Metabolism of Daidzein and Genistein by Gut Bacteria of the Class Coriobacteriia
Abstract
:1. Introduction
2. Materials and Methods
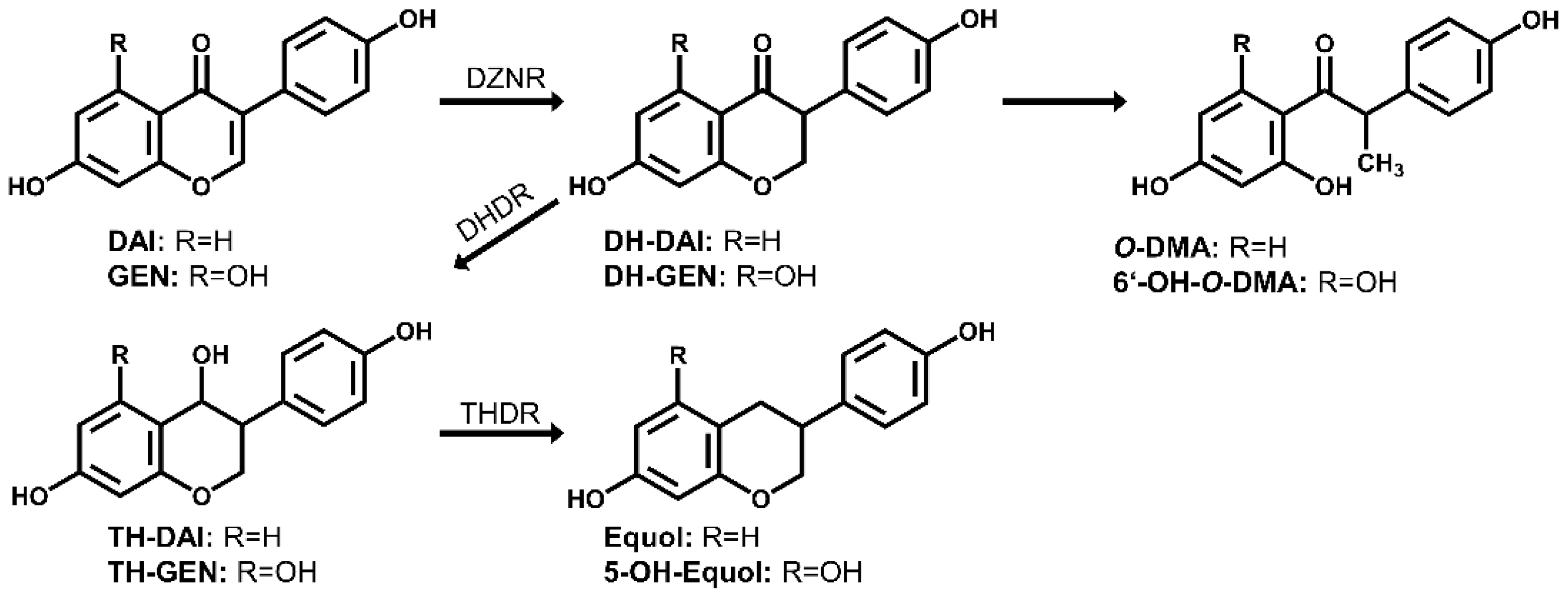
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef] [Green Version]
- Hwang, K.A.; Choi, K.C. Chapter One—Endocrine-Disrupting Chemicals with Estrogenicity Posing the Risk of Cancer Progression in Estrogen-Responsive Organs. In Advances in Molecular Toxicology; Fishbein, J.C., Heilman, J.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 1–33. [Google Scholar]
- Messina, M. Soy and Health Update: Evaluation of the Clinical and Epidemiologic Literature. Nutrients 2016, 8, 754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hüser, S.; Guth, S.; Joost, H.G.; Soukup, S.T.; Köhrle, J.; Kreienbrock, L.; Diel, P.; Lachenmeier, D.W.; Eisenbrand, G.; Vollmer, G.; et al. Effects of isoflavones on breast tissue and the thyroid hormone system in humans: A comprehensive safety evaluation. Arch. Toxicol. 2018, 92, 2703–2748. [Google Scholar] [CrossRef] [Green Version]
- Clavel, T.; Lepage, P.; Charrier, C. The Family Coriobacteriaceae, in The Prokaryotes—Actinobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Mayo, B.; Vázquez, L.; Flórez, A.B. Equol: A Bacterial Metabolite from The Daidzein Isoflavone and Its Presumed Beneficial Health Effects. Nutrients 2019, 11, 2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duda-Chodak, A.; Tarko, T.; Satora, P.; Sroka, P. Interaction of dietary compounds, especially polyphenols, with the intestinal microbiota: A review. Eur. J. Nutr. 2015, 54, 325–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthies, A.; Blaut, M.; Braune, A. Isolation of a Human Intestinal Bacterium Capable of Daidzein and Genistein Conversion. Appl. Environ. Microbiol. 2009, 75, 1740–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthies, A.; Clavel, T.; Gütschow, M.; Engst, W.; Haller, D.; Blaut, M.; Braune, A. Conversion of Daidzein and Genistein by an Anaerobic Bacterium Newly Isolated from the Mouse Intestine. Appl. Environ. Microbiol. 2008, 74, 4847–4852. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.-S.; Nishihata, T.; Kakiuchi, N.; Hattori, M. Biotransformation of C-Glucosylisoflavone Puerarin to Estrogenic (3S)-Equol in Co-culture of Two Human Intestinal Bacteria. Biol. Pharm. Bull. 2008, 31, 1621–1625. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.S.; Chen, W.J.; Adeolu, M.; Chai, Y. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3379–3397. [Google Scholar] [CrossRef] [Green Version]
