]
- Jørgensen, L.V.; Dalgaard, P.; Huss, H.H. Multiple Compound Quality Index for Cold-Smoked Salmon (Salmo Salar) Developed by Multivariate Regression of Biogenic Amines and PH. J. Agric. Food Chem. 2000, 48, 2448–2453. [Google Scholar] [CrossRef]
- Jørgensen, L.V.; Huss, H.H.; Dalgaard, P. Significance of Volatile Compounds Produced by Spoilage Bacteria in Vacuum-Packed Cold-Smoked Salmon (Salmo Salar) Analyzed by GC-MS and Multivariate Regression. J. Agric. Food Chem. 2001, 49, 2376–2381. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, M.-N.; Sanz, J.-J.; Santos, J.-Á.; Otero, A.; Garcia-Lopez, M.-L. Numbers and Types of Microorganisms in Vacuum-Packed Cold-Smoked Freshwater Fish at the Retail Level. Int. J. Food Microbiol. 2002, 77, 161–168. [Google Scholar] [CrossRef]
- Cardinal, M.; Gunnlaugsdottir, H.; Bjoernevik, M.; Ouisse, A.; Luc Vallet, J.; Leroi, F. Sensory Characteristics of Cold-Smoked Atlantic Salmon (Salmo Salar) from European Market and Relationships with Chemical, Physical and Microbiological Measurements. Food Res. Int. 2004, 37, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Rachman, C.; Fourrier, A.; Sy, A.; De La Cochetiere, M.F.; Prevost, H.; Dousset, X. Monitoring of Bacterial Evolution and Molecular Identification of Lactic Acid Bacteria in Smoked Salmon during Storage. Le Lait 2004, 84, 145–154. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, T.C.; Ahrné, S.; Molin, G. The Bacterial Flora of Vacuum-Packed Cold-Smoked Salmon Stored at 7 °C, Identified by Direct 16S RRNA Gene Analysis and Pure Culture Technique. J. Appl. Microbiol. 2007, 103, 109–119. [Google Scholar] [CrossRef]
- Leroi, F. Role of Bacteria in Seafood Products. In Seafood Science: Advances in Chemistry, Technology and Applications; Kim, S.K., Ed.; CRC press: Boca Raton, FL, USA, 2014; pp. 458–482. [Google Scholar]
- Leisner, J.J.; Millan, J.C.; Huss, H.H.; Larsen, L.M. Production of Histamine and Tyramine by Lactic Acid Bacteria Isolated from Vacuum-Packed Sugar-Salted Fish. J. Appl. Bacteriol. 1994, 76, 417–423. [Google Scholar] [CrossRef]
- Lyhs, U.; Lahtinen, J.; Fredriksson-Ahomaa, M.; Hyytiä-Trees, E.; Elfing, K.; Korkeala, H. Microbiological Quality and Shelf-Life of Vacuum-Packaged ‘Gravad’ Rainbow Trout Stored at 3 and 8 °C. Int. J. Food Microbiol. 2001, 70, 221–230. [Google Scholar] [CrossRef]
- Wiernasz, N.; Leroi, F.; Chevalier, F.; Cornet, J.; Cardinal, M.; Rohloff, J.; Passerini, D.; Skırnisdóttir, S.; Pilet, M.-F. Salmon Gravlax Biopreservation with Lactic Acid Bacteria: A Polyphasic Approach to Assessing the Impact on Organoleptic Properties, Microbial Ecosystem and Volatilome Composition. Front. Microbiol. 2020, 10, 3103. [Google Scholar] [CrossRef] [PubMed]
- Chaillou, S.; Chaulot-Talmon, A.; Caekebeke, H.; Cardinal, M.; Christieans, S.; Denis, C.; Hélène Desmonts, M.; Dousset, X.; Feurer, C.; Hamon, E.; et al. Origin and Ecological Selection of Core and Food-Specific Bacterial Communities Associated with Meat and Seafood Spoilage. ISME J. 2015, 9, 1105–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leroi, F.; Cornet, J.; Chevalier, F.; Cardinal, M.; Coeuret, G.; Chaillou, S.; Joffraud, J.-J. Selection of Bioprotective Cultures for Preventing Cold-Smoked Salmon Spoilage. Int. J. Food Microbiol. 2015, 213, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Maillet, A.; Denojean, P.; Bouju-Albert, A.; Scaon, E.; Leuillet, S.; Dousset, X.; Jaffrès, E.; Combrisson, J.; Prévost, H. Characterization of Bacterial Communities of Cold-Smoked Salmon during Storage. Foods 2021, 10, 362. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC A Quality Control Tool for High Throughput Sequence Data; BibSonomy: Hessen, Germany, 2010. [Google Scholar]
- Hannon, G.J. FASTX-Toolkit 2010. Available online: http://hannonlab.cshl.edu/fastx_toolkit/ (accessed on 18 June 2018).
