Mathematical Modelling of Canola Oil Biodegradation and Optimisation of Biosurfactant Production by an Antarctic Bacterial Consortium Using Response Surface Methodology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Shake Flask Culture Conditions
2.2. Biodegradation Kinetics Modelling
Percentage Degradation of WCO and PCO
2.3. Analysis of Biosurfactant Production
2.3.1. Haemolytic Activity
2.3.2. Microbial Adhesion to Hydrocarbons (MATH) Assay
2.3.3. Drop-Collapse Test and Oil Displacement
2.3.4. Emulsification Index (E24)
2.4. Extraction of Biosurfactants
2.5. Experimental Design and Optimisation
2.6. Statistical Analyses
3. Results
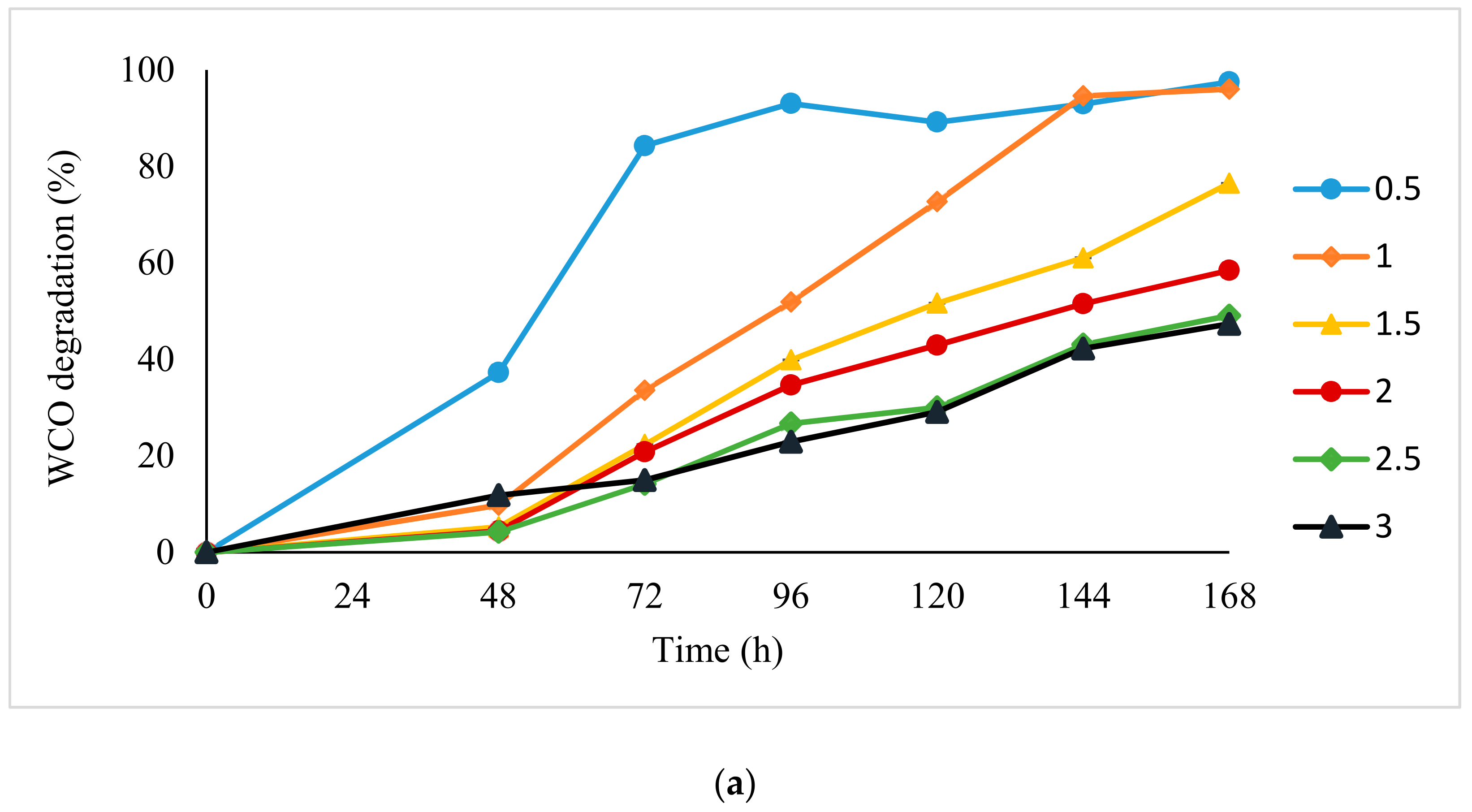
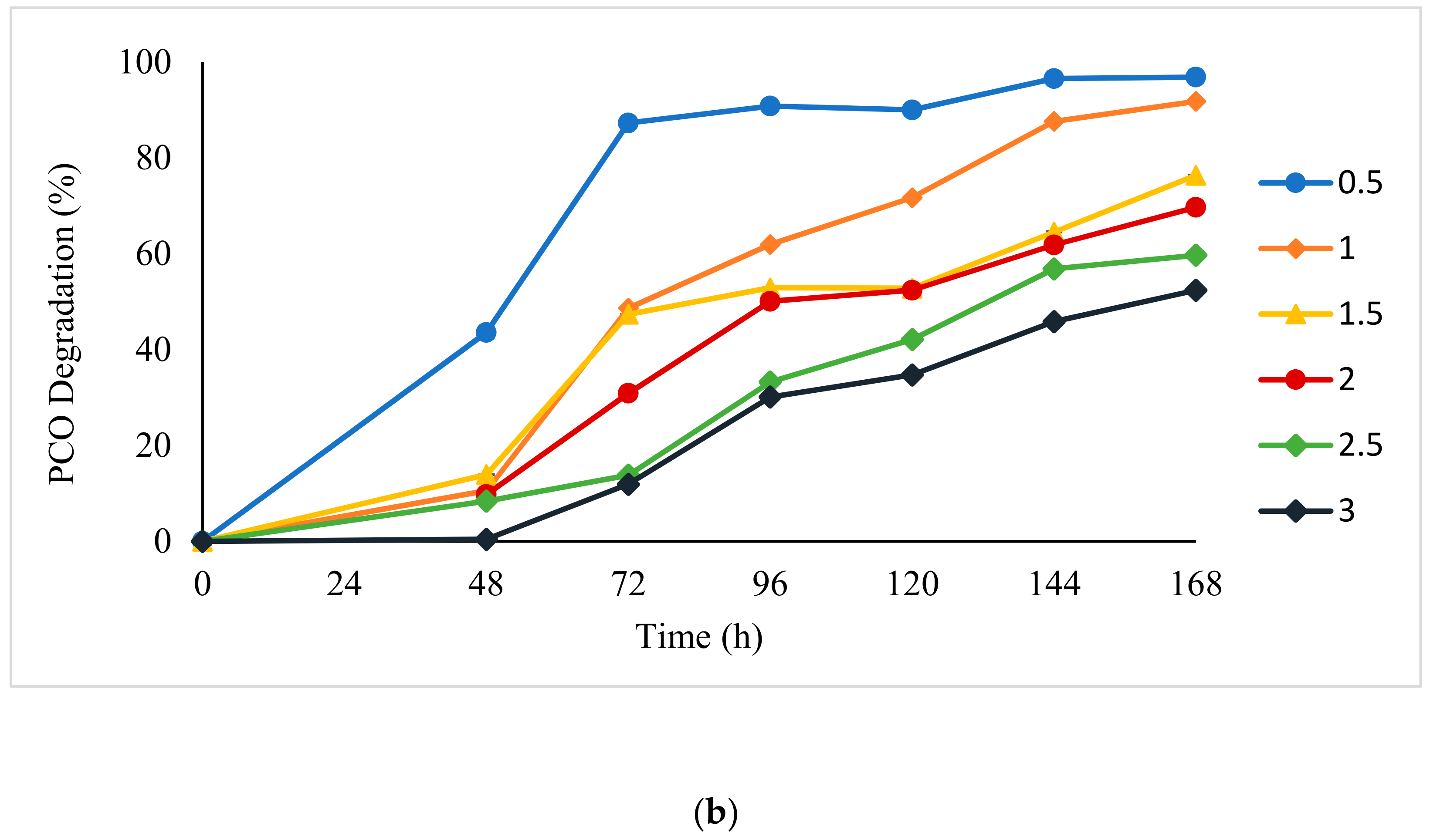
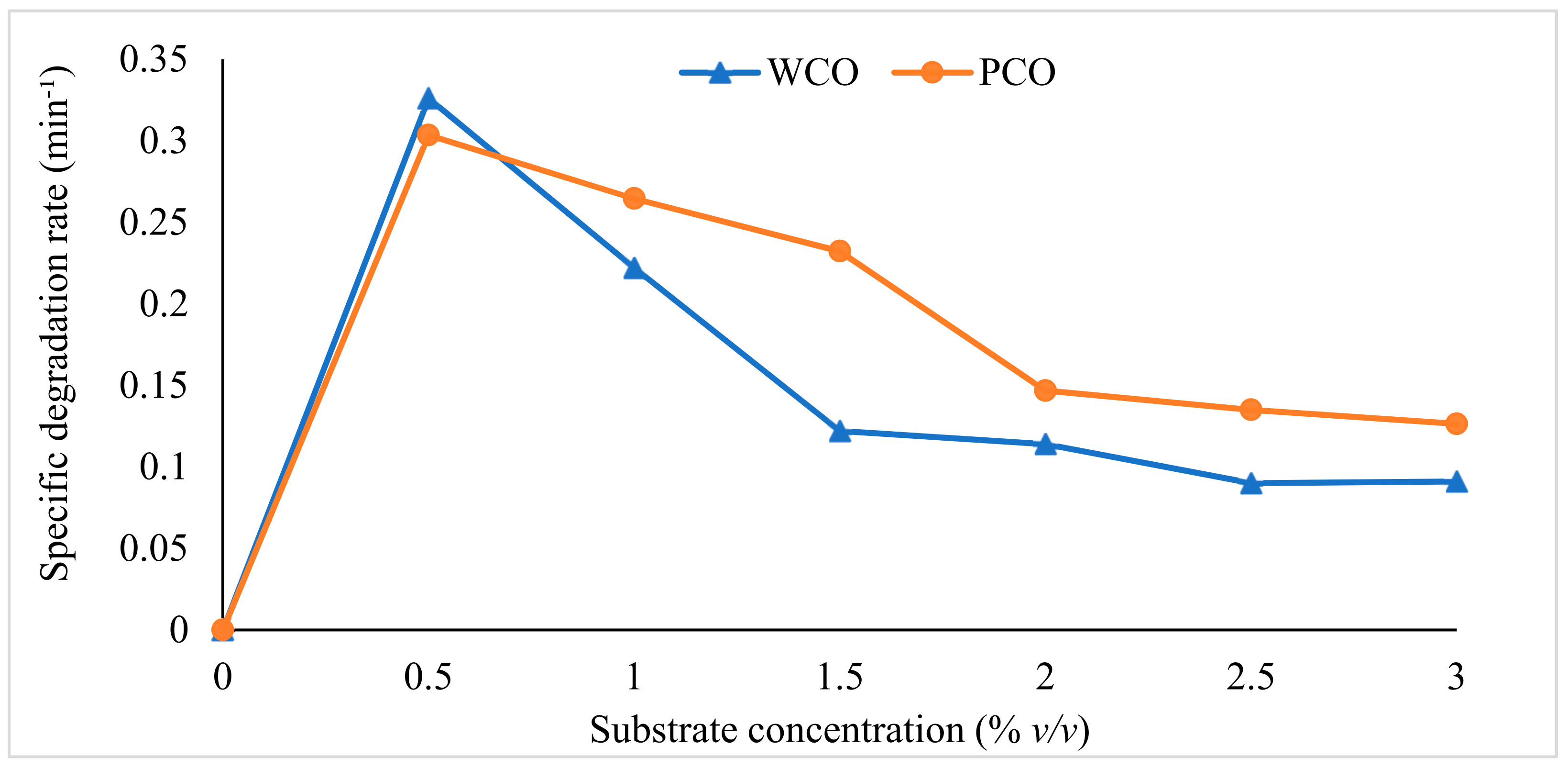
3.1. Degradation Kinetics Modelling
3.2. Preliminary Screening
3.2.1. Haemolytic Test
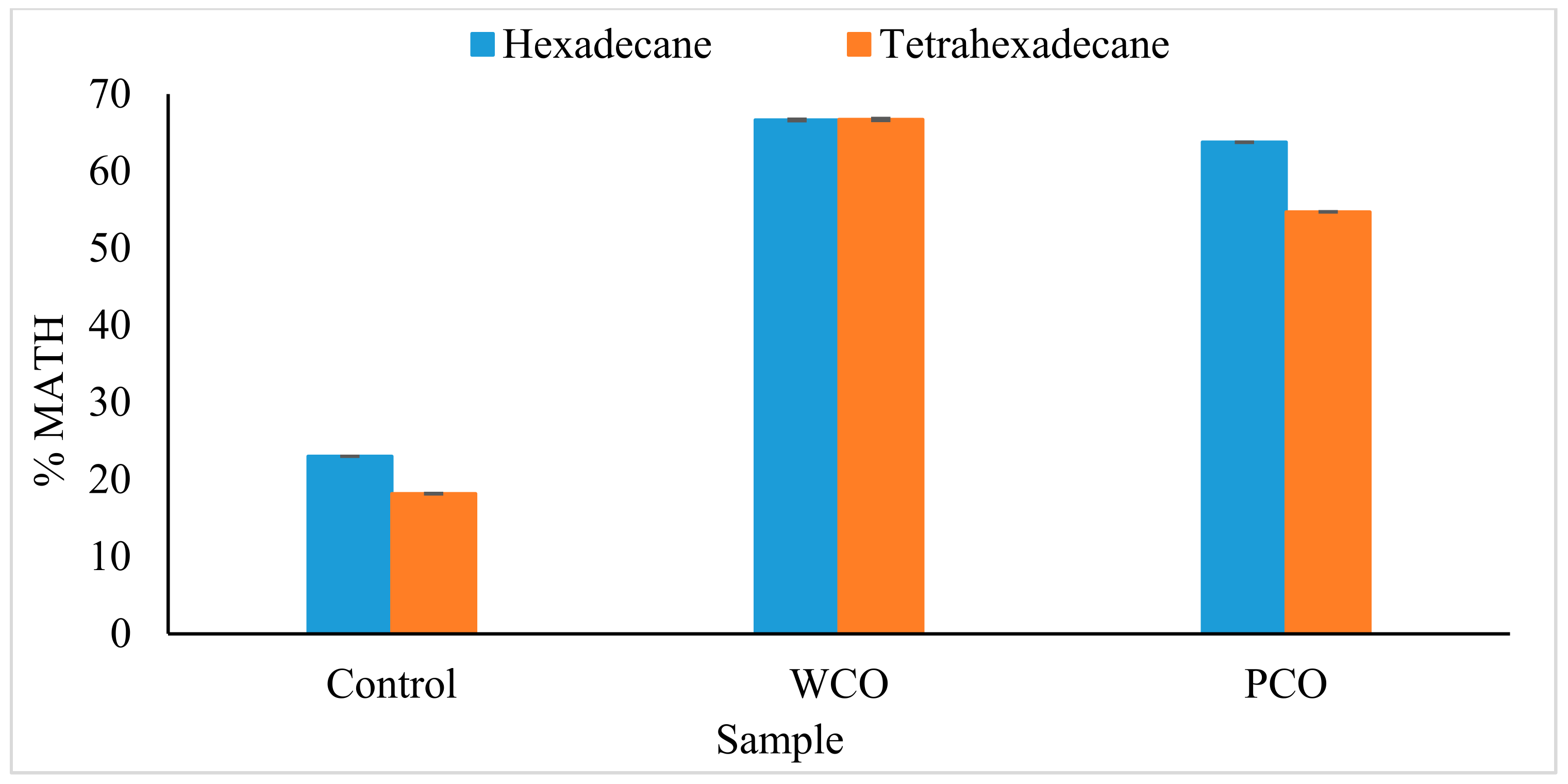
3.2.2. Microbial Adhesion to Hydrocarbons (MATH) Assay
3.2.3. Oil-Spreading and Drop-Collapse Test
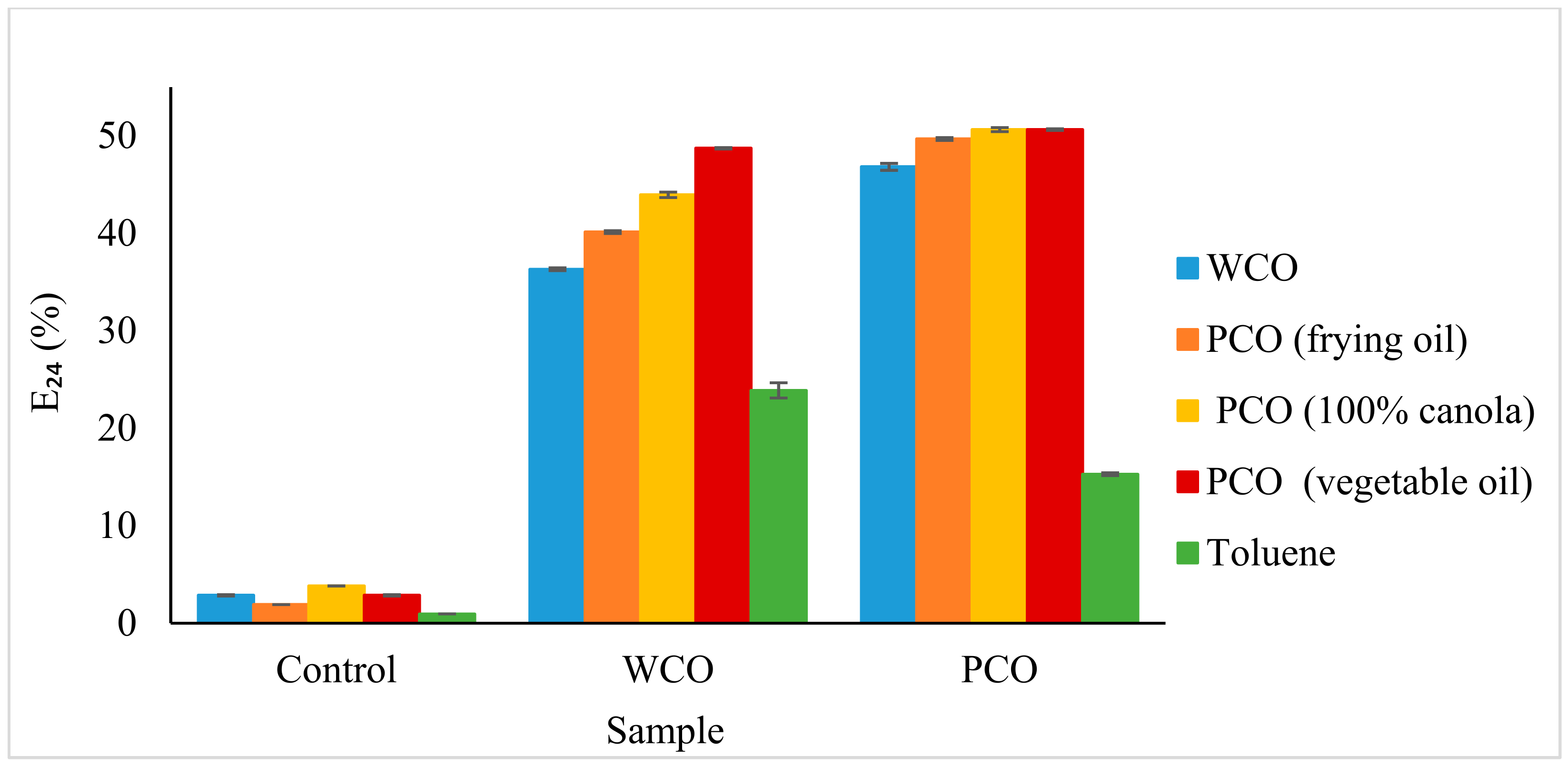
3.2.4. Emulsification Index (E24)
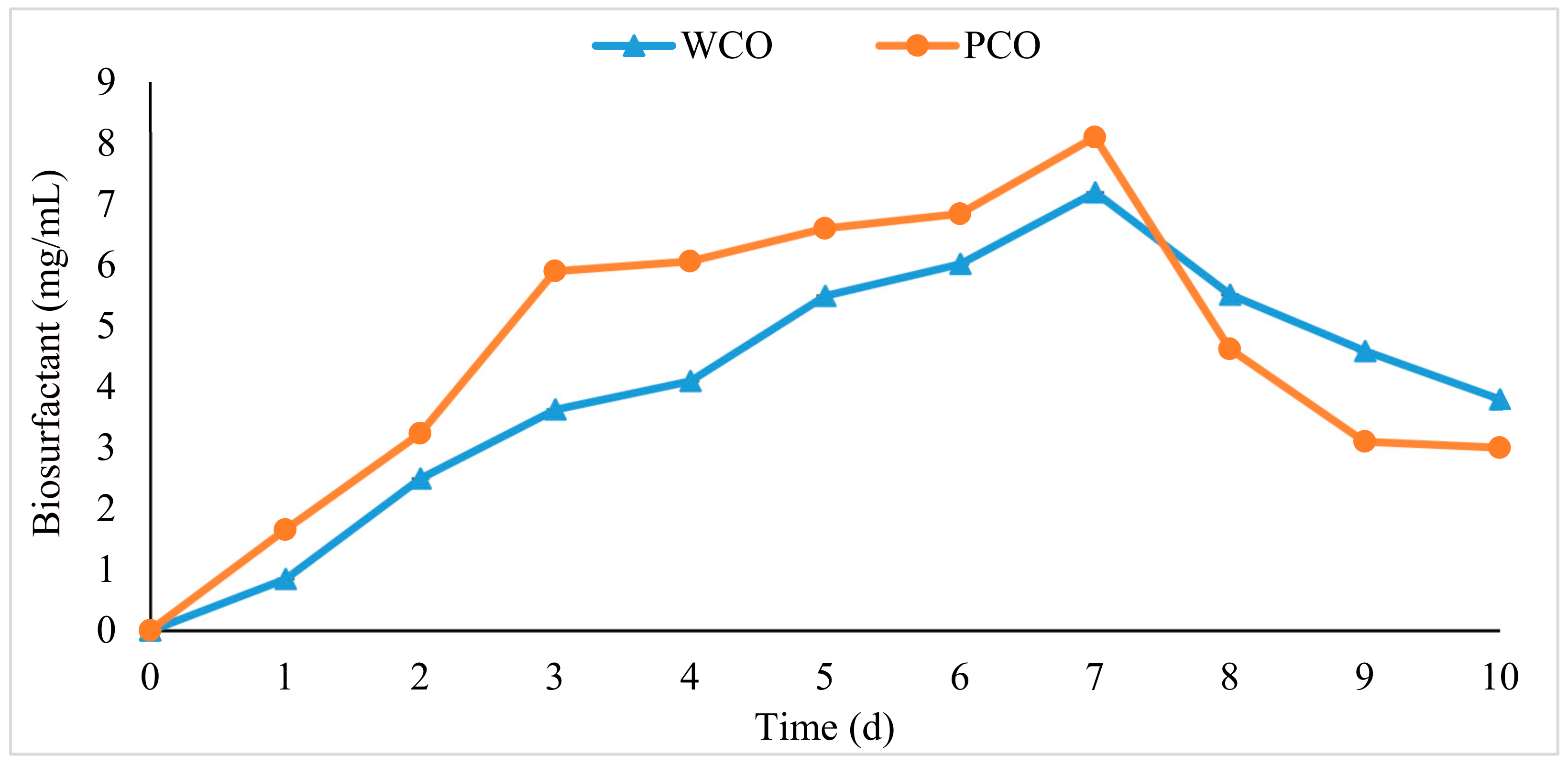
3.3. Measurement of Biosurfactant Production
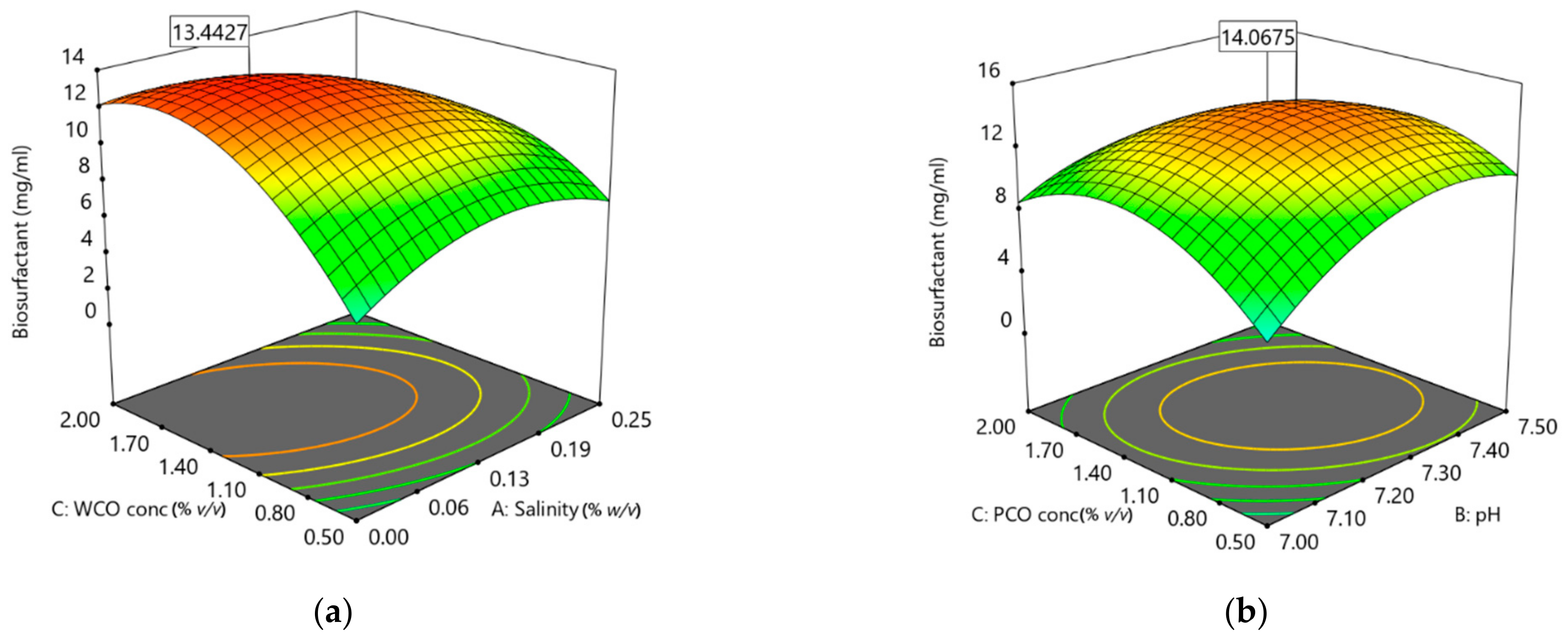
3.4. Optimisation of Biosurfactant Production
4. Discussion
4.1. Mathematical Modelling
4.2. Preliminary Test on Biosurfactant Production
4.3. Biosurfactant Production
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Australian Antarctic Division. Pollution and Waste, Department of the Environmental and Energy. 2012. Available online: https://www.antarctica.gov.au (accessed on 30 December 2018).
- Bharti, P.K.; Sharma, B.; Singh, R.K.; Tyagi, A.K. Waste generation and management in Antarctica. Procedia Environ. Sci. 2016, 35, 40–50. [Google Scholar] [CrossRef]
- Kariminia, S.; Ahmad, S.S.; Hashim, R.; Ismail, Z. Environmental consequences of Antarctic tourism from a global prospective. Procedia Soc. Behav. Sci. 2013, 105, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Cripps, G.C.; Shears, J. The fate in the marine environment of a minor diesel fuel spill from an Antarctic research station. Environ. Monit. Assess. 1996, 46, 221–232. [Google Scholar] [CrossRef]
- Aislabie, J.M.; Balks, M.R.; Fought, J.M.; Waterhouse, E.J. Hydrocarbon spills on Antarctic soils: Effects and management. Environ. Sci. Technol. 2004, 38, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Giudice, A.L.; Bruni, V.; De Domenico, M.; Michaud, L. Psychrophiles-Cold-Adapted Hydrocarbons-Degrading Microorganisms; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1898–1921. [Google Scholar] [CrossRef]
- Koshlaf, E.; Ball, A. Soil bioremediation approaches for petroleum hydrocarbon polluted environments. AIMS Microbiol. 2017, 3, 25–49. [Google Scholar] [CrossRef]
- Miri, S.; Naghdi, M.; Rouissi, T.; Brar, S.K.; Martel, R. Recent biotechnological advances in petroleum hydrocarbons degradation under cold climate conditions: A review. Crit. Rev. Environ. Sci. Technol. 2018, 49, 553–586. [Google Scholar] [CrossRef]
- Zahri, K.N.M.; Zulkharnain, A.; Fuentes, C.; Sabri, S.; Khalil, K.A.; Convey, P.; Ahmad, S.A. The use of response surface methodology as a statistical tool for the optimisation of the waste and pure canola oil biodegradation by Antarctic soil bacteria. Life 2021, 11, 456. [Google Scholar] [CrossRef]
- Sakthiselvan, P.; Meenambiga, S.S.; Madhumathi, R. Kinetic Studies on cell growth, cell growth. IntechOpen 2019, 13. [Google Scholar] [CrossRef] [Green Version]