- Oren, A.; Garrity, G.M. Notification that new names of prokaryotes and new combinations have appeared in volume 63, part 9, of the IJSEM. Int. J. Syst. Evol. Microbiol. 2013, 63, 4371–4373. [Google Scholar] [CrossRef]
- Minamida, K.; Tanaka, M.; Abe, A.; Sone, T.; Tomita, F.; Hara, H.; Asano, K. Production of equol from daidzein by gram-positive rod-shaped bacterium isolated from rat intestine. J. Biosci. Bioeng. 2006, 102, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Maruo, T.; Sakamoto, M.; Ito, C.; Toda, T.; Benno, Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int. J. Syst. Evol. Microbiol. 2008, 58, 1221–1227. [Google Scholar] [CrossRef]
- Clavel, T.; Charrier, C.; Braune, A.; Wenning, M.; Blaut, M.; Haller, D. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 1805–1812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawada, Y.; Yokoyama, S.-I.; Yanase, E.; Niwa, T.; Suzuki, T. The production of S-equol from daidzein is associated with a cluster of three genes in Eggerthella sp. YY7918. Biosci. Microbiota Food Health 2016, 35, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.S.; Kitahara, M.; Sakamoto, M.; Hattori, M.; Benno, Y. Slackia equolifaciens sp. nov., a human intestinal bacterium capable of producing equol. Int. J. Syst. Evol. Microbiol. 2010, 60, 1721–1724. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.N.; Hao, Q.H.; Zhang, H.L.; Zhou, B.; Yu, X.M.; Wang, X.L. Reduction of soy isoflavones by use of Escherichia coli whole-cell biocatalyst expressing isoflavone reductase under aerobic conditions. Lett. Appl. Microbiol. 2016, 63, 111–116. [Google Scholar] [CrossRef]
- Tsuji, H.; Moriyama, K.; Nomoto, K.; Miyanaga, N.; Akaza, H. Isolation and characterization of the equol-producing bacterium Slackia sp. strain NATTS. Arch. Microbiol. 2010, 192, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.; Yasuda, S.; Takahashi, M.; Hayashi, T.; Miyazawa, N.; Sato, I.; Abiru, Y.; Uchiyama, S.; Hishigaki, H. Cloning and Expression of a Novel NADP(H)-Dependent Daidzein Reductase, an Enzyme Involved in the Metabolism of Daidzein, from Equol-Producing Lactococcus Strain 20–92. Appl. Environ. Microbiol. 2010, 76, 5892–5901. [Google Scholar] [CrossRef] [Green Version]
- Shimada, Y.; Takahashi, M.; Miyazawa, N.; Ohtani, T.; Abiru, Y.; Uchiyama, S.; Hishigaki, H. Identification of Two Novel Reductases Involved in Equol Biosynthesis in Lactococcus Strain 20–92. J. Mol. Microbiol. Biotechnol. 2011, 21, 160–172. [Google Scholar] [CrossRef]