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinforma. Oxf. Engl. 2018, 34, 1287–1294. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinforma. Oxf. Engl. 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and Fast Clustering Method for Amplicon-Based Studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2018. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; Elsevier: Boston, MA, USA, 2016. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R package version 2.4-6, 2018; ResearchGate: Berlin, Germany, 2019. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Use R! 2nd ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- ISO 13299. Sensory Analysis, Methodology. General Guidance for Establishing a Sensory Profile; BS ISO 13299:2016: Geneva, Switzerland, 2016. [Google Scholar]
- Macé, S.; Joffraud, J.-J.; Cardinal, M.; Malcheva, M.; Cornet, J.; Lalanne, V.; Chevalier, F.; Sérot, T.; Pilet, M.-F.; Dousset, X. Evaluation of the Spoilage Potential of Bacteria Isolated from Spoiled Raw Salmon (Salmo Salar) Fillets Stored under Modified Atmosphere Packaging. Int. J. Food Microbiol. 2013, 160, 227–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ISO 8589. Sensory Analysis, General Guidance for the Design of Tests Rooms 2014; BS ISO 8589:2014: Geneva, Switzerland, 2014. [Google Scholar]
- Tang, Y.; Horikoshi, M. Ggfortify: Unified Interface to Visualize Statistical Result of Popular R Packages. R J. 2016, 8, 478–489. [Google Scholar] [CrossRef] [Green Version]
- Conway, E.J.; Byrne, A. An Absorption Apparatus for the Micro-Determination of Certain Volatile Substances. Biochem. J. 1933, 27, 419–429. [Google Scholar]
- Wiernasz, N.; Cornet, J.; Cardinal, M.; Pilet, M.-F.; Passerini, D.; Leroi, F. Lactic Acid Bacteria Selection for Biopreservation as a Part of Hurdle Technology Approach Applied on Seafood. Front. Mar. Sci. 2017, 4–19. [Google Scholar] [CrossRef] [Green Version]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data, R package version 3.0.02016; ResearchGate: Berlin, Germany, 2009. [Google Scholar]
- Ghaziri, A.E.; Cariou, V.; Rutledge, D.N.; Qannari, E.M. Analysis of Multiblock Datasets Using ComDim: Overview and Extension to the Analysis of (K + 1) Datasets. J. Chemom. 2016, 30, 420–429. [Google Scholar] [CrossRef]
- Tchandao Mangamana, E.; Cariou, V.; Vigneau, E.; Glèlè Kakaï, R.L.; Qannari, E.M. Unsupervised Multiblock Data Analysis: A Unified Approach and Extensions. Chemom. Intell. Lab. Syst. 2019, 194, 103856. [Google Scholar] [CrossRef]
- de la Hoz, L.; López-Gálvez, D.E.; Fernández, M.; Hierro, E.; Ordóñez, J.A. Use of Carbon Dioxide Enriched Atmospheres in the Refrigerated Storage (2 °C) of Salmon (Salmo Salar) Steaks. Eur. Food Res. Technol. 2000, 210, 179–188. [Google Scholar] [CrossRef]
- Maqsood, S.; Benjakul, S.; Shahidi, F. Emerging Role of Phenolic Compounds as Natural Food Additives in Fish and Fish Products. Crit. Rev. Food Sci. Nutr. 2013, 53, 162–179. [Google Scholar] [CrossRef]
- Leroi, F.; Joffraud, J.J.; Chevalier, F. Effect of Salt and Smoke on the Microbiological Quality of Cold-Smoked Salmon during Storage at 5 Degrees C as Estimated by the Factorial Design Method. J. Food Prot. 2000, 63, 502–508. [Google Scholar] [CrossRef]
- Giménez, B.; Dalgaard, P. Modelling and Predicting the Simultaneous Growth of Listeria Monocytogenes and Spoilage Micro-Organisms in Cold-Smoked Salmon. J. Appl. Microbiol. 2004, 96, 96–109. [Google Scholar] [CrossRef]
- Porsby, C.H.; Vogel, B.F.; Mohr, M.; Gram, L. Influence of Processing Steps in Cold-Smoked Salmon Production on Survival and Growth of Persistent and Presumed Non-Persistent Listeria Monocytogenes. Int. J. Food Microbiol. 2008, 122, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.-A. The Probability of Growth of Listeria Monocytogenes in Cooked Salmon and Tryptic Soy Broth as Affected by Salt, Smoke Compound, and Storage Temperature. Food Microbiol. 2009, 26, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Dalgaard, P.; Gram, L.; Huss, H.H. Spoilage and Shelf-Life of Cod Fillets Packed in Vacuum or Modified Atmospheres. Int. J. Food Microbiol. 1993, 19, 283–294. [Google Scholar] [CrossRef]
- Jørgensen, L.V.; Huss, H.H.; Dalgaard, P. The Effect of Biogenic Amine Production by Single Bacterial Cultures and Metabiosis on Cold-Smoked Salmon. J. Appl. Microbiol. 2000, 89, 920–934. [Google Scholar] [CrossRef]
- Fletcher, G.C.; Summers, G.; Corrigan, V.K.; Johanson, M.R.; Hedderley, D. Optimizing Gas Mixtures for Modified Atmosphere Packaging of Fresh King Salmon (Oncorhynchus Tshawytscha). J. Aquat. Food Prod. Technol. 2005, 13, 5–28. [Google Scholar] [CrossRef]
- Götz, F.; Bannerman, T.; Schleifer, K.-H. The Genera Staphylococcus and Macrococcus. In The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 5–75. ISBN 978-0-387-30744-2. [Google Scholar]