- Pacwa-Plociniczak, M.; Plaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef]
- Kubicki, S.; Bollinger, A.; Katzke, N.; Jaeger, K.; Loeschcke, A.; Thies, S. Marine biosurfactants: Biosynthesis, structural diversity and biotechnology applications. Mar. Drugs 2019, 17, 408. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, S.; Zulkharnain, A.; Zahri, K.N.M.; Lee, G.L.Y.; Convey, P.; Gomez-Fuentes, C.; Sabri, S.; Khalil, K.; Alias, S.; Gonzalez-Rocha, G.; et al. Effect of heavy metals and other xenobiotics on biodegradation of waste canola oil by cold-adapted Rhodococcus sp. strain AQ5-07. Rev. Mex. Ing. Quím. 2020, 19, 1041–1052. [Google Scholar] [CrossRef] [Green Version]
- Zahri, K.N.M.; Zulkharnain, A.; Gomez-Fuentes, C.; Sabri, S.; Ahmad, S.A. Study of growth kinetics of Antarctic bacterial community for biodegradation of waste canola oil. Desalin. Water Treat. 2021, 213, 128–138. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef] [Green Version]
- Haldane, J.S.B. Enzymes; Longmans, Green & Co, MIT Press: Cambridge, UK, 1930. [Google Scholar]
- Edward, V.H. The influence of high substrate concentration on microbial kinetics. Biotechnol. Bioeng. 1970, 12, 679–712. [Google Scholar] [CrossRef]
- Aiba, S.; Shoda, M.; Nagatani, M. Kinetics of product inhibition in alcohol fermentation. Biotechnol. Bioeng. 1968, 10, 845–864. [Google Scholar] [CrossRef]
- Yano, T.; Koga, S. Dynamic behavior the chemostat subject to substrate inhibition. Biotechnol. Bioeng. 1969, 11, 139–153. [Google Scholar] [CrossRef]
- Shahaby, A.F.; Alharthi, A.A.; El Tarras, A.E. Bioremediation of petroleum oil by potential biosurfactant-producing bacteria using gravimetric assay. Int. J. Microbiol. Appl. Sci. 2015, 4, 390–403. [Google Scholar]
- Patowary, K.; Patowary, R.; Kalita, M.C.; Deka, S. Development of an efficient bacterial consortium for the potential remediation of hydrocarbons from contaminated sites. Front. Microbiol. 2016, 14, 1092. [Google Scholar] [CrossRef]
- Priyadharshini, A.D.; Latha, D. Biosurfactant production from Bacillus sp. and its application in the medical field. Int. Res. J. Pharm. Biosci. 2016, 3, 1–8. [Google Scholar]
- Nayarisseri, A.; Singh, P.; Singh, S.K. Screening, isolation and characterisation of biosurfactant producing Bacillus subtilis strain ANSKLAB03. J. Biomed. Inform. 2018, 14, 304–318. [Google Scholar] [CrossRef]
- Rani, M.; Weadge, J.T.; Jabaji, S. Isolation and characterisation of biosurfactant-producing bacteria from oil well batteries with antimicrobial activities against food-borne and plant pathogens. Front. Microbiol. 2020, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Chittepu, O.R. Isolation and characterisation of biosurfactant producing bacteria from groundnut oil cake dumping site for the control of foodborne pathogens. Grain Oil Sci. Technol. 2019, 2, 15–20. [Google Scholar] [CrossRef]
- Sohail, R.; Jamil, N. Isolation of biosurfactant producing bacteria from Potwar oil fields: Effect of non-fossil fuel based carbon sources. Green Process Synth. 2020, 9, 77–86. [Google Scholar] [CrossRef]
- Almeida, D.G.; Soares da Silva, R.C.F.; Luna, J.M.; Rufino, R.D.; Santos, V.A.; Sarubbo, L.A. Response surface methodology for optimising the production of biosurfactant by Candida tropicalis on industrial waste substrate. Front. Microbiol. 2017, 8, 157. [Google Scholar] [CrossRef] [Green Version]
