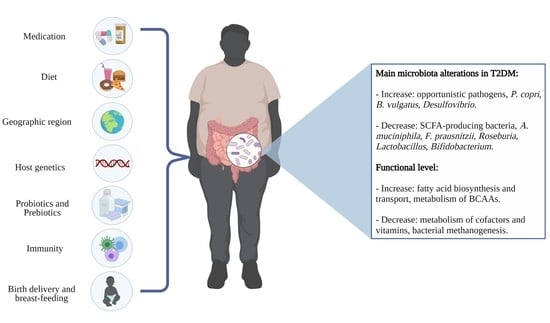
Characteristics of the Gut Microbiota and Potential Effects of Probiotic Supplements in Individuals with Type 2 Diabetes mellitus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Method
2.2. Eligibility Criteria and Study Selection
3. Gut Microbiota and Association with T2DM
4. Microbial Metabolites and Components Linked to T2DM
4.1. Low-Grade Inflammation
4.2. Short-Chain Fatty Acids
4.3. Trimethylamine N-Oxide
4.4. Imidazole Propionate
4.5. Branched-Chain Amino Acids
4.6. Tryptophan Metabolites
4.7. Bile Acids
5. Effects of Probiotics Supplements in T2DM
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kerner, W.; Bruckel, J. Definition, classification and diagnosis of diabetes mellitus. Exp. Clin. Endocrinol. Diabetes 2014, 122, 384–386. [Google Scholar] [CrossRef] [Green Version]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37 (Suppl. 1), S81–S90. [Google Scholar] [CrossRef] [Green Version]
- American Diabetes Association. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2020. Diabetes Care 2020, 43, S14–S31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbari, M.; Hassan-Zadeh, V. The inflammatory effect of epigenetic factors and modifications in type 2 diabetes. Inflammopharmacology 2020, 28, 345–362. [Google Scholar] [CrossRef]
- Hameed, I.; Masoodi, S.R.; Mir, S.A.; Nabi, M.; Ghazanfar, K.; Ganai, B.A. Type 2 diabetes mellitus: From a metabolic disorder to an inflammatory condition. World J. Diabetes 2015, 6, 598–612. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martin, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Kolb, H.; Martin, S. Environmental/lifestyle factors in the pathogenesis and prevention of type 2 diabetes. BMC Med. 2017, 15, 131. [Google Scholar] [CrossRef]
- Williams, R.; Karuranga, S.; Malanda, B.; Saeedi, P.; Basit, A.; Besancon, S.; Bommer, C.; Esteghamati, A.; Ogurtsova, K.; Zhang, P.; et al. Global and regional estimates and projections of diabetes-related health expenditure: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract. 2020, 162, 108072. [Google Scholar] [CrossRef] [Green Version]
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; da Rocha Fernandes, J.D.; Ohlrogge, A.W.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef]
- Esser, N.; Legrand-Poels, S.; Piette, J.; Scheen, A.J.; Paquot, N. Inflammation as a link between obesity, metabolic syndrome and type 2 diabetes. Diabetes Res. Clin. Pract. 2014, 105, 141–150. [Google Scholar] [CrossRef] [Green Version]
- Bicsak, T.A.; Walsh, B.; Fineman, M. Metformin-associated lactic acidosis: Moving towards a new paradigm? Diabetes Obes. Metab. 2017, 19, 1499–1501. [Google Scholar] [CrossRef] [Green Version]
- Dujic, T.; Causevic, A.; Bego, T.; Malenica, M.; Velija-Asimi, Z.; Pearson, E.R.; Semiz, S. Organic cation transporter 1 variants and gastrointestinal side effects of metformin in patients with Type 2 diabetes. Diabet. Med. 2016, 33, 511–514. [Google Scholar] [CrossRef] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Verges, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Munoz-Garach, A.; Diaz-Perdigones, C.; Tinahones, F.J. Gut microbiota and type 2 diabetes mellitus. Endocrinol. Nutr. 2016, 63, 560–568. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heianza, Y.; Sun, D.; Li, X.; DiDonato, J.A.; Bray, G.A.; Sacks, F.M.; Qi, L. Gut microbiota metabolites, amino acid metabolites and improvements in insulin sensitivity and glucose metabolism: The POUNDS Lost trial. Gut 2019, 68, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858. [Google Scholar] [CrossRef]