- Shimada, Y.; Takahashi, M.; Miyazawa, N.; Abiru, Y.; Uchiyama, S.; Hishigaki, H. Identification of a Novel Dihydrodaidzein Racemase Essential for Biosynthesis of Equol from Daidzein in Lactococcus sp. Strain 20–92. Appl. Environ. Microbiol. 2012, 78, 4902–4907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schröder, C.; Matthies, A.; Engst, W.; Blaut, M.; Braune, A. Identification and Expression of Genes Involved in the Conversion of Daidzein and Genistein by the Equol-Forming Bacterium Slackia isoflavoniconvertens. Appl. Environ. Microbiol. 2013, 79, 3494–3502. [Google Scholar] [CrossRef] [Green Version]
- Tsuji, H.; Moriyama, K.; Nomoto, K.; Akaza, H. Identification of an Enzyme System for Daidzein-to-Equol Conversion in Slackia sp. Strain NATTS. Appl. Environ. Microbiol. 2012, 78, 1228–1236. [Google Scholar] [CrossRef] [Green Version]
- Flórez, A.B.; Vázquez, L.; Rodríguez, J.; Redruello, B.; Mayo, B. Transcriptional Regulation of the Equol Biosynthesis Gene Cluster in Adlercreutzia equolifaciens DSM19450T. Nutrients 2019, 11, 993. [Google Scholar] [CrossRef] [Green Version]
- Setchell, K.D.R.; Clerici, C.; Lephart, E.D.; Cole, S.J.; Heenan, C.; Castellani, D.; Wolfe, B.E.; Nechemias-Zimmer, L.; Brown, N.M.; Lund, T.D.; et al. S-Equol, a potent ligand for estrogen receptor β, is the exclusive enantiomeric form of the soy isoflavone metabolite produced by human intestinal bacterial flora. Am. J. Clin. Nutr. 2005, 81, 1072–1079. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, S.-I.; Suzuki, T. Isolation and Characterization of a Novel Equol-Producing Bacterium from Human Feces. Biosci. Biotechnol. Biochem. 2008, 72, 2660–2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danylec, N.; Stoll, D.A.; Dötsch, A.; Huch, M. Draft Genome Sequences of Type Strains of Gordonibacter faecihominis, Paraeggerthella hongkongensis, Parvibacter caecicola, Slackia equolifaciens, Slackia faecicanis, and Slackia isoflavoniconvertens. Microbiol. Resour. Announc. 2019, 8, e01532-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez, L.; Guadamuro, L.; Giganto, F.; Mayo, B.; Flórez, A.B. Development and Use of a Real-Time Quantitative PCR Method for Detecting and Quantifying Equol-Producing Bacteria in Human Faecal Samples and Slurry Cultures. Front. Microbiol. 2017, 8, 1155. [Google Scholar] [CrossRef] [PubMed]
- Bode, L.M.; Bunzel, D.; Huch, M.; Cho, G.-S.; Ruhland, D.; Bunzel, M.; Bub, A.; Franz, C.M.A.P.; Kulling, S.E. In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota. Am. J. Clin. Nutr. 2013, 97, 295–309. [Google Scholar] [CrossRef]
- Koonin, E.V.; Galperin, M.Y. Sequence—Evolution—Function: Computational Approaches in Comparative Genomics; Kluwer Academic: Boston, MA, USA, 2003. [Google Scholar]