- Slepecky, R.A.; Hemphill, H.E. The Genus Bacillus—Nonmedical. In The Prokaryotes: Volume 4: Bacteria: Firmicutes, Cyanobacteria; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 530–562. ISBN 978-0-387-30744-2. [Google Scholar]
- Thapa, N.; Pal, J.; Tamang, J.P. Phenotypic Identification and Technological Properties of Lactic Acid Bacteria Isolated from Traditionally Processed Fish Products of the Eastern Himalayas. Int. J. Food Microbiol. 2006, 107, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Andrighetto, C.; Lombardi, A.; Ferrati, M.; Guidi, A.; Corrain, C.; Arcangeli, G. Lactic Acid Bacteria Biodiversity in Italian Marinated Seafood Salad and Their Interactions on the Growth of Listeria Monocytogenes. Food Control 2009, 20, 462–468. [Google Scholar] [CrossRef]
- Macé, S.; Cornet, J.; Chevalier, F.; Cardinal, M.; Pilet, M.-F.; Dousset, X.; Joffraud, J.-J. Characterisation of the Spoilage Microbiota in Raw Salmon (Salmo Salar) Steaks Stored under Vacuum or Modified Atmosphere Packaging Combining Conventional Methods and PCR–TTGE. Food Microbiol. 2012, 30, 164–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaro, B.; Hernandez, I. Evolution of the Indigenous Microbiota in Modified Atmosphere Packaged Atlantic Horse Mackerel (Trachurus Trachurus) Identified by Conventional and Molecular Methods. Int. J. Food Microbiol. 2013, 167, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Sivertsvik, M.; Jeksrud, W.K.; Rosnes, J.T. A Review of Modified Atmosphere Packaging of Fish and Fishery Products—Significance of Microbial Growth, Activities and Safety. Int. J. Food Sci. Technol. 2002, 37, 107–127. [Google Scholar] [CrossRef]
- Bouletis, A.D.; Arvanitoyannis, I.S.; Hadjichristodoulou, C. Application of Modified Atmosphere Packaging on Aquacultured Fish and Fish Products: A Review. Crit. Rev. Food Sci. Nutr. 2017, 57, 2263–2285. [Google Scholar] [CrossRef]
- Silbande, A.; Adenet, S.; Chopin, C.; Cornet, J.; Smith-Ravin, J.; Rochefort, K.; Leroi, F. Effect of Vacuum and Modified Atmosphere Packaging on the Microbiological, Chemical and Sensory Properties of Tropical Red Drum (Sciaenops Ocellatus) Fillets Stored at 4 °C. Int. J. Food Microbiol. 2018, 266, 31–41. [Google Scholar] [CrossRef]
- Leroi, F. Occurrence and Role of Lactic Acid Bacteria in Seafood Products. Food Microbiol. 2010, 27, 698–709. [Google Scholar] [CrossRef] [Green Version]
- Matamoros, S.; Leroi, F.; Cardinal, M.; Gigout, F.; Kasbi Chadli, F.; Cornet, J.; Prévost, H.; Pilett, M.F. Psychrotrophic Lactic Acid Bacteria Used to Improve the Safety and Quality of Vacuum-Packaged Cooked and Peeled Tropical Shrimp and Cold-Smoked Salmon. J. Food Prot. 2009, 72, 365–374. [Google Scholar] [CrossRef]
- Ghanbari, M.; Jami, M.; Domig, K.J.; Kneifel, W. Seafood Biopreservation by Lactic Acid Bacteria—A Review. LWT—Food Sci. Technol. 2013, 54, 315–324. [Google Scholar] [CrossRef]
- Stohr, V.; Joffraud, J.J.; Cardinal, M.; Leroi, F. Spoilage Potential and Sensory Profile Associated with Bacteria Isolated from Cold-Smoked Salmon. Food Res. Int. 2001, 34, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Joffraud, J.-J.; Cardinal, M.; Cornet, J.; Chasles, J.-S.; Léon, S.; Gigout, F.; Leroi, F. Effect of Bacterial Interactions on the Spoilage of Cold-Smoked Salmon. Int. J. Food Microbiol. 2006, 112, 51–61. [Google Scholar] [CrossRef]
- Saraoui, T.; Leroi, F.; Björkroth, J.; Pilet, M.F. Lactococcus Piscium: A Psychrotrophic Lactic Acid Bacterium with Bioprotective or Spoilage Activity in Food-a Review. J. Appl. Microbiol. 2016, 121, 907–918. [Google Scholar] [CrossRef] [Green Version]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.K.; Schmidt, T.M. RrnDB: Improved Tools for Interpreting RRNA Gene Abundance in Bacteria and Archaea and a New Foundation for Future Development. Nucleic Acids Res. 2015, 43, D593–D598. [Google Scholar] [CrossRef] [Green Version]
- Dondero, M.; Cisternas, F.; Carvajal, L.; Simpson, R. Changes in Quality of Vacuum-Packed Cold-Smoked Salmon (Salmo Salar) as a Function of Storage Temperature. Food Chem. 2004, 87, 543–550. [Google Scholar] [CrossRef]
- Gram, L.; Huss, H.H. Microbiological Spoilage of Fish and Fish Products. Int. J. Food Microbiol. 1996, 33, 121–137. [Google Scholar] [CrossRef]
- Biji, K.B.; Ravishankar, C.N.; Venkateswarlu, R.; Mohan, C.O.; Gopal, T.K.S. Biogenic Amines in Seafood: A Review. J. Food Sci. Technol. 2016, 53, 2210–2218. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, C.A.L.; Conte-Junior, C.A. Chapter 6—Detection of Biogenic Amines: Quality and Toxicity Indicators in Food of Animal Origin. In Food Control and Biosecurity; Holban, A.M., Grumezescu, A.M., Eds.; Handbook of Food Bioengineering; Academic Press: Cambridge, MA, USA, 2018; pp. 225–257. ISBN 978-0-12-811445-2. [Google Scholar]
- Council Regulation (EC) n° 2074/2005 Rules N° 2074/2005 of 5 December 2005 Laying down Implementing Measures for Certain Products under Regulation. 2005. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32005R2074 (accessed on 8 October 2018).