- Anandaraj, B.; Thivakaran, P. Isolation and production of biosurfactant producing organism from oil spilled oil. J. Biosci. Technol. 2010, 1, 120–126. [Google Scholar]
- Maan, P.; Bharti, A.K.; Gautam, S.; Dutt, D. Screening of important factors for xylanase and cellulose production from the fungus C. cinerea RM-1 NFCCI-3086 through placket-Burman experimental design. Bioresources 2016, 11, 8268–8276. [Google Scholar] [CrossRef] [Green Version]
- Sadhukhan, B.; Mondal, N.K.; Chattoraj, S. Optimisation using central composite design (CCD) and the desirability function for sorption of methylene blue from aqueous solution onto Lemna major. Karbala Int. J. Modern Sci. 2016, 2, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Huang, P.T.; Zhang, K.Y.; Ding, F.R. Isolation of biosurfactant producers, optimisation and properties of biosurfactant produced by Acinetobacter sp. from petroleum-contaminated soil. J. Appl. Microbiol. 2012, 112, 660–671. [Google Scholar] [CrossRef]
- Kumar, S.; Mathur, A.; Signh, V.; Nandy, S.; Khare, S.K.; Negi, S. Bioremediation of waste cooking oil using a novel lipase produced by Penicillium chrysogenum SNP5 grown in solid medium containing waste grease. Biosour. Technol. 2012, 120, 300–304. [Google Scholar] [CrossRef]
- Malcolmson, L.; Vaisey-Genser, M. Canola Oil: Performance Properties of Canola Oil; Canola Council of Canada: Winnipeg, MB, Canada, 2001. [Google Scholar]
- Choe, E.; Min, D.B. Chemistry of deep-fat frying oils. J. Food Sci. 2007, 72, 77–86. [Google Scholar] [CrossRef]
- Przybylski, R.; Mag, T.; Eskin, N.A.M.; McDonald, B.E. Canola Oil: Physical and Chemical Properties; John Wiley and Sons Inc.: New York, NY, USA, 2005. [Google Scholar]
- Batista, A.C.F.; Viera, A.T.; Rodrigues, H.S.; Silva, T.A.; Assumcao, R.M.N.; Beluomini, M.A.; Rezende, H.P.; Hernandez-Terrones, M.G. Production and physiochemical characterisation of methylic and ethylic biodiesel from canola oil. Brazillian J. Biosyst, Eng. 2014, 8, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Chipasa, K.B.; Medrzycka, K. Behavior of lipids in biological wastewaster treatment processes. J. Ind. Microbiol. Biotechnol. 2006, 33, 635–645. [Google Scholar] [CrossRef]
- Sugimori, D.; Utsue, T. A study of the efficiency of edible oils degraded in alkaline conditions by Pseudomonas aeruginosa SS-219 and Acinetobacter sp. SS-192 bacteria isolated from Japanese soil. World J. Microbiol. Biotechnol. 2011, 28, 841–848. [Google Scholar] [CrossRef]
- Affandi, I.Z.; Suratman, N.H.; Abdullah, S.; Ahamd, W.A.; Zakaria, Z.A. Degradation of oil and grease from high-strength industrial effluents using locally isolated aerobic biosurfactant-producing bacteria. Int. Biodeter. Biodegr. 2014, 95, 33–40. [Google Scholar] [CrossRef]
- Ibrahim, S.; Zahri, K.N.M.; Convey, P.; Khalil, K.A.; Gomez-Fuentes, C.; Zulkharnain, A.; Alias, S.A.; Gonzalez-Rocha, G.; Ahmad, S.A. Optimisation of biodegradation conditions for waste canola oil by cold-adapted Rhodococcus sp. AQ5-07 from Antarctica. Electron. J. Biotechnol. 2020, 48, 1–12. [Google Scholar] [CrossRef]
- Aggary, S.E.; Solomon, B.O.; Audu, T.O.K. Substrate utilization and inhibition kinetics: Batch degradation of phenol by indigenous monoculture of Pseudomonas aeruginosa. Int. J. Biotechnol. Mol. Biol. Res. 2010, 1, 22–30. [Google Scholar]
- Aggary, S.E.; Solomon, B.O.; Layokun, S.K. Kinetics of batch microbial degradation of phenols by indigenous binary mixed culture of Pseudomonas aeruginosa and Pseudomonas fluorescence. African J. Biotechnol. 2008, 7, 2417–2423. [Google Scholar] [CrossRef]
