Allelic Variation at Glutenin Loci (Glu-1, Glu-2 and Glu-3) in a Worldwide Durum Wheat Collection and Its Effect on Quality Attributes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
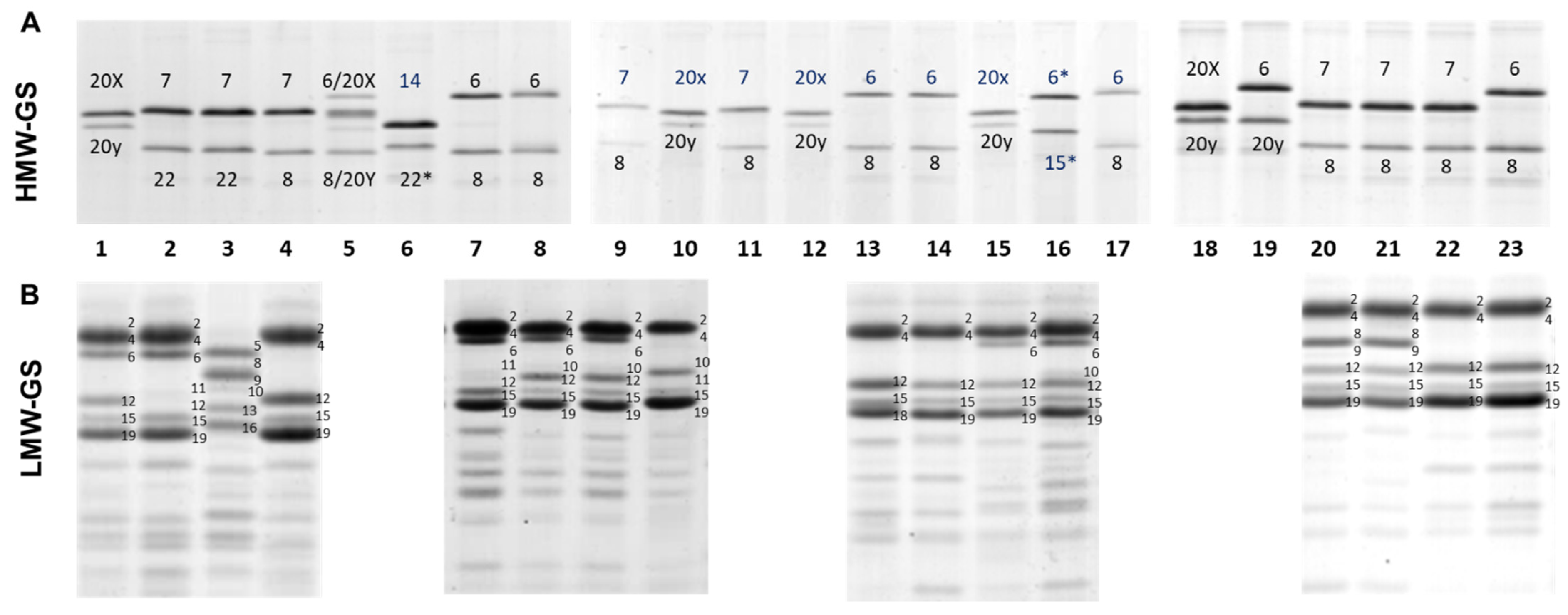
2.2. Glutenin Characterisation and Haplotype Analysis
2.3. KASP Markers
2.4. Field Experiments
2.5. Phenotypic Evaluations
2.6. Statistical Analyses
3. Results
3.1. High-Molecular-Weight Glutenin Subunits (HMW-GS)
3.2. Low-Molecular-Weight Glutenin Subunits (LMW-GS)
3.3. Effect of HMW-GS and LMW-GS on Gluten Strength and Protein Content
3.4. HMW-GS and LMW-GS Distribution along Different Periods and Origins
3.5. Quality Traits Variation over Different Breeding Periods
3.6. Glutenin Haplotype Frequency, Distribution and Effect
3.7. Haplotype Effect on Additional Quality Parameters in Argentinian Genotypes
4. Discussion
4.1. Glutenin Allelic and Haplotype Variation and Its Effect on GPC and the SDSS Test
4.2. Variability in Quality Traits between Breeding Periods and Its Relationship with Allelic Variation at Glutenin Loci
4.3. Haplotype Effect on Additional Quality Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matsuo, R.R. Durum wheat: Its unique pasta-making properties. In Wheat: Production, Properties and Quality; Bushuk, W., Rasper, V.F., Eds.; Springer: Boston, MA, USA, 1994; pp. 169–178. [Google Scholar] [CrossRef]
- Unión de Industriales Fideeros de la República Argentina (UIFRA). Pastas Secas, Estadísticas Sectoriales. Unión De Industriales Fideeros De La República Argentina. 2020, pp. 1–12. Available online: https://uifra.org.ar/wp-content/uploads/2020/04/dossier2020v2.pdf (accessed on 20 September 2021).