- Yaribeygi, H.; Sathyapalan, T.; Sahebkar, A. Molecular mechanisms by which GLP-1 RA and DPP-4i induce insulin sensitivity. Life Sci. 2019, 234, 116776. [Google Scholar] [CrossRef]
- Doumatey, A.P.; Adeyemo, A.; Zhou, J.; Lei, L.; Adebamowo, S.N.; Adebamowo, C.; Rotimi, C.N. Gut Microbiome Profiles Are Associated With Type 2 Diabetes in Urban Africans. Front. Cell. Infect. Microbiol. 2020, 10, 63. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Tremaroli, V.; Schmidt, C.; Lundqvist, A.; Olsson, L.M.; Kramer, M.; Gummesson, A.; Perkins, R.; Bergstrom, G.; Backhed, F. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab. 2020, 32, 379–390 e373. [Google Scholar] [CrossRef]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Sircana, A.; Framarin, L.; Leone, N.; Berrutti, M.; Castellino, F.; Parente, R.; De Michieli, F.; Paschetta, E.; Musso, G. Altered Gut Microbiota in Type 2 Diabetes: Just a Coincidence? Curr. Diabetes Rep. 2018, 18, 98. [Google Scholar] [CrossRef]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, D.; Fang, Z.; Jie, Z.; Qiu, X.; Zhang, C.; Chen, Y.; Ji, L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE 2013, 8, e71108. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Salamon, D.; Sroka-Oleksiak, A.; Kapusta, P.; Szopa, M.; Mrozinska, S.; Ludwig-Slomczynska, A.H.; Wolkow, P.P.; Bulanda, M.; Klupa, T.; Malecki, M.T.; et al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on nextgeneration sequencing of the 16S rRNA gene fragment. Pol. Arch. Intern. Med. 2018, 128, 336–343. [Google Scholar] [CrossRef]
- Li, Q.; Chang, Y.; Zhang, K.; Chen, H.; Tao, S.; Zhang, Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci. Rep. 2020, 10, 5450. [Google Scholar] [CrossRef]
- Wang, J.; Li, W.; Wang, C.; Wang, L.; He, T.; Hu, H.; Song, J.; Cui, C.; Qiao, J.; Qing, L.; et al. Enterotype Bacteroides Is Associated with a High Risk in Patients with Diabetes: A Pilot Study. J. Diabetes Res. 2020, 2020, 6047145. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Larsen, N.; Vogensen, F.K.; van den Berg, F.W.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sorensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [Green Version]
- Schneeberger, M.; Everard, A.; Gomez-Valades, A.G.; Matamoros, S.; Ramirez, S.; Delzenne, N.M.; Gomis, R.; Claret, M.; Cani, P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Lazarevic, V.; Gaia, N.; Johansson, M.; Stahlman, M.; Backhed, F.; Delzenne, N.M.; Schrenzel, J.; Francois, P.; Cani, P.D. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014, 8, 2116–2130. [Google Scholar] [CrossRef] [PubMed]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [Green Version]
- Markowiak-Kopec, P.; Slizewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intestinal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef]
- Koh, A.; Molinaro, A.; Stahlman, M.; Khan, M.T.; Schmidt, C.; Manneras-Holm, L.; Wu, H.; Carreras, A.; Jeong, H.; Olofsson, L.E.; et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell 2018, 175, 947–961 e917. [Google Scholar] [CrossRef] [Green Version]
- Zeisel, S.H.; Warrier, M. Trimethylamine N-Oxide, the Microbiome, and Heart and Kidney Disease. Annu. Rev. Nutr. 2017, 37, 157–181. [Google Scholar] [CrossRef]