- Mpongwana, N.; Ntwampe, S.K.O.; Omodanisi, E.I.; Chisi, B.S.; Razanamahandry, L.C.; Dlangamandla, C.; Mukansi, M.R. Bio-kinetics of simultaneous nitrification and aerobic denitrification (SNaD) by a cyanide-degrading bacterium under cyanide-laden conditions. Appl. Sci. 2020, 10, 4823. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Zhang, Y.; Zhang, J.; Li, J.; Wang, S.; Chen, G. Isolation and characterisation of Acinetobacter sp. JQ1004 and evaluation of its inhibitory kinetics by free ammonia. Desalin. Water Treat. 2019, 147, 316–325. [Google Scholar] [CrossRef]
- Tazdait, D.; Abdi, N.; Grib, H.; Lounici, H.; Pauss, A.; Mameri, N. Comparison of different models of substrate inhibition in aerobic batch biodegradation of malathion. Turkish J. Eng. Environ. Sci. 2013, 37, 221–230. [Google Scholar] [CrossRef]
- Morikawa, M.; Daido, H.; Takao, T.; Murata, S.; Shimonishi, Y.; Imanaka, T. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J. Bacteriol. 1993, 175, 6459–6466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adebajo, S.O.; Akintokun, A.K.; Balogun, S.A. Screening and characterisation of biosurfactant producing bacteria from soil samples in Ogun-State, Nigeria. Microbiol. Res. J. Int. 2017, 18, 1–12. [Google Scholar]
- Shalel, S.; Streichman, S.; Marmur, A. The use of hemolysis kinetics to evaluate erythrocyte-bound surfactant. Colloids Surf. B. 2003, 27, 215–222. [Google Scholar] [CrossRef]
- Shalel, S.; Streichman, S.; Marmur, A. The mechanism of hemolysis by surfactants: Effect of solution composition. J. Colloid Interface Sci. 2002, 252, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Manaargadoo-Catin, M.; Ali-Cherif, A.; Pougnas, J.; Perrin, C. Hemolysis by surfactants—A review. Adv. Colloid Interface Sci. 2016, 228, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shoeb, E.; Ahmed, A.; Akhter, J.; Badar, U.; Siddqui, K.; Ansari, F.A.; Waqar, M.; Imtiza, S.; Akhtar, N.; Shaikh, Q.A.; et al. Screening and characterisation of biosurfactant-producing bacteria isolated from the Arabian Sea coast of Karachi. Turkish J. Biol. 2015, 39, 210–216. [Google Scholar] [CrossRef]
- Jain, D.; Collins-Thompson, D.; Lee, H.; Trevors, J.T. A drop-collapsing test for screening surfactant-producing microorganisms. J. Microbiol. Methods. 1991, 13, 271–279. [Google Scholar] [CrossRef]
- Mulligan, C.; Cooper, D.; Neufeld, R. Selection of microbes producing biosurfactants in media without hydrocarbons. J. Fermentation Technol. 1984, 62, 311–314. [Google Scholar]
- Rosenberg, E.; DeLong, E.F.; Lory, S.; Stackebrandt, E.; Thompson, F. The Prokaryotes: Applied Bacteriology and Biotechnology; Springer: Berlin, Germany, 2013. [Google Scholar]
- Kaczorek, E.; Pacholak, A.; Zdarta, A.; Smulek, W. The impact of biosurfactants on microbial cell properties leading to hydrocarbon bioavailability increase. Colloids Interfaces 2018, 2, 35. [Google Scholar] [CrossRef] [Green Version]
- Thavasi, R.; Nambaru, V.R.M.S.; Jayalakshmi, S.; Balasubramanian, T.; Banat, I.M. Biosurfactant production by Pseudomonas aeruginosa from renewable resources. Indian J. Microbiol. 2011, 51, 30–36. [Google Scholar] [CrossRef] [Green Version]
- Pendse, A.; Aruna, K. Use of various screening methods for isolation of potential biosurfactant producing microorganism from oil-contaminated soil samples. J. Pharm. Res. 2018, 12, 599–605. [Google Scholar]