- Motzo, R.; Giunta, F.; Fois, S.; Deidda, M. Evoluzione Varietale e Qualità in Frumento Duro (Triticum turgidum subsp. durum): Dalle Vecchie Popolazioni Alle Attuali Cultivar; Dipartimento di Scienze Agronomiche e Genetica Vegetale Agraria, Facoltà di Agraria, Università degli Studi di Sassari: Sassari, Italy, 2001; p. 30. [Google Scholar]
- Matsuo, R.R.; Bradley, J.W.; Irvine, G.M. Effect of protein content on the cooking quality of spaghetti. Cereal Chem. 1972, 49, 707. [Google Scholar]
- Dexter, J.E.; Matsuo, R.R. Relationship between Durum Wheat Protein Properties and Pasta Dough Rheology and Spaghetti Cooking Quality. J. Agric. Food Chem. 1980, 8, 899–902. [Google Scholar] [CrossRef]
- Boggini, G.; Pogna, N.E. The Breadmaking Quality and Storage Protein Composition of Italian Durum Wheat. J. Cereal Sci. 1989, 9, 131–138. [Google Scholar] [CrossRef]
- Matsuo, R.R.; Irvine, G.M. Effect of gluten on the cooking quality of spaghetti. Cereal Chem. 1970, 47, 173–180. [Google Scholar]
- Impiglia, A.; Nachit, M.; Lafiandra, D.; Porceddu, E. Effect of gliadin and glutenin components on gluten strength in durum wheat. In Durum Wheat Quality in the Mediterranean Region; Di Fonzo, N., Kaan, F., Nachit, M., Eds.; Options Méditerranéennes: Série, A. Séminaires Méditerranéens n 22; CIHEAM: Zaragoza, Spain, 1995; pp. 167–172. [Google Scholar]
- Troccoli, A.; Borrelli, G.M.; De Vita, P.; Fares, C.; Di Fonzo, N. Durum Wheat Quality: A Multidisciplinary Concept. Mini Review. J. Cereal Sci. 2000, 32, 99–113. [Google Scholar] [CrossRef]
- Osborne, T.B. The Proteins of the Wheat Kernel; Carnegie Inst Washington Publication: Washington, WA, USA, 1907; Volume 84, pp. 1–119. [Google Scholar]
- Dexter, J.E.; Matsuo, R.R. Influence of protein content on some durum wheat quality parameters. Can. J. Plant Sci. 1977, 57, 717–727. [Google Scholar] [CrossRef]
- Payne, P.I.; Holt, L.M.; Lawrence, G.J.; Law, C.N. The genetics of gliadin and glutenin, the major storage proteins of the wheat endosperm. Plant Foods Hum. Nutr. 1982, 31, 229–241. [Google Scholar] [CrossRef]
- Porceddu, E. Durum wheat quality in the Mediterranean countries. In Durum Wheat Quality in the Mediterranean Region; Di Fonzo, N., Kaan, F., Nachit, M., Eds.; Options Méditerranéennes: Série, A. Séminaires Méditerranéens n 22; CIHEAM: Zaragoza, Spain, 1995; pp. 11–21. [Google Scholar]
- Shepherd, K.W. Chromosomal control of endosperm proteins in wheat and rye. In Proceedings of the 3rd International Wheat Genetic Symptoms, Australian Academy of Sciences, Canberra, Australia, 5–9 August 1968; pp. 86–96. [Google Scholar]
- Joppa, L.R.; Khan, K.; Williams, N.D. Chromosomal location of genes for gliadin polypeptides in durum wheat Triticum turgidum. Theor. Appl. Genet. 1983, 64, 289–293. [Google Scholar] [CrossRef]
- Pogna, N.; Lafiandra, D.; Feillet, P.; Autran, J.C. Evidence for direct casual effect of low molecular weight subunits of glutenins on gluten viscoelasticity in durum wheats. J. Cereal Sci. 1988, 7, 211–214. [Google Scholar] [CrossRef]