- Minamida, K.; Ota, K.; Nishimukai, M.; Tanaka, M.; Abe, A.; Sone, T.; Tomita, F.; Hara, H.; Asano, K. Asaccharobacter celatus gen. nov., sp. nov., isolated from rat caecum. Int. J. Syst. Evol. Microbiol. 2008, 58, 1238–1240. [Google Scholar] [CrossRef]
- Uchiyama, S.; Ueno, T.; Suzuki, T. Identification of a Newly Isolated Equol-Producing Lactic Acid Bacterium from the Human Feces. J. Intest. Microbiol. 2007, 21, 217–220. [Google Scholar]
- Yokoyama, S.-I.; Oshima, K.; Nomura, I.; Hattori, M.; Suzuki, T. Complete Genomic Sequence of the Equol-Producing Bacterium Eggerthella sp. Strain YY7918, Isolated from Adult Human Intestine. J. Bacteriol. 2011, 193, 5570–5571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez, L.; Flórez, A.B.; Redruello, B.; Mayo, B. Metabolism of Soy Isoflavones by Intestinal Bacteria: Genome Analysis of an Adlercreutzia equolifaciens Strain That Does Not Produce Equol. Biomolecules 2020, 10, 950. [Google Scholar] [CrossRef]
- Elsawi, Z.; Togo, A.H.; Beye, M.; Dubourg, G.; Andrieu, C.; Armsrtong, N.; Richez, M.; Di Pinto, F.; Bittar, F.; Labas, N.; et al. Hugonella massiliensis gen. nov., sp. nov., genome sequence, and description of a new strictly anaerobic bacterium isolated from the human gut. MicrobiologyOpen 2017, 6, e00458. [Google Scholar] [CrossRef] [Green Version]
- Han, K.-I.; Kim, J.-S.; Lee, K.C.; Eom, M.K.; Suh, M.K.; Kim, H.S.; Park, S.-H.; Lee, J.H.; Kang, S.W.; Park, J.-E.; et al. Lee1,4, Senegalimassilia faecalis sp. nov., an anaerobic actinobacterium isolated from human faeces, and emended description of the genus Senegalimassilia. Int. J. Syst. Evol. Microbiol. 2020, 70, 1684–1690. [Google Scholar] [CrossRef] [PubMed]
- Clavel, T.; Duck, W.; Charrier, C.; Wenning, M.; Elson, C.; Haller, D. Enterorhabdus caecimuris sp. nov., a member of the family Coriobacteriaceae isolated from a mouse model of spontaneous colitis, and emended description of the genus Enterorhabdus Clavel et al. 2009. Int. J. Syst. Evol. Microbiol. 2010, 60, 1527–1531. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Peirotén, Á.; Gaya, P.; Álvarez, I.; Landete, J.M. Production of O-desmethylangolensin, tetrahydrodaidzein, 6′-hydroxy-O-desmethylangolensin and 2-(4-hydroxyphenyl)-propionic acid in fermented soy beverage by lactic acid bacteria and Bifidobacterium strains. Food Chem. 2020, 318, 126521. [Google Scholar] [CrossRef] [PubMed]



| T (h) | Daidzein | DH-Daidzein | Equol | O-DMA | |
|---|---|---|---|---|---|
| Control (medium without bacteria) # | 0 | 75.47 ± 5.05 | - | - | - |
| 24 | 77.54 ± 3.05 | - | - | - | |