- Sumathi, R.; Yogananth, N. Isolation and identification of biosurfactant producing Pseudomonas aeruginosa from marine sediment samples and its antimicrobial properties. Int. J. Adv. Res. Biol. Sci. 2016, 3, 200–212. [Google Scholar]
- Akintokun, A.K.; Adebajo, S.O.; Akinremi, C.A. Potential biosurfactant-producing bacteria from pharmaceutical wastewater using simple screening methods in south-west Nigeria. App. Environ. Res. 2017, 39, 41–54. [Google Scholar] [CrossRef]
- Syahriansyah, U.K.M.; Hamzah, A. Determination of optimum conditions and stability study of biosurfactant produced by Bacillus subtilis UKMP-4M5. Malaysian J. Anal. Sci. 2016, 20, 986–1000. [Google Scholar]
- Mouafo, T.H.; Mbawala, A.; Ndjouenkeu, R. Effect of different carbon sources on biosurfactants production by three strains of Lactobacillus spp. Biomed. Res. Int. 2018, 2018, 5034783. [Google Scholar] [CrossRef] [Green Version]
- Ghasemi, A.; Moosavi-Nasab, M.; Setoodeh, P.; Mesbahi, G.; Yousefi, G. Biosurfactant production by lactic acid bacterium Pediococcus dextrinicus SHU1593 grown on different carbon source: Strain screening followed by product characterisation. Sci. Rep. 2019, 9, 5287. [Google Scholar] [CrossRef] [Green Version]
- Walter, V.; Syldatk, C.; Hausmann, R. Screening concepts for the isolation of biosurfactant producing microorganisms. Screening concepts for the isolation of biosurfactant producing microorganisms. In Madame Curie Bioscience Database; Landes Bioscience: Austin, TX, USA, 2010. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6189/ (accessed on 20 February 2021).
- Amodu, O.S.; Ntwampe, S.K.; Ojumu, T.V. Emulsification of hydrocarbons by biosurfactant: Exclusive use of agrowaste. Bioresources 2014, 9, 3508–3525. [Google Scholar] [CrossRef]
- Abdulrasheed, M.; Zakaria, N.N.; Roslee, A.F.A.; Shukor, M.Y.; Zulkharnain, A.; Napis, S.; Convey, P.; Alias, S.A.; Gonzalez-Rocha, G.; Ahmad, S.A. Biodegradation of diesel oil by cold-adapted bacterial strains of Arthrobacter spp. from Antarctica. Antarct. Sci. 2020, 32, 341–353. [Google Scholar] [CrossRef]
- Elazzazy, A.M.; Abdelmoneim, T.S.; Almaghrabi, O.A. Isolation and characterisation of biosurfactant production under extreme environmental conditions by alkali-halo-thermophilic bacteria from Saudi Arabia. Saudi J. Biol. Sci. 2015, 22, 466–475. [Google Scholar] [CrossRef] [Green Version]
- Khopade, A.; Biao, R.; Liu, X.; Mahadik, K.; Zhang, L.; Kokare, C. Production and stability studies of the biosurfactant isolated from marine Nocardiopsis sp. B4. Desalination 2012, 285, 198–204. [Google Scholar] [CrossRef]
- Rodrigues, L.; Banat, I.M.; Teixeira, J.; Oliveira, R. Biosurfactants: Potential applications in medicine. J. Antimicrob. Chemother. 2006, 57, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Arora, J.S. Chapter 20—Additional Topics on OPTIMUM Design; Academic Press; Elsevier Inc.: Iowa City, IA, USA, 2012. [Google Scholar]
- Souza, E.C.; de Azevedo, P.O.S.; Dominguez, J.M.; Converti, A.; Oliveira, R.P.S. Influence of temperature and pH on the production of biosurfactant, bacteriocin and lactic acid by Lactococcus lactis CECT-4434. J. Food 2017, 15, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Ikhwani, A.Z.N.; Nurlaila, H.S.; Ferdinand, F.D.K.; Fachria, R.; Hasan, A.E.Z.; Yani, M.; Setyawati, I. Preliminary study: Optimisation of pH and salinity for biosurfactant production from Pseudomonas aeruginosa in diesel fuel and crude oil medium. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Bogor, Indonesia, 4 November 2016; IOP Publishing: Bristol, UK, 2017; Volume 58, p. 012056. [Google Scholar] [CrossRef]