- Mutaguchi, Y.; Kasuga, K.; Kojima, I. Production of d-Branched-Chain Amino Acids by Lactic Acid Bacteria Carrying Homologs to Isoleucine 2-Epimerase of Lactobacillus buchneri. Front. Microbiol. 2018, 9, 1540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tett, A.; Huang, K.D.; Asnicar, F.; Fehlner-Peach, H.; Pasolli, E.; Karcher, N.; Armanini, F.; Manghi, P.; Bonham, K.; Zolfo, M.; et al. The Prevotella copri Complex Comprises Four Distinct Clades Underrepresented in Westernized Populations. Cell Host Microbe 2019, 26, 666–679 e667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, W.; Xie, G.; Jia, W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, J.Y.L.; Ferrell, J.M. Bile Acids as Metabolic Regulators and Nutrient Sensors. Annu. Rev. Nutr. 2019, 39, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef]
- Yu, L.; Li, Y.; Du, C.; Zhao, W.; Zhang, H.; Yang, Y.; Sun, A.; Song, X.; Feng, Z. Pattern Recognition Receptor-Mediated Chronic Inflammation in the Development and Progression of Obesity-Related Metabolic Diseases. Mediat. Inflamm. 2019, 2019, 5271295. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef]
- Tanimura, N.; Saitoh, S.; Matsumoto, F.; Akashi-Takamura, S.; Miyake, K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem. Biophys. Res. Commun. 2008, 368, 94–99. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Ding, X.; Wu, G.; Lam, Y.Y.; Wang, X.; Fu, H.; Xue, X.; Lu, C.; Ma, J.; et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taesuwan, S.; Cho, C.E.; Malysheva, O.V.; Bender, E.; King, J.H.; Yan, J.; Thalacker-Mercer, A.E.; Caudill, M.A. The metabolic fate of isotopically labeled trimethylamine-N-oxide (TMAO) in humans. J. Nutr. Biochem. 2017, 45, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; You, T.; Li, J.; Pan, T.; Xiang, L.; Han, Y.; Zhu, L. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: A systematic review and meta-analysis of 11 prospective cohort studies. J. Cell. Mol. Med. 2018, 22, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.; Wang, Z.; Li, X.S.; Fan, Y.; Li, D.S.; Wu, Y.; Hazen, S.L. Increased Trimethylamine N-Oxide Portends High Mortality Risk Independent of Glycemic Control in Patients with Type 2 Diabetes Mellitus. Clin. Chem. 2017, 63, 297–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Liu, X.; Xu, J.; Xue, C.; Xue, Y.; Wang, Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 2014, 118, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.; Dou, P.; Gao, M.; Kong, X.; Li, C.; Liu, Z.; Huang, T. Assessment of Causal Direction Between Gut Microbiota-Dependent Metabolites and Cardiometabolic Health: A Bidirectional Mendelian Randomization Analysis. Diabetes 2019, 68, 1747–1755. [Google Scholar] [CrossRef]
- Molinaro, A.; Bel Lassen, P.; Henricsson, M.; Wu, H.; Adriouch, S.; Belda, E.; Chakaroun, R.; Nielsen, T.; Bergh, P.O.; Rouault, C.; et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat. Commun. 2020, 11, 5881. [Google Scholar] [CrossRef]
- Asghari, G.; Farhadnejad, H.; Teymoori, F.; Mirmiran, P.; Tohidi, M.; Azizi, F. High dietary intake of branched-chain amino acids is associated with an increased risk of insulin resistance in adults. J. Diabetes 2018, 10, 357–364. [Google Scholar] [CrossRef] [Green Version]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef] [Green Version]