| 48 | 79.11 ± 0.59 | - | - | - | |
| 72 | 78.13 ± 2.04 | - | - | - | |
| Adlercreutzia equolifaciens subsp. celatus DSM 18785T | 0 | 77.80 ± 1.31 | - | - | - |
| 24 | 5.04 ± 4.44 | 18.49 ± 11.57 | 44.00 ± 4.41 | - | |
| 48 | 15.70 ± 3.00 | 23.15 ± 1.78 | 29.08 ± 4.13 | - | |
| 72 | 61.61 ± 4.49 | 10.99 ± 1.70 | 2.58 ± 1.31 | - | |
| Adlercreutzia equolifaciens subsp. equolifaciens DSM 19450T | 0 | 72.48 ± 7.86 | - | - | - |
| 24 | 24.03 ± 3.55 | 26.23 ± 1.48 | 19.39 ± 1.08 | - | |
| 48 | 2.06 ± 0.73 | 11.02 ± 2.56 | 51.09 ± 3.75 | - | |
| 72 | 6.04 ± 3.87 | 26.80 ± 1.10 | 32.77 ± 4.37 | - | |
| Adlercreutzia mucosicola DSM 19490T | 0 | 78.68 ± 2.69 | - | - | - |
| 24 | in three samples * | 0.41 ± 0.62 | 64.55 ± 0.79 | - | |
| 48 | in three samples * | 0.48 ± 0.74 | 64.55 ± 0.58 | - | |
| 72 | 1.79 ± 0.32 | 2.23 ± 0.59 | 62.70 ± 0.28 | - | |
| ‘Hugonella massiliensis’ DSM 101782T | 0 | 77.20 ± 1.32 | - | - | - |
| 24 | - | in three samples * | 63.44 ± 4.60 | - | |
| 48 | in three samples * | in three samples * | 64.93 ± 1.89 | - | |
| 72 | in three samples * | in three samples * | 64.61 ± 2.14 | - | |
| Senegalimassilia faecalis KGMB 04484T | 0 | 52.36 ± 28.73 | - | - | - |
| 24 | 69.24 ± 4.44 | in one sample * | - | - | |
| 48 | 68.36 ± 1.98 | in one sample * | - | - | |
| 72 | 66.52 ± 2.35 | in two samples * | - | - | |
| Slackia equolifaciens DSM 24851T # | 0 | 67.70 ± 8.34 | - | - | - |
| 24 | 52.34 ± 3.73 | 18.11 ± 1.40 | 5.42 ± 1.72 | - | |
| 48 | 19.89 ± 3.87 | 33.91 ± 0.51 | 16.49 ± 3.46 | - | |
| 72 | 22.23 ± 4.51 | 32.70 ± 0.21 | 15.48 ± 3.62 | - | |
| Slackia exigua DSM 15923T | 0 | 79.09 ± 0.70 | - | - | - |
| 24 | 22.51 ± 2.11 | - | - | 42.20 ± 1.91 | |
| 48 | 24.16 ± 3.38 | - | - | 43.29 ± 2.19 | |
| 72 | 23.25 ± 3.64 | - | - | 43.44 ± 2.24 | |
| Slackia isoflavoniconvertens DSM 22006T | 0 | 78.27 ± 3.82 | - | - | - |
| 24 | 11.70 ± 8.46 | 17.35 ± 5.72 | 37.79 ± 4.21 | - | |
| 48 | 1.29 ± 0.19 | 10.95 ± 3.65 | 52.32 ± 5.25 | - | |
| 72 | 4.73 ± 1.16 | 17.88 ± 1.37 | 43.72 ± 3.49 | - |
| T (h) | Genistein | DH-Genistein | 5-OH-Equol | 6′OH-O-DMA | |
|---|---|---|---|---|---|
| Control (medium without bacteria) # | 0 | 81.17 ± 0.80 | - | - | - |
| 24 | 79.05 ± 1.44 | - | - | - | |
| 48 | 74.91 ± 4.86 | - | - | - | |
| 72 | 75.78 ± 3.52 | - | - | - | |
| Adlercreutzia equolifaciens subsp. celatus DSM 18785T | 0 | 78.21 ± 9.11 | - | - | - |
| 24 | 4.32 ± 5.06 | 59.40 ± 0.81 | 2.05 ± 1.65 | - | |
| 48 | 14.67 ± 1.85 | 48.38 ± 3.19 | 1.78 ± 1.43 | - | |
| 72 | 42.33 ± 6.17 | 25.88 ± 6.96 | 1.49 ± 1.15 | - | |