- Hamzah, A.; Sabturani, N.; Radiman, S. Screening and optimisation of biosurfactant production by the hydrocarbon-degrading bacteria. Sains Malays. 2013, 42, 615–623. [Google Scholar]
- Ahmad, Z.; Zhang, X.; Imran, M.; Zhong, H.; Andleeb, S.; Zulekha, R.; Liu, G.; Ahmad, I.; Coulon, F. Production, functional stability, and effect of rhamnolipid biosurfactant from Klebsilla sp. on phenanthrene degradation in various medium system. Ecotoxicol. Environ. Safety 2021, 207, 111514. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef] [Green Version]
- Dhasayan, A.; Selvin, J.; Kiran, S. Biosurfactant production from marine bacteria associated with sponge Callyspongia diffusa. 3 Biotech 2015, 5, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Hisham, N.H.M.B.; Ibrahim, M.F.; Ramli, N.; Abd-Aziz, S. Production of biosurfactant produced from used cooking oil by Bacillus sp., HIP3 for heavy metals removal. Molecules 2019, 24, 2617. [Google Scholar] [CrossRef] [Green Version]
- Roslee, A.F.A.; Zakaria, N.N.; Convey, P.; Zulkharnain, A.; Lee, G.L.Y.; Gomez-Fuentes, C.; Ahmad, S.A. Statistical optimisation of growth conditions and diesel degradation by the Antarctic bacterium, Rhodococcus sp. strain AQ5-07. Extremophiles 2020, 24, 277–291. [Google Scholar] [CrossRef]
- Phulpoto, I.A.; Yu, Z.; Hu, B.; Wang, Y.; Ndayisenga, F.; Li, J.; Liang, H.; Qazi, M.A. Production and characterisation of surfactin-like biosurfactant produced by novel strain Bacillus nealsonii S2MT and its potential for oil contaminated soil remediation. Microbial Cell Fact. 2020, 19, 145. [Google Scholar] [CrossRef]
- Kemmitt, S.J.; Wright, D.; Goulding, K.W.T.; Jones, D.L. pH regulation of carbon and nitrogen dynamics in two agricultural soils. Soil Biol. Biochem. 2006, 38, 898–911. [Google Scholar] [CrossRef]
- Jin, Q.; Kirk, M.F. pH as a primary control in environmental microbiology: 1. Thermodynamic perspective. Front. Environ. Sci. 2018, 6, 21. [Google Scholar] [CrossRef]
- Banat, I.M.; Satpute, S.K.; Cameotra, S.S.; Patil, R.; Nyayanit, N.V. Cost effective technologies and renewable substrates for biosurfactants’ production. Front. Microbiol. 2014, 5, 697. [Google Scholar] [CrossRef] [Green Version]
- Mubarak, M.Q.E.; Jufri, S.H.M.; Zahar, S.M.N.S.; Kalil, M.S.; Hamid, A.A.; Isa, M.H.M. Kinetics of surfactin production by Bacillus subtilis in a 5 L stirred-tan bioreactor. Sains Malays. 2017, 46, 1541–1548. [Google Scholar] [CrossRef]
- Heryani, H.; Putra, M.D. Kinetic study and modelling of biosurfactant production using Bacillus sp. Electron. J. Biotechnol. 2017, 27, 49–54. [Google Scholar] [CrossRef]
- Karlapudi, A.P.; Venkateswarulu, T.C.; Jahnavi, T.; Lohit, K.; Bharath, K.R.; Vijaya, R.D.; Vidya, P. Role of biosurfactants in bioremediation of oil pollution-a review. Petroleum 2018, 4, 241–249. [Google Scholar] [CrossRef]
- Bognolo, G. Biosurfactants as emulsifying agents for hydrocarbons. Colloids Surf. A Physicochem. Eng. Asp. 1999, 152, 41–52. [Google Scholar] [CrossRef]
- Bai, N.; Wang, S.; Abuduaini, R.; Zhang, M.; Zhu, X.; Zhao, Y. Rhamnolipid-aided biodegradation of carbendazim by Rhodococcus sp. D-1: Characteristics, products, and phytotoxicity. Sci. Total Environ. 2017, 590–591, 343–351. [Google Scholar] [CrossRef]