- Masson, F.; Talon, R.; Montel, M.C. Histamine and Tyramine Production by Bacteria from Meat Products. Int. J. Food Microbiol. 1996, 32, 199–207. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved Screening Procedure for Biogenic Amine Production by Lactic Acid Bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Olafsdóttir, G.; Jonsdottir, R.; Lauzon, H.L.; Luten, J.; Kristbergsson, K. Characterization of Volatile Compounds in Chilled Cod (Gadus Morhua) Fillets by Gas Chromatography and Detection of Quality Indicators by an Electronic Nose. J. Agric. Food Chem. 2005, 53, 10140–10147. [Google Scholar] [CrossRef] [PubMed]
- Varlet, V.; Knockaert, C.; Prost, C.; Serot, T. Comparison of Odor-Active Volatile Compounds of Fresh and Smoked Salmon. J. Agric. Food Chem. 2006, 54, 3391–3401. [Google Scholar] [CrossRef] [PubMed]
- Varlet, V.; Prost, C.; Serot, T. Volatile Aldehydes in Smoked Fish: Analysis Methods, Occurence and Mechanisms of Formation. Food Chem. 2007, 105, 1536–1556. [Google Scholar] [CrossRef]
- Jónsdóttir, R.; Ólafsdóttir, G.; Chanie, E.; Haugen, J.-E. Volatile Compounds Suitable for Rapid Detection as Quality Indicators of Cold Smoked Salmon (Salmo Salar). Food Chem. 2008, 109, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Joffraud, J.J.; Leroi, F.; Roy, C.; Berdagué, J.L. Characterisation of Volatile Compounds Produced by Bacteria Isolated from the Spoilage Flora of Cold-Smoked Salmon. Int. J. Food Microbiol. 2001, 66, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Jaffrès, E.; Lalanne, V.; Macé, S.; Cornet, J.; Cardinal, M.; Sérot, T.; Dousset, X.; Joffraud, J.-J. Sensory Characteristics of Spoilage and Volatile Compounds Associated with Bacteria Isolated from Cooked and Peeled Tropical Shrimps Using SPME-GC-MS Analysis. Int. J. Food Microbiol. 2011, 147, 195–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardinal, M.; Berdague, J.L.; Dinel, V.; Knockaert, C.; Vallet, J.L. Effect of various smoking techniques on the nature of volatile compounds and on the sensory characteristics of salmon meat. Sci. Aliment. Fr. 1997, 17, 679–696. [Google Scholar]
- Guillen, M.D.; Errecalde, M.C.; Salmeron, J.; Casas, C. Headspace Volatile Components of Smoked Swordfish (Xiphias Gladius) and Cod (Gadus Morhua) Detected by Means of Microextraction and Gas Chromatography-Mass Spectrometry. Food Chem. 2006, 94, 151–156. [Google Scholar] [CrossRef]
- Wierckx, N.; Koopman, F.; Ruijssenaars, H.J.; de Winde, J.H. Microbial Degradation of Furanic Compounds: Biochemistry, Genetics, and Impact. Appl. Microbiol. Biotechnol. 2011, 92, 1095–1105. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-K.; Kim, M.-S.; Jung, M.-J.; Nam, Y.-D.; Park, E.-J.; Roh, S.W.; Bae, J.-W. Brachybacterium Squillarum Sp. Nov., Isolated from Salt-Fermented Seafood. Int. J. Syst. Evol. Microbiol. 2011, 61, 1118–1122. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xie, Q.-Y.; Shi, W.; Li, L.; An, J.-Y.; Zhao, Y.-M.; Hong, K. Brachybacterium Huguangmaarense Sp. Nov., Isolated from Lake Sediment. Int. J. Syst. Evol. Microbiol. 2014, 64, 1673–1678. [Google Scholar] [CrossRef] [Green Version]