- Newgard, C.B.; An, J.; Bain, J.R.; Muehlbauer, M.J.; Stevens, R.D.; Lien, L.F.; Haqq, A.M.; Shah, S.H.; Arlotto, M.; Slentz, C.A.; et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009, 9, 311–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, C.; Nakamura, K.; Wada, K.; Tsuji, M.; Tamai, Y.; Kawachi, T. Branched-chain amino acid intake and the risk of diabetes in a Japanese community: The Takayama study. Am. J. Epidemiol. 2013, 178, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jennis, M.; Cavanaugh, C.R.; Leo, G.C.; Mabus, J.R.; Lenhard, J.; Hornby, P.J. Microbiota-derived tryptophan indoles increase after gastric bypass surgery and reduce intestinal permeability in vitro and in vivo. Neurogastroenterol. Motil. 2018, 30, e13178. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37 e26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Mello, V.D.; Paananen, J.; Lindstrom, J.; Lankinen, M.A.; Shi, L.; Kuusisto, J.; Pihlajamaki, J.; Auriola, S.; Lehtonen, M.; Rolandsson, O.; et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci. Rep. 2017, 7, 46337. [Google Scholar] [CrossRef] [PubMed]
- Chimerel, C.; Emery, E.; Summers, D.K.; Keyser, U.; Gribble, F.M.; Reimann, F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014, 9, 1202–1208. [Google Scholar] [CrossRef] [Green Version]
- Holst, J.J. The physiology of glucagon-like peptide 1. Physiol. Rev. 2007, 87, 1409–1439. [Google Scholar] [CrossRef]
- Pathak, P.; Xie, C.; Nichols, R.G.; Ferrell, J.M.; Boehme, S.; Krausz, K.W.; Patterson, A.D.; Gonzalez, F.J.; Chiang, J.Y.L. Intestine farnesoid X receptor agonist and the gut microbiota activate G-protein bile acid receptor-1 signaling to improve metabolism. Hepatology 2018, 68, 1574–1588. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E.; Merenstein, D.J.; Reid, G.; Gibson, G.R.; Rastall, R.A. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.J.; Lin, T.L.; Tsai, Y.L.; Wu, T.R.; Lai, W.F.; Lu, C.C.; Lai, H.C. Next generation probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Ouwehand, A.C. A review of dose-responses of probiotics in human studies. Benef. Microbes 2017, 8, 143–151. [Google Scholar] [CrossRef]
- Mazloom, Z.; Yousefinejad, A.; Dabbaghmanesh, M.H. Effect of probiotics on lipid profile, glycemic control, insulin action, oxidative stress, and inflammatory markers in patients with type 2 diabetes: A clinical trial. Iran. J. Med Sci. 2013, 38, 38–43. [Google Scholar]
- Firouzi, S.; Majid, H.A.; Ismail, A.; Kamaruddin, N.A.; Barakatun-Nisak, M.Y. Effect of multi-strain probiotics (multi-strain microbial cell preparation) on glycemic control and other diabetes-related outcomes in people with type 2 diabetes: A randomized controlled trial. Eur. J. Nutr. 2017, 56, 1535–1550. [Google Scholar] [CrossRef]
- Sabico, S.; Al-Mashharawi, A.; Al-Daghri, N.M.; Yakout, S.; Alnaami, A.M.; Alokail, M.S.; McTernan, P.G. Effects of a multi-strain probiotic supplement for 12 weeks in circulating endotoxin levels and cardiometabolic profiles of medication naive T2DM patients: A randomized clinical trial. J. Transl. Med. 2017, 15, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobini, R.; Tremaroli, V.; Stahlman, M.; Karlsson, F.; Levin, M.; Ljungberg, M.; Sohlin, M.; Berteus Forslund, H.; Perkins, R.; Backhed, F.; et al. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: A randomized controlled trial. Diabetes Obes. Metab. 2017, 19, 579–589. [Google Scholar] [CrossRef]
- Hsieh, M.C.; Tsai, W.H.; Jheng, Y.P.; Su, S.L.; Wang, S.Y.; Lin, C.C.; Chen, Y.H.; Chang, W.W. The beneficial effects of Lactobacillus reuteri ADR-1 or ADR-3 consumption on type 2 diabetes mellitus: A randomized, double-blinded, placebo-controlled trial. Sci. Rep. 2018, 8, 16791. [Google Scholar] [CrossRef] [PubMed]