- Lawrence, G.J.; Shepherd, K.W. Variation in Glutenin Protein Subunits of Wheat. Aust. J. Bioi. Sci. 1980, 33, 221–234. [Google Scholar] [CrossRef]
- Payne, P.I. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Ann. Rev. Plant Physiol. 1987, 38, 141–153. [Google Scholar] [CrossRef]
- Jackson, E.A.; Holt, L.M.; Payne, P.I. Glu-B2, a storage protein locus controlling the D group of LMW glutenin subunits in bread wheat (Triticum aestivum). Genet. Res. 1985, 46, 11–17. [Google Scholar] [CrossRef]
- Shewry, P.; Tatham, A.S.; Forde, J.; Kreis, M.; Miflin, B.J. The Classification and Nomenclature of Wheat Gluten Proteins: A Reassessment. J. Cereal Sci. 1986, 4, 97–106. [Google Scholar] [CrossRef]
- Harberd, N.P.; Bartels, D.; Thompson, R.D. DNA restriction-fragment variation in the gene family encoding high molecular weight (HMW) glutenin subunits of wheat. Biochem. Genet. 1986, 24, 579–596. [Google Scholar] [CrossRef] [PubMed]
- Damidaux, R.; Grignac, P.; Feillet, P. Relation applicable en sélection entre l’électrophoregramme des gliadines et les proprietés viscoélastiques du gluten de Triticum durum Desf. C. R. Acad. Sci. Paris 1978, 287, 701–704. [Google Scholar]
- Payne, P.I.; Jackson, E.A.; Holt, L.M. The association between γ-gliadin 45 and gluten strength in durum wheat varieties: A direct causal effect or the result of genetic linkage? J. Cereal Sci. 1984, 2, 73–81. [Google Scholar] [CrossRef]
- Autran, J.-C.; Laignelet, B.; Morel, M.-H. Characterization and quantification of low molecular weight glutenins in durum wheats. Biochimie 1987, 69, 699–711. [Google Scholar] [CrossRef]
- Levy, A.A.; Galili, G.; Feldman, M. Polymorphism and genetic control of high molecular weight glutenin subunits in wild tetraploid wheat Triticum turgidum var. dicoccoides. Heredity 1988, 61, 63–72. [Google Scholar] [CrossRef]
- Xu, S.S.; Khan, K.; Klindworth, D.L.; Faris, J.D.; Nygard, G. Chromosomal location of genes for novel glutenin subunits and gliadins in wild emmer wheat (Triticum turgidum L. var. dicoccoides). Theor. Appl. Genet. 2004, 108, 1221–1228. [Google Scholar] [CrossRef]
- Branlard, G.; Autran, C.; Monneveux, P. High molecular weight glutenin subunit in durum wheat (T. durum). Theor. Appl. Genet. 1989, 78, 353–358. [Google Scholar] [CrossRef]
- Caballero, L.; Martín, M.A.; Alvarez, J.B. Allelic variation for the high- and low-molecular-weight glutenin subunits in wild diploid wheat (Triticum urartu) and its comparison with durum wheats. Aust. J. Agric. Res. 2008, 59, 906–910. [Google Scholar] [CrossRef]
- Alvarez, J.B.; Guzmán, C. Recovery of Wheat Heritage for Traditional Food: Genetic Variation for High Molecular Weight Glutenin Subunits in Neglected/Underutilized Wheat. Agronomy 2019, 9, 755. [Google Scholar] [CrossRef]