| Adlercreutzia equolifaciens subsp. equolifaciens DSM 19450T | 0 | 78.17 ± 6.66 | - | - | - |
| 24 | 29.04 ± 26.03 | 41.61 ± 19.58 | - | - | |
| 48 | 2.15 ± 0.92 | 59.16 ± 1.17 | 1.42 ± 1.10 | - | |
| 72 | 14.99 ± 5.16 | 45.90 ± 4.00 | 1.83 ± 0.22 | - | |
| Adlercreutzia mucosicola DSM 19490T | 0 | 79.21 ± 1.15 § | - § | - § | - § |
| 24 | 0.75 ± 0.99 § | 48.76 ± 7.63 § | 10.02 ± 3.54 § | - § | |
| 48 | 0.72 ± 0.95 § | 26.77 ± 10.20 § | 20.17 ± 4.81 § | - § | |
| 72 | 7.22 ± 0.58 § | 16.89 ± 9.19 § | 20.33 ± 4.20 § | - § | |
| ‘Hugonella massiliensis’ DSM 101782T | 0 | 84.05 ± 2.44 | - | - | - |
| 24 | 1.94 ± 3.27 | 16.99 ± 14.19 | 27.36 ± 10.17 | - | |
| 48 | in one sample * | 0.91 ± 1.48 | 33.93 ± 2.40 | - | |
| 72 | in one sample * | in three samples * | 32.28 ± 2.80 | - | |
| Senegalimassilia faecalis KGMB 04484T | 0 | 90.96 ± 4.59 | - | - | - |
| 24 | 67.93 ± 8.13 | - | - | - | |
| 48 | 71.16 ± 0.37 | - | - | - | |
| 72 | 71.57 ± 1.36 | in three samples * | - | - | |
| Slackia equolifaciens DSM 24851T # | 0 | 70.47 ± 7.56 | - | - | - |
| 24 | 42.99 ± 2.42 | 29.83 ± 1.63 | - | - | |
| 48 | 25.58 ± 0.99 | 42.12 ± 1.65 | - | - | |
| 72 | 34.66 ± 0.22 | 33.97 ± 0.33 | - | - | |
| Slackia exigua DSM 15923T | 0 | 73.80 ± 3.44 | - | - | - |
| 24 | 31.44 ± 10.95 | - | - | 12.74 ± 1.23 | |
| 48 | 10.14 ± 12.55 § | - § | - § | 5.08 ± 0.68 § | |
| 72 | 20.52 ± 16.91 § | - § | - § | - § | |
| Slackia isoflavoniconvertens DSM 22006T | 0 | 75.31 ± 6.70 § | - § | - § | - § |
| 24 | 36.60 ± 12.03 | 39.13 ± 9.73 | - | - | |
| 48 | 7.17 ± 2.25 | 59.27 ± 1.68 | - | - | |
| 72 | 9.13 ± 0.41 | 55.71 ± 0.11 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soukup, S.T.; Stoll, D.A.; Danylec, N.; Schoepf, A.; Kulling, S.E.; Huch, M. Metabolism of Daidzein and Genistein by Gut Bacteria of the Class Coriobacteriia. Foods 2021, 10, 2741. https://doi.org/10.3390/foods10112741
Soukup ST, Stoll DA, Danylec N, Schoepf A, Kulling SE, Huch M. Metabolism of Daidzein and Genistein by Gut Bacteria of the Class Coriobacteriia. Foods. 2021; 10(11):2741. https://doi.org/10.3390/foods10112741
Chicago/Turabian StyleSoukup, Sebastian Tobias, Dominic Alexander Stoll, Nicolas Danylec, Alena Schoepf, Sabine Emma Kulling, and Melanie Huch. 2021. "Metabolism of Daidzein and Genistein by Gut Bacteria of the Class Coriobacteriia" Foods 10, no. 11: 2741. https://doi.org/10.3390/foods10112741
APA StyleSoukup, S. T., Stoll, D. A., Danylec, N., Schoepf, A., Kulling, S. E., & Huch, M. (2021). Metabolism of Daidzein and Genistein by Gut Bacteria of the Class Coriobacteriia. Foods, 10(11), 2741. https://doi.org/10.3390/foods10112741