- Kobyliak, N.; Falalyeyeva, T.; Mykhalchyshyn, G.; Kyriienko, D.; Komissarenko, I. Effect of alive probiotic on insulin resistance in type 2 diabetes patients: Randomized clinical trial. Diabetes Metab. Syndr. 2018, 12, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Sabico, S.; Al-Mashharawi, A.; Al-Daghri, N.M.; Wani, K.; Amer, O.E.; Hussain, D.S.; Ahmed Ansari, M.G.; Masoud, M.S.; Alokail, M.S.; McTernan, P.G. Effects of a 6-month multi-strain probiotics supplementation in endotoxemic, inflammatory and cardiometabolic status of T2DM patients: A randomized, double-blind, placebo-controlled trial. Clin. Nutr. 2019, 38, 1561–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalili, L.; Alipour, B.; Asghari Jafar-Abadi, M.; Faraji, I.; Hassanalilou, T.; Mesgari Abbasi, M.; Vaghef-Mehrabany, E.; Alizadeh Sani, M. The Effects of Lactobacillus casei on Glycemic Response, Serum Sirtuin1 and Fetuin-A Levels in Patients with Type 2 Diabetes Mellitus: A Randomized Controlled Trial. Iran. Biomed. J. 2019, 23, 68–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palacios, T.; Vitetta, L.; Coulson, S.; Madigan, C.D.; Lam, Y.Y.; Manuel, R.; Briskey, D.; Hendy, C.; Kim, J.N.; Ishoey, T.; et al. Targeting the Intestinal Microbiota to Prevent Type 2 Diabetes and Enhance the Effect of Metformin on Glycaemia: A Randomised Controlled Pilot Study. Nutrients 2020, 12, 2041. [Google Scholar] [CrossRef]
- Van Hemert, S.; Ormel, G. Influence of the Multispecies Probiotic Ecologic® BARRIER on Parameters of Intestinal Barrier Function. Food Nutr. Sci. 2014, 5, 1739. [Google Scholar] [CrossRef] [Green Version]
- Karczewski, J.; Troost, F.J.; Konings, I.; Dekker, J.; Kleerebezem, M.; Brummer, R.J.; Wells, J.M. Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G851–G859. [Google Scholar] [CrossRef] [Green Version]
- Hummel, S.; Veltman, K.; Cichon, C.; Sonnenborn, U.; Schmidt, M.A. Differential targeting of the E-Cadherin/beta-Catenin complex by gram-positive probiotic lactobacilli improves epithelial barrier function. Appl. Environ. Microbiol. 2012, 78, 1140–1147. [Google Scholar] [CrossRef] [Green Version]
- Kocsis, T.; Molnar, B.; Nemeth, D.; Hegyi, P.; Szakacs, Z.; Balint, A.; Garami, A.; Soos, A.; Marta, K.; Solymar, M. Probiotics have beneficial metabolic effects in patients with type 2 diabetes mellitus: A meta-analysis of randomized clinical trials. Sci. Rep. 2020, 10, 11787. [Google Scholar] [CrossRef]
- Sun, J.; Buys, N.J. Glucose- and glycaemic factor-lowering effects of probiotics on diabetes: A meta-analysis of randomised placebo-controlled trials. Br. J. Nutr. 2016, 115, 1167–1177. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, Y.; Fei, X. Effect of probiotics on glucose metabolism in patients with type 2 diabetes mellitus: A meta-analysis of randomized controlled trials. Medicina 2016, 52, 28–34. [Google Scholar] [CrossRef] [PubMed]

| Sample Size | Age (y) | Sex | Technique | Microbiota Modifications | Functional Modifications | Study |
|---|---|---|---|---|---|---|
| 183 T2D 185 Controls (Chinese) | 13–86 | Women (153) Men (209) | Metagenomic sequencing | Increased in T2D: A. muciniphila, Bacteroides caccae, Bacteroides Intestinalis, C. hathewayi, Clostridium ramosum, C. symbiosum, Desulfovibrio sp., Eggerthella lenta, and Escherichia coli Decreased in T2D: Clostridiales sp. SS3/4, Eubacterium rectale, F. prausnitzii, Roseburia intestinalis, and Roseburia inulinivorans. | Increased in T2D: membrane transport of sugars, branched-chain amino acid (BCAA) transport, methane metabolism, xenobiotics degradation and metabolism, and sulphate reduction Increased in control: bacterial chemotaxis, flagellar assembly, butyrate biosynthesis, and metabolism of cofactors and vitamins. | [23] |