- Wallace, J.; Bainotti, C.; Nisi, M.M.; Formica, B.; Seghezzo, M.L.; Molfese, E.; Jensen, C.; Nisi, J.; Helguera, M. Variabilidad genética de proteínas de reserva en trigos candeales argentinos y su interacción con la calidad industrial. AgriScientia 2003, 20, 19–27. [Google Scholar] [CrossRef]
- Lerner, S.E.; Cogliatti, M.; Ponzio, N.R.; Seghezzo, M.L.; Molfese, E.R.; Rogers, W.J. Genetic variation for grain protein components and industrial quality of durum wheat cultivars sown in Argentina. J. Cereal Sci. 2004, 40, 161–166. [Google Scholar] [CrossRef]
- Roncallo, P.F.; Larsen, A.O.; Achilli, A.L.; Saint Pierre, C.; Gallo, C.A.; Dreisigacker, S.; Echenique, V. Linkage disequilibrium patterns, population structure and diversity analysis in a worldwide durum wheat collection including Argentinian genotypes. BMC Genom. 2021, 22, 233. [Google Scholar] [CrossRef] [PubMed]
- Peña, R.J.; Gonzalez-Santoyo, J.; Cervantes, F. Relationship between Glu-D1/Glu-B3 allelic combinations and bread-making quality-related parameters commonly used in wheat breeding. In The Gluten Proteins, Proceedings of the 8th Gluten Workshop, Viterbo, Italy, 8–10 September 2003; Lafiandra, D., Masci, S., D’Ovidio, R., Eds.; Royal Society of Chemistry: Cambridge, UK, 2003; pp. 156–157. [Google Scholar] [CrossRef]
- Maryami, Z.; Huertas-García, A.B.; Azimi, M.R.; Hernández-Espinosa, N.; Payne, T.; Cervantes, F.; Govindan, V.; Ibba, M.I.; Guzman, C. Variability for Glutenins, Gluten Quality, Iron, Zinc and Phytic Acid in a Set of One Hundred and Fifty-Eight Common Wheat Landraces from Iran. Agronomy 2020, 10, 1797. [Google Scholar] [CrossRef]
- Payne, P.I.; Lawrence, G.J. Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1, and Glu-D1 which code for high-molecular-weight subunits of glutenin in hexaploid wheat. Cereal Res. Commun. 1983, 11, 29–35. [Google Scholar]
- Nieto-Taladriz, M.T.; Ruiz, M.; Martínez, M.C.; Vazquez, J.F.; Carrillo, J.M. Variation and classification of B low-molecular-weight glutenin subunit alleles in durum wheat. Theor. Appl. Genet. 1997, 95, 1155–1160. [Google Scholar] [CrossRef]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.M.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc. Nat. Acad. Sci. USA 1984, 81, 8014–8019. [Google Scholar] [CrossRef]
- Patterson, H.D.; Williams, E.R. A new class of resolvable incomplete block designs. Biometrika 1976, 63, 83–92. [Google Scholar] [CrossRef]
- Irvine, G.; Bradley, J.; Martin, G. A farinograph technique for macaroni doughs. Cereal Chem. 1961, 38, 153–164. [Google Scholar]
- Molfese, E.; Astiz, V.; Seghezzo, M.L. Evaluación de la calidad del trigo candeal (Triticum turgidum L. subsp. durum) en los programas de mejoramiento de Argentina. RIA 2017, 43, 303–311. [Google Scholar]
- SAS Institute INC. Base SAS 9.2 Procedures Guide; SAS Institute INC: Cary, NC, USA, 2010. [Google Scholar]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, C.W. InfoStat Versión 2016; Grupo InfoStat (FCA, Universidad Nacional de Córdoba): Cordoba, Argentina, 2016; Available online: http://www.infostat.com.ar (accessed on 2 September 2021).