- Kaur, G.; Kumar, N.; Mual, P.; Kumar, A.; Kumar, R.M.; Mayilraj, S. Brachybacterium Aquaticum Sp. Nov., a Novel Actinobacterium Isolated from Seawater. Int. J. Syst. Evol. Microbiol. 2016, 66, 4705–4710. [Google Scholar] [CrossRef] [PubMed]
- Tak, E.J.; Kim, P.S.; Hyun, D.-W.; Kim, H.S.; Lee, J.-Y.; Kang, W.; Sung, H.; Shin, N.-R.; Kim, M.-S.; Whon, T.W.; et al. Phenotypic and Genomic Properties of Brachybacterium Vulturis Sp. Nov. and Brachybacterium Avium Sp. Nov. Front. Microbiol. 2018, 9–20. [Google Scholar] [CrossRef]
- Aranda, S.; Montes-Borrego, M.; Landa, B.B. Purple-Pigmented Violacein-Producing Duganella Spp. Inhabit the Rhizosphere of Wild and Cultivated Olives in Southern Spain. Microb. Ecol. 2011, 62, 446–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madhaiyan, M.; Poonguzhali, S.; Saravanan, V.S.; Hari, K.; Lee, K.-C.; Lee, J.-S. Duganella Sacchari Sp. Nov. and Duganella Radicis Sp. Nov., Two Novel Species Isolated from Rhizosphere of Field-Grown Sugar Cane. Int. J. Syst. Evol. Microbiol. 2013, 63, 1126–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haack, F.S.; Poehlein, A.; Kröger, C.; Voigt, C.A.; Piepenbring, M.; Bode, H.B.; Daniel, R.; Schäfer, W.; Streit, W.R. Molecular Keys to the Janthinobacterium and Duganella Spp. Interaction with the Plant Pathogen Fusarium Graminearum. Front. Microbiol. 2016, 7, 1668. [Google Scholar] [CrossRef] [Green Version]
- An, S.-Y.; Asahara, M.; Goto, K.; Kasai, H.; Yokota, A. Terribacillus Saccharophilus Gen. Nov., Sp. Nov. and Terribacillus Halophilus Sp. Nov., Spore-Forming Bacteria Isolated from Field Soil in Japan. Int. J. Syst. Evol. Microbiol. 2007, 57, 51–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Jiang, L.; Guo, C.; Yang, S.S. Terribacillus Aidingensis Sp. Nov., a Moderately Halophilic Bacterium. Int. J. Syst. Evol. Microbiol. 2010, 60, 2940–2945. [Google Scholar] [CrossRef] [Green Version]
- Lu, P.; Lei, M.; Xiao, F.; Zhang, L.; Wang, Y. Complete Genome Sequence of Terribacillus Aidingensis Strain MP602, a Moderately Halophilic Bacterium Isolated from Cryptomeria Fortunei in Tianmu Mountain in China. Genome Announc. 2015, 3, e0012-15. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.D. Spelaeicoccus Albus Gen. Nov., Sp. Nov., an Actinobacterium Isolated from a Natural Cave. Int. J. Syst. Evol. Microbiol. 2013, 63, 3958–3963. [Google Scholar] [CrossRef]
- Rattray, F.P.; Fox, P.F. Aspects of Enzymology and Biochemical Properties of Brevibacterium Linens Relevant to Cheese Ripening: A Review. J. Dairy Sci. 1999, 82, 891–909. [Google Scholar] [CrossRef]