| Run | Standard Deviation | A | B | C | D |
|---|---|---|---|---|---|
| 1 | 3 | 0.00 | 7.50 | 15.00 | 0.50 |
| 2 | 6 | 0.25 | 7.50 | 15.00 | 0.50 |
| 3 | 5 | 0.25 | 7.50 | 10.00 | 2.00 |
| 4 | 2 | 0.25 | 7.50 | 10.00 | 2.00 |
| 5 | 10 | 0.25 | 7.00 | 10.00 | 0.50 |
| 6 | 4 | 0.25 | 7.00 | 15.00 | 2.00 |
| 7 | 8 | 0.00 | 7.00 | 15.00 | 2.00 |
| 8 | 11 | 0.00 | 7.50 | 10.00 | 0.50 |
| 9 | 7 | 0.00 | 7.50 | 15.00 | 2.00 |
| 10 | 9 | 0.00 | 7.00 | 10.00 | 2.00 |
| 11 | 12 | 0.00 | 7.00 | 10.00 | 0.50 |
| 12 | 1 | 0.25 | 7.00 | 15.00 | 0.50 |
| Substrate | A | B | C | Predicted (mg/mL) |
|---|---|---|---|---|
| WCO | 0.00 | 7.35 | 2.0 | 12.65 |
| PCO | 0.05 | 7.27 | 1.76 | 10.94 |
| Model | R2 | Adj R2 | DF | RMSE | AICc | SSE | AF | BF |
|---|---|---|---|---|---|---|---|---|
| WCO | ||||||||
| Haldane | 0.985 | 0.992 | 4 | 0.017 | −54.814 | 0.001 | 0.999 | 0.999 |
| Yano | 0.985 | 0.992 | 3 | 0.019 | −47.758 | 0.001 | 0.999 | 0.999 |
| Teissier–Edwards | 0.978 | 0.989 | 4 | 0.030 | −45.251 | 0.004 | 1.000 | 1.000 |
| Aiba | 0.977 | 0.988 | 4 | 0.020 | −51.869 | 0.002 | 0.999 | 0.999 |
| Monod | 0.910 | 0.954 | 5 | 0.035 | −46.654 | 0.006 | 1.000 | 1.000 |
| PCO | ||||||||
| Haldane | 0.983 | 0.991 | 4 | 0.016 | −54.821 | 0.001 | 0.999 | 0.999 |
| Yano | 0.984 | 0.992 | 3 | 0.019 | −47.821 | 0.001 | 0.999 | 0.999 |
| Teissier–Edwards | 0.981 | 0.991 | 4 | 0.017 | −53.802 | 0.001 | 0.999 | 0.999 |
| Aiba | 0.981 | 0.990 | 4 | 0.016 | −53.553 | 0.001 | 0.999 | 0.999 |
| Sample | Diameter (mm) | |
|---|---|---|
| Oil Spreading Test | Drop Collapse Test | |
| Control | 5.67 | 6.67 |
| WCO | 23.00 | 9.33 |
| PCO | 31.17 | 9.00 |
| Source | Sum of Squares | DF | F Value | Prob > F |
|---|---|---|---|---|
| Model | 82.83 | 4 | 17.06 | 0.0078 ** |
| A | 9.82 | 1 | 14.16 | 0.0197 * |
| B | 8.43 | 1 | 12.16 | 0.0252 * |
| C | 2.21 | 1 | 3.18 | 0.1491 |
| D | 28.13 | 1 | 40.56 | 0.0031 ** |
| Residual | 2.77 | 7 | ||
| Cor Total | 85.61 | 11 | ||
| R-squared | 0.968 | Pred R-squared | 0.745 | |
| Adj R-squared | 0.911 | Adeq Precision | 11.58 |
| Source | Sum of Squares | DF | F Value | Prob > F |
|---|---|---|---|---|
| Model | 54.83 | 4 | 32.72 | 0.0023 ** |
| A | 11.33 | 1 | 47.34 | 0.0023 ** |
| B | 1.96 | 1 | 8.17 | 0.0460 * |
| C | 0.54 | 1 | 2.25 | 0.2082 |
| D | 7.68 | 1 | 32.07 | 0.0048 ** |
| Residual | 0.96 | 7 | ||
| Cor Total | 55.79 | 11 | ||
| R-squared | 0.993 | Pred R-squared | 0.861 | |
| Adj R-squared | 0.953 | Adeq Precision | 18.02 |
| Run | Standard Deviation | A | B | C | WCO | PCO | ||
|---|---|---|---|---|---|---|---|---|
| Actual | Predicted | Actual | Predicted | |||||
| 1 | 17 | 0.13 | 7.25 | 1.25 | 13.80 | 12.95 | 6.73 | 5.29 |
| 2 | 8 | 0.25 | 7.50 | 2.00 | 3.02 | 4.06 | 13.50 | 13.80 |
| 3 | 13 | 0.13 | 7.25 | −0.01 | 1.31 | 1.37 | 12.46 | 13.98 |
| 4 | 18 | 0.13 | 7.25 | 1.25 | 10.36 | 12.99 | 1.71 | 4.55 |
| 5 | 20 | 0.13 | 7.25 | 1.25 | 11.73 | 12.93 | 3.50 | 4.17 |
| 6 | 2 | 0.25 | 7.00 | 0.50 | 2.43 | 4.04 | 7.27 | 6.81 |
| 7 | 3 | 0.00 | 7.50 | 0.50 | 3.15 | 4.04 | 4.40 | 4.98 |
| 8 | 12 | 0.13 | 7.67 | 1.25 | 5.44 | 6.19 | 5.40 | 6.31 |
| 9 | 11 | 0.13 | 6.83 | 1.25 | 3.27 | 2.24 | 13.50 | 13.90 |
| 10 | 4 | 0.25 | 7.50 | 0.50 | 5.27 | 3.69 | 14.30 | 14.00 |
| 11 | 14 | 0.13 | 7.25 | 2.51 | 6.88 | 6.53 | 3.95 | 4.73 |
| 12 | 15 | 0.13 | 7.25 | 1.25 | 13.87 | 12.90 | 4.23 | 4.15 |
| 13 | 19 | 0.13 | 7.25 | 1.25 | 13.96 | 13.00 | 14.76 | 14.99 |