- Miller, R.G. Simultaneous Statistical Inference, 2nd ed.; Springer: New York, NY, USA, 1981. [Google Scholar] [CrossRef]
- Bautista, M.G.; Smith, D.W.; Steiner, R.L. A Cluster-Based approach to Means Separation. J. Agric. Biol. Environ. Stat. 1997, 2, 179–197. [Google Scholar] [CrossRef]
- Di Rienzo, J.A.; Guzman, A.W.; Casanoves, F. A multiple-comparisons method based on the distribution of the root node distance of a binary tree. JABES 2002, 7, 129–142. [Google Scholar] [CrossRef]
- Kosmolak, F.G.; Dexter, J.E.; Matsuo, R.R.; Leisle, D.; Marchylo, B.A. A relationship between durum wheat quality and gliadin electrophoregrams. Can. J. Plant Sci. 1980, 60, 427–432. [Google Scholar] [CrossRef]
- Carrillo, J.M.; Vazquez, J.F.; Orellana, J. Relationship between gluten strength and glutenin proteins in durum wheat cultivars. Plant Breed 1990, 104, 325–333. [Google Scholar] [CrossRef]
- Carrillo, J.M.; Martínez, M.C.; Brites, M.C.; Nieto-Taladriz, M.T.; Vázquez, J.F. Proteins and quality in durum wheat (Triticum turgidum L. var. durum). In Durum Wheat Quality in the Mediterranean Region: New Challenges; Royo, C., Nachit, M., Di Fonzo, N., Araus, J.L., Eds.; Options Méditerranéennes: Série, A. Séminaires Méditerranéens; n. 40; CIHEAM: Zaragoza, Spain, 2000; pp. 463–467. [Google Scholar]
- Nazco, R.; Peña, R.J.; Ammar, K.; Villegas, D.; Crossa, J.; Moragues, M.; Royo, C. Variability in glutenin subunit composition of Mediterranean durum wheat germplasm and its relationship with gluten strength. J. Agric. Sci. 2014, 152, 379–393. [Google Scholar] [CrossRef]
- De Santis, M.A.; Giuliani, M.M.; Giuzio, L.; De Vita, P.; Lovegrove, A.; Shewry, P.R.; Flagella, Z. Differences in gluten protein composition between old and modern durum wheat genotypes in relation to 20th century breeding in Italy. Eur. J. Agron. 2017, 87, 19–29. [Google Scholar] [CrossRef]
- Martínez, M.C.; Ruiz, M.; Carrillo, J.M. Effect of different prolamin alleles on durum wheat quality properties. J. Cereal Sci. 2005, 41, 123–131. [Google Scholar] [CrossRef]
- De Vita, P.; Nicosia, O.L.D.; Nigro, F.; Platani, C.; Riefolo, C.; Di Fonzo, N.; Cattivelli, L. Breeding progress in morpho-physiological, agronomical and qualitative traits of durum wheat cultivars released in Italy during the 20th century. Eur. J. Agron. 2007, 26, 39–53. [Google Scholar] [CrossRef]
- Vázquez, J.F.; Ruiz, M.; Nieto-Taladriz, M.T.; Albuquerque, M.M. Effects on gluten strength of low Mr glutenin subunits coded by alleles at Glu-A3 and Glu-B3 loci in durum wheat. J. Cereal Sci. 1996, 24, 125–130. [Google Scholar] [CrossRef]
- Chegdali, Y.; Ouabbou, H.; Essamadi, A.; Cervantes, F.; Ibba, M.I.; Guzmán, C. Assessment of the glutenin subunits diversity in a durum wheat (T. turgidum ssp. durum) collection from Morocco. Agronomy 2020, 10, 957. [Google Scholar] [CrossRef]
- Hernández-Espinosa, N.; Payne, T.; Huerta-Espino, J.; Cervantes, F.; Gonzalez-Santoyo, H.; Ammar, K.; Guzmán, C. Preliminary characterization for grain quality traits and high and low molecular weight glutenins subunits composition of durum wheat landraces from Iran and Mexico. J. Cereal Sci. 2019, 88, 47–56. [Google Scholar] [CrossRef]