- Onraedt, A.; Soetaert, W.; Vandamme, E. Industrial Importance of the Genus Brevibacterium. Biotechnol. Lett. 2005, 27, 527–533. [Google Scholar] [CrossRef]
- Srilekha, V.; Krishna, G.; Seshasrinivas, V.; Charya, M.A.S. Antibacterial and Anti-Inflammatory Activities of Marine Brevibacterium Sp. Res. Pharm. Sci. 2017, 12, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.D.; Siddiqi, M.Z.; Liu, Q.; Muhammad Shafi, S.; Durrani, Y.; Lee, S.Y.; Kang, M.-S.; Im, W.T. Brevibacterium Hankyongi Sp. Nov., Isolated from Compost. Int. J. Syst. Evol. Microbiol. 2018, 68, 2783–2788. [Google Scholar] [CrossRef] [PubMed]
- Valles, C.; Fournier, P.-E.; Raoult, D.; Cadoret, F. “Brevibacterium Ihuae” Sp. Nov., Isolated from a Stool Sample of a Healthy 25-Year-Old Woman. New Microbes New Infect. 2018, 21, 49–50. [Google Scholar] [CrossRef] [PubMed]
- Wauters, G.; De Baere, T.; Willems, A.; Falsen, E.; Vaneechoutte, M. Description of Comamonas Aquatica Comb. Nov. and Comamonas Kerstersii Sp. Nov. for Two Subgroups of Comamonas Terrigena and Emended Description of Comamonas Terrigena. Int. J. Syst. Evol. Microbiol. 2003, 53, 859–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.-N.; Zhang, J.; Chen, Q.; He, J.; Li, Q.-F.; Li, S.-P. Comamonas Jiangduensis Sp. Nov., a Biosurfactant-Producing Bacterium Isolated from Agricultural Soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 2168–2173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, D.; Xie, C.; Huang, Y.; Sun, J.; Zhang, W. Description of Comamonas Serinivorans Sp. Nov., Isolated from Wheat Straw Compost. Int. J. Syst. Evol. Microbiol. 2014, 64, 4141–4146. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Zhu, Y.; Wang, X.; Sakenova, N.; Yang, Z.; Wang, H.; Li, G.; He, J.; Huang, D.; Cai, Y.; et al. Draft Genome Sequence of the Bacterium Comamonas Aquatica CJG. Genome Announc. 2016, 4, e01186-16. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.; Soo Kim, P.; Hyun, D.-W.; Lee, J.-Y.; Sik Kim, H.; Joon Oh, S.; Shin, N.-R.; Bae, J.-W. Comamonas Piscis Sp. Nov., Isolated from the Intestine of a Korean Rockfish, Sebastes Schlegelii. Int. J. Syst. Evol. Microbiol. 2016, 66, 780–785. [Google Scholar] [CrossRef]
- Veress, A.; Wilk, T.; Kiss, J.; Papp, P.P.; Olasz, F. Two Draft Genome Sequences of Sphingobacterium Sp. Strains Isolated from Honey. Genome Announc. 2017, 5, e01364-17. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, S.; Mukhopadhyay, S.K.; Gauri, S.S.; Dey, S. Sphingobactan, a New α-Mannan Exopolysaccharide from Arctic Sphingobacterium Sp. IITKGP-BTPF3 Capable of Biological Response Modification. Int. Immunopharmacol. 2018, 60, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, D.K.; Kim, J. Sphingobacterium Terrae Sp. Nov., Isolated from Oil-Contaminated Soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 609–615. [Google Scholar] [CrossRef]