| 53 T2D 49 IGT 43 Controls (Swedish) | 69–72 | Women (145) | Metagenomic sequencing | Increased in T2D: Clostridium clostridioforme, former-Lactobacillus gasseri, and Streptococcus mutans Decreased in T2D: Roseburia, Clostridium spp., Eubacterium eligens, Coriobacteriaceae, and Bacteroides intestinalis. | Increased in T2D: starch and glucose metabolism, fructose and mannose metabolism, ABC transporters for amino acids, ions and simple sugars, fatty acid biosynthesis, and cysteine and methionine metabolism Increased in control: flagellar assembly, and riboflavin metabolism. | [25] |
| 13 T2D 64 Prediabetes 44 Controls (Chinese) | 52–55 | Not available | 16S rRNA V3-V5 region | Increased in T2D: Clostridiales, Dorea, Prevotella, Collinsella, and Ruminococcus Decreased in T2D: Bacteroides, A. muciniphila, F. prausnitzii, Haemophilus parainfluenzae, and Roseburia | - | [26] |
| 75 T2M 291 Controls (Danish) | 50–66 | Women (187) Men (179) | Metagenomic sequencing | Increased in T2D: Prevotella copri and Bacteroides vulgatus Decreased in T2D: Roseburia, Bifidobacterium, Faecalibacterium, Oscillibacter, Coprococcus, and Butyrivibrio | Increased in T2D: lipopolysaccharide and BCAA biosynthesis Decreased in T2D: BCAA transport into bacterial cells, methanogenesis, and pyruvate oxidation. | [27] |
| 22 T1D 23 T2D 23 Controls (Polish) | 20–65 | Women (40) Men (28) | 16S rRNA | Increased in T2D: Ruminococcus, Enterobacteriaceae, and Verrucomicrobia Decreased in T2D: Bacteroides, Roseburia, and Faecalibacterium (n.s.) | - | [28] |
| 20 T2D 40 Controls (Chinese) | 20–60 | Women (42) Men (18) | 16S rRNA V4-V5 region | Increased in T2D: Dorea, Fusobacterium, and F. prausnitzii Decreased in T2D: Parabacteroides, Akkermansia, Bifidobacterium, and Streptococcus | Increased in T2D: butyrate production via transferase, methanol conversion, and pentose phosphate pathway Decreased in T2D: tyrosine degradation, leucine degradation, and anaerobic fatty acid beta-oxidation | [29] |
| 98 T2D 193 Controls (Africans) | 41–70 | Not available | 16S rRNA V4 region | Increased in T2D: Desulfovibrio piger, Prevotella, Eubacterium, and Peptostreptococcus Decreased in T2D: Collinsella, Ruminococcus lactaris, Anaerostipes, Epulopiscium, and Clostridium | Increased in T2D: proteasome pathway Decreased in T2D: none | [21] |
| 134 T2D 37 Controls (Chinese) | 45–67 | Women (92) Men (79) | 16S rRNA V3-V4 region | Increased in T2D: Prevotella, Dialister, and Sutterella Decreased in T2D: Bacteroides, Bifidobacterium, Clostridium XIVa, Parabacteroides, Staphylococcus, Granulicatella, Porphyromonas, Clostridium XI, Blautia, Anaerostipes, Clostridium XVIII, Fusicatenibacter, Enterococcus, Clostridium IV, Eggerthella, and Flavonifractor. | - | [30] |
| 46 T2D 75 CGI 178 IGT 189 IFG 523 Controls (Swedish) | 57–61 | Women (568) Men (443) | Metagenomic sequencing | Increased in T2D: Coprococcus eutactus, Clostridiales bacterium, and Lachnospiraceae bacterium Decreased in T2D: Clostridium sp., C. hathewayi, Clostridium bolteae, C. symbiosum, and Roseburia faecis | Increased in T2D: two-component systems, phosphotransferase systems, fructose and mannose metabolism, pentose phosphate pathway, bacterial biosynthesis of branched-chain amino acids, and metabolism of the B-group vitamins biotin and thiamine. Decreased in T2D: bacterial methanogenesis, glycolysis, peptidoglycan biosynthesis, vancomycin resistance, and DNA replication and transcription. | [22] |