| 14 | 7 | 0.00 | 7.50 | 2.00 | 12.97 | 11.57 | 3.83 | 2.92 |
| 15 | 10 | 0.34 | 7.25 | 1.25 | 5.64 | 5.51 | 4.32 | 4.31 |
| 16 | 6 | 0.25 | 7.00 | 2.00 | 3.35 | 2.67 | 3.77 | 1.99 |
| 17 | 9 | −0.09 | 7.25 | 1.25 | 9.23 | 9.06 | 4.69 | 5.43 |
| 18 | 1 | 0.00 | 7.00 | 0.50 | 1.6 | 0.77 | 10.69 | 8.78 |
| 19 | 16 | 0.13 | 7.25 | 1.25 | 13.91 | 13.50 | 6.20 | 4.71 |
| 20 | 5 | 0.00 | 7.00 | 2.00 | 4.75 | 6.54 | 15.63 | 14.99 |
| Source | Sum of Squares | DF | F Value | Prob > F |
|---|---|---|---|---|
| Model | 54.83 | 9 | 16.08 | <0.0001 *** |
| A | 11.33 | 1 | 5.69 | 0.0383 * |
| B | 1.96 | 1 | 6.94 | 0.0249 * |
| C | 0.54 | 1 | 12.05 | 0.0060 ** |
| AB | 7.68 | 1 | 2.46 | 0.1481 |
| AC | 12.47 | 1 | 9.54 | 0.0115 * |
| BC | 25.50 | 1 | 0.57 | 0.4670 |
| A2 | 9.45 | 1 | 21.54 | 0.0009 *** |
| B2 | 0.96 | 1 | 51.34 | <0.0001 *** |
| C2 | 0.55 | 1 | 54.45 | <0.0001 *** |
| Residual | 0.41 | 10 | ||
| Lack of Fit | 55.79 | 5 | 1.28 | 0.3962 |
| Pure Error | 54.83 | 5 | ||
| Cor Total | 11.33 | 19 | ||
| R-squared | 0.935 | Pred R-squared | 0.638 | |
| Adj R-squared | 0.877 | Adeq Precision | 10.521 |
| Source | Sum of Squares | DF | F Value | Prob > F |
|---|---|---|---|---|
| Model | 377.91 | 9 | 14.81 | 0.0001 *** |
| A | 1.91 | 1 | 0.67 | 0.4306 |
| B | 11.72 | 1 | 4.13 | 0.0694 |
| C | 0.06 | 1 | 0.02 | 0.8895 |
| AB | 3.30 | 1 | 1.17 | 0.3058 |
| AC | 4.26 | 1 | 1.50 | 0.2482 |
| BC | 14.84 | 1 | 5.24 | 0.0452 * |
| A2 | 152.00 | 1 | 53.61 | <0.0001 *** |
| B2 | 99.82 | 1 | 35.20 | 0.0001 *** |
| C2 | 156.55 | 1 | 55.21 | <0.0001 *** |
| Residual | 28.35 | 10 | ||
| Lack of Fit | 22.16 | 5 | 3.57 | 0.0942 |
| Pure Error | 6.20 | 5 | ||
| Cor Total | 406.26 | 19 | ||
| R-squared | 0.930 | Pred R-squared | 0.558 | |
| Adj R-squared | 0.867 | Adeq Precision | 10.071 |
| Substrate | Biosurfactant (mg/mL) | p Value | |
|---|---|---|---|
| Expected Value | Actual Value | ||
| WCO | 12.65 | 11.97 | 0.787 |
| PCO | 10.94 | 10.78 | 0.954 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahri, K.N.M.; Khalil, K.A.; Gomez-Fuentes, C.; Zulkharnain, A.; Sabri, S.; Convey, P.; Lim, S.; Ahmad, S.A. Mathematical Modelling of Canola Oil Biodegradation and Optimisation of Biosurfactant Production by an Antarctic Bacterial Consortium Using Response Surface Methodology. Foods 2021, 10, 2801. https://doi.org/10.3390/foods10112801
Zahri KNM, Khalil KA, Gomez-Fuentes C, Zulkharnain A, Sabri S, Convey P, Lim S, Ahmad SA. Mathematical Modelling of Canola Oil Biodegradation and Optimisation of Biosurfactant Production by an Antarctic Bacterial Consortium Using Response Surface Methodology. Foods. 2021; 10(11):2801. https://doi.org/10.3390/foods10112801
Chicago/Turabian StyleZahri, Khadijah Nabilah Mohd, Khalilah Abdul Khalil, Claudio Gomez-Fuentes, Azham Zulkharnain, Suriana Sabri, Peter Convey, Sooa Lim, and Siti Aqlima Ahmad. 2021. "Mathematical Modelling of Canola Oil Biodegradation and Optimisation of Biosurfactant Production by an Antarctic Bacterial Consortium Using Response Surface Methodology" Foods 10, no. 11: 2801. https://doi.org/10.3390/foods10112801
APA StyleZahri, K. N. M., Khalil, K. A., Gomez-Fuentes, C., Zulkharnain, A., Sabri, S., Convey, P., Lim, S., & Ahmad, S. A. (2021). Mathematical Modelling of Canola Oil Biodegradation and Optimisation of Biosurfactant Production by an Antarctic Bacterial Consortium Using Response Surface Methodology. Foods, 10(11), 2801. https://doi.org/10.3390/foods10112801