- Kaur, M.; Singh, H.; Sharma, S.; Mishra, S.; Tanuku, N.R.S.; Pinnaka, A.K. Sphingobacterium Bovisgrunnientis Sp. Nov., Isolated from Yak Milk. Int. J. Syst. Evol. Microbiol. 2018, 68, 636–642. [Google Scholar] [CrossRef]
- Niu, X.; Cui, W.; Cui, M.; Zhang, X.; Zhang, S.; Xu, B.; Gao, M. Sphingobacterium Solani Sp. Nov., Isolated from Potato Stems. Int. J. Syst. Evol. Microbiol. 2018, 68, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Chatterjee, S. Psychrotolerant Sphingobacterium Kitahiroshimense LT-2 Isolated from Dhundi Glacier, Himachal Pradesh: Origin Prediction and Future Application. Indian J. Microbiol. 2018, 58, 234–238. [Google Scholar] [CrossRef]
- Van Le, V.; Padakandla, S.R.; Kim, H.; Chae, J.-C. Sphingobacterium Praediipecoris Sp. Nov. Isolated from Effluent of a Dairy Manure Treatment Plant. Arch. Microbiol. 2018, 200, 1481–1486. [Google Scholar] [CrossRef]
- Yim, K.J.; Cha, I.-T.; Lee, H.-W.; Song, H.S.; Kim, K.-N.; Lee, S.-J.; Nam, Y.-D.; Hyun, D.-W.; Bae, J.-W.; Rhee, S.-K.; et al. Halorubrum Halophilum Sp. Nov., an Extremely Halophilic Archaeon Isolated from a Salt-Fermented Seafood. Antonie Van Leeuwenhoek 2014, 105, 603–612. [Google Scholar] [CrossRef]
- Corral, P.; de la Haba, R.R.; Sánchez-Porro, C.; Amoozegar, M.A.; Papke, R.T.; Ventosa, A. Halorubrum Persicum Sp. Nov., an Extremely Halophilic Archaeon Isolated from Sediment of a Hypersaline Lake. Int. J. Syst. Evol. Microbiol. 2015, 65, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Minegishi, H.; Echigo, A.; Shimane, Y.; Kamekura, M.; Itoh, T.; Ohkuma, M.; Takahashi-Ando, N.; Fukushima, Y.; Yoshida, Y.; et al. Halorubrum Gandharaense Sp. Nov., an Alkaliphilic Haloarchaeon from Commercial Rock Salt. Int. J. Syst. Evol. Microbiol. 2015, 65, 2345–2350. [Google Scholar] [CrossRef]
- Sánchez-Nieves, R.; Facciotti, M.T.; Saavedra-Collado, S.; Dávila-Santiago, L.; Rodríguez-Carrero, R.; Montalvo-Rodríguez, R. Draft Genome Sequence of Halorubrum Tropicale Strain V5, a Novel Halophilic Archaeon Isolated from the Solar Salterns of Cabo Rojo, Puerto Rico. Genom. Data 2016, 7, 284–286. [Google Scholar] [CrossRef] [Green Version]
- Mou, Y.-Z.; Qiu, X.-X.; Zhao, M.-L.; Cui, H.-L.; Oh, D.; Dyall-Smith, M.L. Halohasta Litorea Gen. Nov. Sp. Nov., and Halohasta Litchfieldiae Sp. Nov., Isolated from the Daliang Aquaculture Farm, China and from Deep Lake, Antarctica, Respectively. Extrem. Life Extrem. Cond. 2012, 16, 895–901. [Google Scholar] [CrossRef] [PubMed]



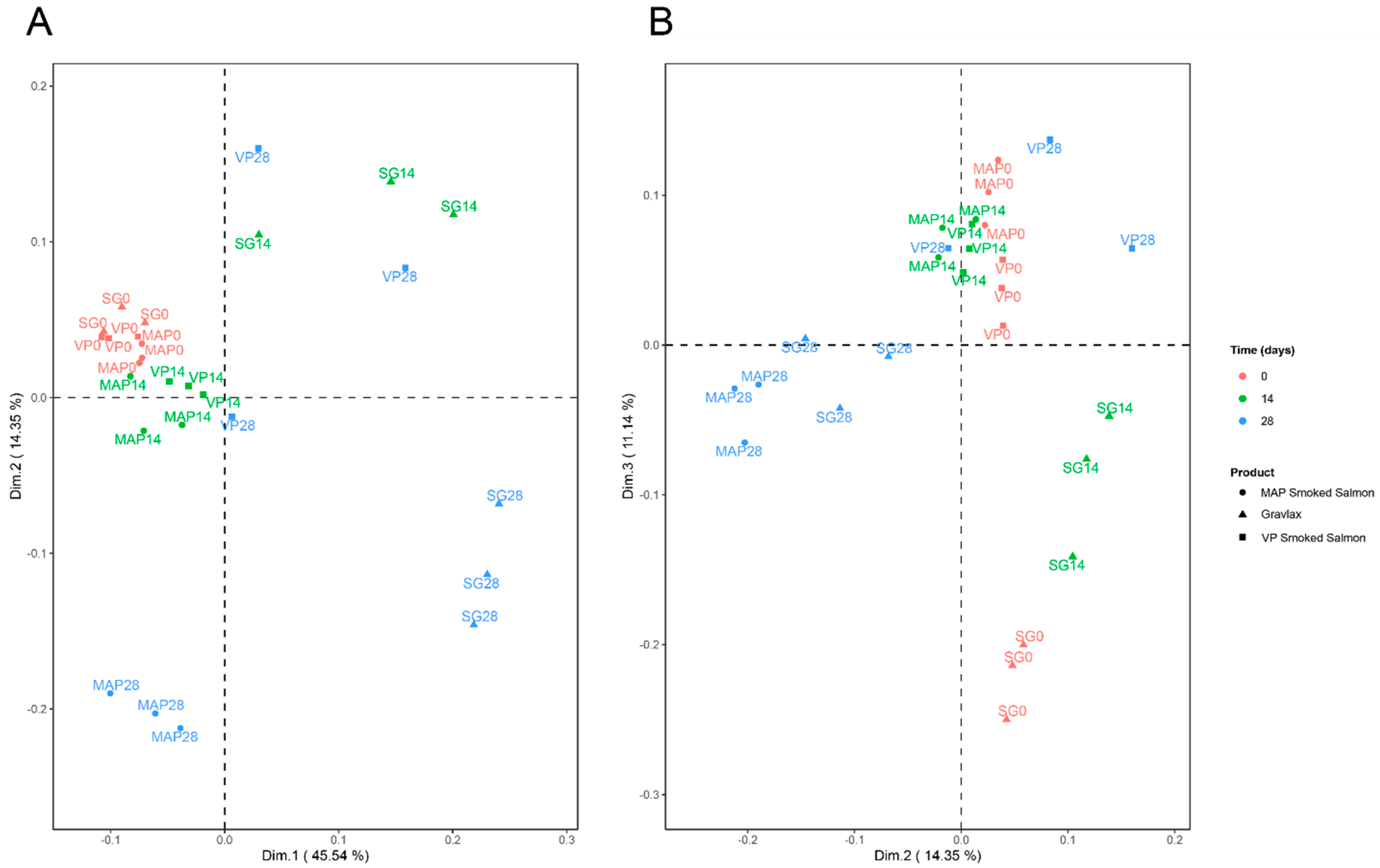
| Sampling Date (days) | 0 | 7 | 14 | 21 | 28 | 35 | 42 | 49 |
|---|---|---|---|---|---|---|---|---|
| SG | ||||||||
| pH | 5.87 ± 0.10 | 5.93 ± 0.02 | 5.62 ± 0.04 | 5.81 ± 0.03 | 5.77 ± 0.02 | 5.84 ± 0.02 | - | - |
| TVBN | 12.0 ± 1.8 | 15.1 ± 0.9 | 22.2 ± 5.3 | 24.9 ± 4.9 | 28.2 ± 1.0 | 23.1 ± 1.3 | - | - |
| Cadaverine | 2.0 ± 0.1 | 6.0 ± 3.6 | 294.2 ± 224.1 | 275.8 ± 30.6 | 404.1 ± 79.4 | 73.5 ± 58.2 | - | - |