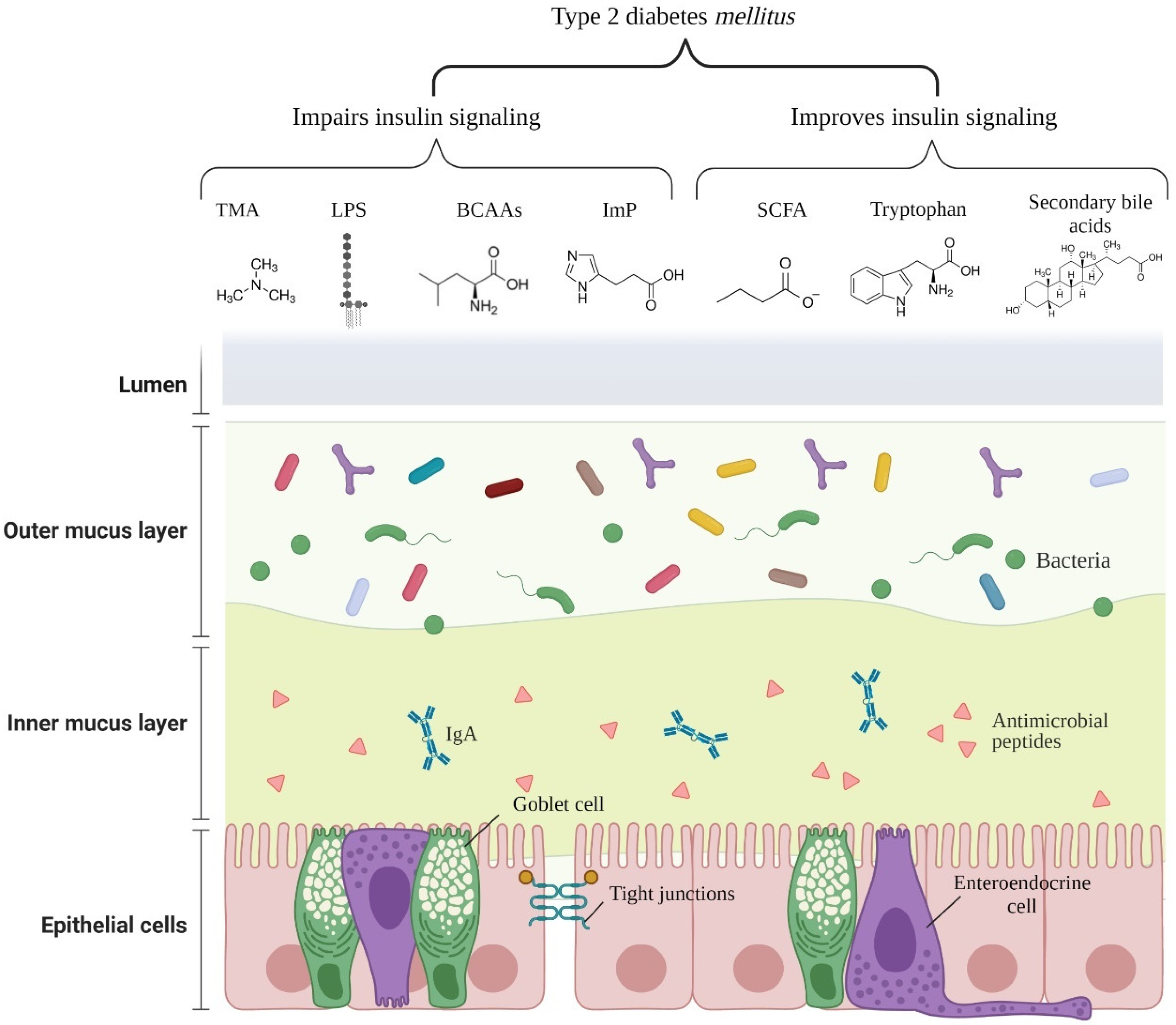
| Metabolite | Producing Bacteria (Genus or Species) | Mechanism of T2DM Risk | Reference |
|---|---|---|---|
| SCFA (acetate, propionate, and butyrate) | Akkermansia, Ruminococcus, Faecalibacterium prausnitzii, Eubacterium, Roseburia, Blautia, Coprococcus, Anaerostipes, and others | - Increases epithelial barrier function by regulation of TJP; - Reduces the passage of LPS, improving inflammation; - Stimulates the secretion of PYY and GLP-1 from L-cells in a GPR41 and GPR43 dependent manner; - Reduces appetite, insulin secretion, plasma glucose levels, and slow gastric emptying through stimulation of GLP-1 and GLP-2 secretion. | [39,40,41] |
| Imidazole propionate | Imidazole propionate: Eggerthella lenta, Streptococcus mutans, Aerococcus urinae, Brevibacillus laterosporus, and others | - Impairs glucose tolerance and insulin signaling by activating the p38γ-p62-mTORC1 pathway. | [42] |
| TMAO/TMA | Desulfovibrio desulfuricans, Providencia, E. coli, Klebsiella pneumoniae, Sporosarcina, and others. | - Exacerbates blockage of the insulin signaling pathway and promotes inflammation in adipose tissue. | [43] |
| Branched-chain amino acids | former-Lactobacillus, Weissella, Leuconostoc, P. copri, and B. vulgatus | - Promotes insulin resistance through serine phosphorylation of IRS-1; by persistent activation of mTORC1/S6K1. | [27,44,45] |
| Bile acids Secondary bile acids | Secondary bile acids: Ruminococcus, Bifidobacterium, Bacteroides, Clostridium, former-Lactobacillus, Eubacterium, Listeria, and others. | - Ligands of nuclear receptors, such as VDR, PXR, and FXR, induce TGR5 expression and regulate insulin and glucose sensitivity. | [46,47] |
| Tryptophan metabolites Tryptamine | Clostridium bartlettii, Clostridium sporogenes, Ruminococcus gnavus, Bacteroides ovatus, Lactobaccilus acidophilus,Limosilactobacillus reuteri, Bifidobacterium fragilis, Bifidobacterium bifidum, and others. | - Improves intestinal epithelial barrier function by the activation of PXR; - Stimulates insulin secretion, supresses appetite, and slows gastric emptying by stimulating GLP-1 secretion; - Promotes gastrointestinal motility by stimulating serotonin release; - Anti-inflammatory and anti-oxidative effects in the systemic circulation. | [48] |
| Sample Size | Design | Duration | Intervention | Metabolic Outcomes | Microbiota Modifications | Functional Modifications | Study |
|---|---|---|---|---|---|---|---|
| Placebo (18) Intervention (16) | Single-blind clinical trial | 6 weeks | Probiotic: 3000mg/day of L. acidophilus, L. bulgaricus, L. bifidum, and L. casei | Probiotic: n.s. | - | - | [77] |
| Placebo (53) Intervention (48) | Randomized, double-blind, parallel-group, controlled clinical trial | 12 weeks | Probiotic: 3 × 1010 CFU/day of L. acidophilus, L. casei, L. lactis, B. bifidum, B. longum, and B. infantis | Probiotic: ↓ HbA1C, FI, HOMA-IR | Probiotic: ↑ Bifidobacterium spp., former-Lactobacillus spp. | - | [78] |
| Placebo (39) Intervention (39) | Randomized, single-centre, double-blind, placebo-controlled | 12 weeks | Probiotic: 1010 CFU/day of B. bifidum W23, B. lactis W52, L. acidophilus W37, L. brevis W63, L. casei W56, L. salivarius W24, L. lactis W19, and L. lactis W58 | Probiotic: ↓ HOMA-IR, FBG, Insulin, C-peptide, TG, LDL-c, WHR | - | - | [79] |
| Placebo (15) Intervention (29) | Randomized, double-blind, placebo-controlled trial | 12 weeks | Probiotic: L. reuteri DSM 17938 LD: 108 CFU/day HD: 1010 CFU/day | Probiotic: HD: ↑ ISI, DCA LD: ↑ unconjugated bile acids | Probiotic: ↑ L. reuteri | - | [80] |
| Placebo (22) Intervention (46) | Randomized, double-blind, placebo-controlled trial | 9 months (6 month intervention) | Group 1: 4 × 109 CFU/day of probiotic L. reuteri ADR-1 Group 2: 2 × 1010 CFU/day heat-killed L. reuteri ADR-3 | Group 1: ↓ HbA1C, TC Group 2: ↓ SBP, IL-1β | Group 1: ↑ L. reuteri Group 2: ↑ Bifidobacterium | - | [81] |
| Placebo (22) Intervention (31) | Randomized, double-blind, single-centre, clinical trial | 8 weeks | Probiotic: 1 sachet (10g)/day of 14 probiotic strains of former-Lactobacillus + Lactococcus (6 × 1010 CFU/g), Bifidobacterium (1 × 1010/g), Propionibacterium (3 × 1010/g), Acetobacter (1 × 106/g) genera | Probiotic: ↓ HOMA-IR, TNF- α, IL-1β, WC | - | - | [82] |
| Placebo (30) Intervention (31) | Randomized, single-centre, double-blind, placebo-controlled clinical trial | 6 months | Probiotic: 1010 CFU/day of B. bifidum W23, B. lactis W52, L. acidophilus W37, L. brevis W63, L. casei W56, L. salivarius W24, L. lactis W19 and L. lactis W58 | Probiotic: ↓ HOMA-IR, FBG, Insulin, C-peptide, TG, TC, TC/HDL, CRP, TNF-α, IL-6, resistin, endotoxin ↑ adiponectin | - | - | [83] |
| Placebo (20) Intervention (20) | Randomized, parallel-group, placebo-controlled trial | 8 weeks | Probiotic: 108 CFU/day of L. casei | Probiotic: ↓ FBG, HOMA-IR, Insulin, fetuin-A, weight, BMI, WC ↑ SIRT1 | - | - | [84] |
| Placebo (30) Intervention (30) | Randomized, double-blind, single-centre, placebo-controlled pilot trial | 12 weeks | Probiotic: 2 × 1011 CFU/day of L. plantarum Lp-115, L. bulgaricus Lb-64, L. gasseri Lg-36, B. breve Bb-03, B. animalis sbsp. lactis Bi-07, B. bifidum Bb-06, S. thermophilus St-21, and S. boulardii DBVPG 6763 | Probiotic: ↑ plasma butyrate Subgroup (metformin): ↓ FBG, HbA1C, insulin resistance, and zonulin ↑ plasma butyrate | Probiotic: n.s. beta diversity ↓ P. copri, Flavonifractor plautii ↑ B. breve, B. caccae, Bacteroidales bacterium ph8, A. muciniphila, C. hathewayi Subgroup (metformin): ↓ Bactoides uniformis ↑ B. breve, B. caccae, Anaerotruncus colihominis | Subgroup (metformin): pyruvate fermentation to butanoate, and Bifidobacterium shunt pathways | [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ballan, R.; Saad, S.M.I. Characteristics of the Gut Microbiota and Potential Effects of Probiotic Supplements in Individuals with Type 2 Diabetes mellitus. Foods 2021, 10, 2528. https://doi.org/10.3390/foods10112528
Ballan R, Saad SMI. Characteristics of the Gut Microbiota and Potential Effects of Probiotic Supplements in Individuals with Type 2 Diabetes mellitus. Foods. 2021; 10(11):2528. https://doi.org/10.3390/foods10112528
Chicago/Turabian StyleBallan, Rafael, and Susana Marta Isay Saad. 2021. "Characteristics of the Gut Microbiota and Potential Effects of Probiotic Supplements in Individuals with Type 2 Diabetes mellitus" Foods 10, no. 11: 2528. https://doi.org/10.3390/foods10112528
APA StyleBallan, R., & Saad, S. M. I. (2021). Characteristics of the Gut Microbiota and Potential Effects of Probiotic Supplements in Individuals with Type 2 Diabetes mellitus. Foods, 10(11), 2528. https://doi.org/10.3390/foods10112528