| Tyramine | 2.0 ± 0.1 | 2.0 ± 0.1 | 65.3 ± 60.9 | 42.2 ± 12.9 | 63.5 ± 15.3 | 13.0 ± 12.0 | - | - |
| Histamine | 0.7 ± 0.6 | 1.3 ± 0.6 | 116.8 ± 104.3 | 57.7 ± 7 | 77.3 ± 24.0 | 17.1 ± 12.7 | - | - |
| MAP CSS | ||||||||
| pH | 5.68 ± 0.02 | 5.81 ± 0.01 | 5.53 ± 0.07 | 5.70 ± 0.02 | 5.65 ± 0.04 | 5.63 ± 0.02 | - | - |
| TVBN | 14.4 ± 0.7 | 14.6 ± 1.0 | 14.2 ±1.9 | 14.8 ± 1.0 | 13.3 ± 2.6 | 16.8 ± 2.1 | - | - |
| Cadaverine | 2.3 ± 2.1 | 4.7 ± 0.6 | 9.0 ± 5.6 | 6.7 ± 2.0 | 6.2 ± 1.0 | 4.4 ± 0.4 | - | - |
| Tyramine | 0.0 | 0.0 | 3.3 ± 3.1 | 0.2 ± 0.4 | 0.3 ± 0.5 | 6.2 ± 2.7 | - | - |
| Histamine | 3.7 ± 3.2 | 6.0 ± 1.0 | 8.0 ± 2.0 | 5.3 ± 0.6 | 5.5 ± 1.5 | 5.2 ± 0.7 | - | - |
| VP CSS | ||||||||
| pH | 5.63 ± 0.03 | 5.81 ± 0.02 | 5.47 ± 0.04 | - | 5.64 ± 0.07 | 5.76 ± 0.01 | 5.60 ± 0.05 | 5.57 ± 0.013 |
| TVBN | 14.5 ± 1.6 | 17.1 ± 1.3 | 17.5 ± 1.3 | - | 21.0 ± 3.7 | 24.7 ± 2.5 | 24.6 ± 3.6 | 25.9 ± 5.0 |
| Cadaverine | 4.0 ± 1.0 | 4.0 ± 0.1 | 4.6 ± 0.9 | - | 153.4 ± 160.5 | 185.0 ± 186.3 | 185.3 ± 281.7 | 218.9 ± 187.9 |
| Tyramine | 0.0 | 0.0 | 0.0 | - | 26.7 ± 36.0 | 37.8 ± 43.9 | 49.5 ± 78.5 | 38.8 ± 33.7 |
| Histamine | 6.7 ± 1.2 | 5.7 ± 0.6 | 5.6 ± 0.2 | - | 33.4 ± 29.6 | 56.3 ± 61.9 | 10.8 ± 6.0 | 59.4 ± 45.2 |
| %expl | cum%expl | |
|---|---|---|
| Dim.1 | 45.56 | 45.56 |
| Dim.2 | 14.35 | 59.91 |
| Dim.3 | 11.15 | 71.06 |
| Dim.4 | 6.45 | 77.51 |
| Dim.5 | 4.15 | 81.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiernasz, N.; Gigout, F.; Cardinal, M.; Cornet, J.; Rohloff, J.; Courcoux, P.; Vigneau, E.; Skírnisdottír, S.; Passerini, D.; Pilet, M.-F.; et al. Effect of the Manufacturing Process on the Microbiota, Organoleptic Properties and Volatilome of Three Salmon-Based Products. Foods 2021, 10, 2517. https://doi.org/10.3390/foods10112517
Wiernasz N, Gigout F, Cardinal M, Cornet J, Rohloff J, Courcoux P, Vigneau E, Skírnisdottír S, Passerini D, Pilet M-F, et al. Effect of the Manufacturing Process on the Microbiota, Organoleptic Properties and Volatilome of Three Salmon-Based Products. Foods. 2021; 10(11):2517. https://doi.org/10.3390/foods10112517
Chicago/Turabian StyleWiernasz, Norman, Frédérique Gigout, Mireille Cardinal, Josiane Cornet, Jens Rohloff, Philippe Courcoux, Evelyne Vigneau, Sigurlaug Skírnisdottír, Delphine Passerini, Marie-France Pilet, and et al. 2021. "Effect of the Manufacturing Process on the Microbiota, Organoleptic Properties and Volatilome of Three Salmon-Based Products" Foods 10, no. 11: 2517. https://doi.org/10.3390/foods10112517
APA StyleWiernasz, N., Gigout, F., Cardinal, M., Cornet, J., Rohloff, J., Courcoux, P., Vigneau, E., Skírnisdottír, S., Passerini, D., Pilet, M.-F., & Leroi, F. (2021). Effect of the Manufacturing Process on the Microbiota, Organoleptic Properties and Volatilome of Three Salmon-Based Products. Foods, 10(11), 2517. https://doi.org/10.3390/foods10112517