- Chacón, E.A.; Vázquez, F.J.; Giraldo, P.; Carrillo, J.M.; Benavente, E.; Rodríguez-Quijano, M. Allelic Variation for Prolamins in Spanish Durum Wheat Landraces and Its Relationship with Quality Traits. Agronomy 2020, 10, 136. [Google Scholar] [CrossRef]
- Pasqualone, A.; Piarulli, L.; Mangini, G.; Gadaleta, A.; Blanco, A.; Simeone, R. Quality characteristics of parental lines of wheat mapping populations. Agric. Food Sci. 2015, 24, 118–127. [Google Scholar] [CrossRef]
- Ammar, K.; Kronstad, W.E.; Morris, C.F. Breadmaking Quality of Selected Durum Wheat Genotypes and Its Relationship with High Molecular Weight Glutenin Subunits Allelic Variation and Gluten Protein Polymeric Composition. Cereal Chem. 2000, 77, 230–236. [Google Scholar] [CrossRef]
- Bellil, I.; Hamdi, O.; Khelifi, D. Diversity of five glutenin loci within durum wheat (Triticum turgidum L. ssp. durum (Desf.) Husn.) germplasm grown in Algeria. Plant Breed 2014, 133, 179–183. [Google Scholar] [CrossRef]
- Carrillo, J.M. Variability for glutenin proteins in Spanis h durum wheat landraces. In Durum Wheat Quality in the Mediterranean Region; Di Fonzo, N., Kaan, F., Nachit, M., Eds.; Options Méditerranéennes: Série, A. Séminaires Méditerranéens n. 22; CIHEAM: Zaragoza, Spain, 1995; pp. 143–147. [Google Scholar]
- Subira, J.; Peña, R.J.; Álvaro, F.; Ammar, K.; Ramdani, A.; Royo, C. Breeding progress in the pasta-making quality of durum wheat cultivars released in Italy and Spain during the 20th Century. Crop. Pasture Sci. 2014, 65, 16–26. [Google Scholar] [CrossRef]
- Hamdi, W.; Bellil, I.; Branlard, G.; Douadi, K. Genetic Variation and Geographical Diversity for Seed Storage Proteins of Seventeen Durum Wheat Populations Collected in Algeria. Not. Bot. Horti Agrobot. Cluj-Napoca 2010, 38, 22–32. [Google Scholar] [CrossRef]
- Vallega, V. High-molecular-weight glutenin subunit composition ofItalian Triticum durum cultivars and spaghetti cooking quality. Cer. Res. Comm. 1986, 14, 251–257. [Google Scholar]
- Mcintosh, R.A.; Yamazaki, Y.; Dubcovsky, J.; Rogers, W.J.; Morris, C.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat. 2013. Available online: https://shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp (accessed on 31 August 2021).
- Ruiz, M.; Bernal, G.; Giraldo, P. An update of low molecular weight glutenin subunits in durum wheat relevant to breeding for quality. J. Cereal Sci. 2018, 83, 236–244. [Google Scholar] [CrossRef]
- Nazco, R.; Peña, R.J.; Ammar, K.; Villegas, D.; Crossa, J.; Royo, C. Durum wheat (Triticum durum Desf.) Mediterranean landraces as sources of variability for allelic combinations at Glu-1/Glu-3 loci affecting gluten strength and pasta cooking quality. Genet. Resour. Crop. Evol. 2014, 61, 1219–1236. [Google Scholar] [CrossRef]
- Martínez-Cruz, E.; Espitia-Rangel, E.; Villaseñor-Mir, H.E.; Molina-Galán, J.D.; Benítez-Riquelme, I.; Santacruz-Varela, A.; Peña-Bautista, R.J. Diversidad genética de gluteninas y gliadinas en trigos harineros (Triticum aestivum L.) mexicanos. Agrociencia 2010, 44, 187–195. [Google Scholar]
- Howell, T.; Hale, I.; Jankuloski, L.; Bonafede, M.; Gilbert, M.; Dubcovsky, J. Mapping a region within the 1RS.1BL translocation in common wheat affecting grain yield and canopy water status. Theor. Appl. Genet. 2014, 127, 2695–2709. [Google Scholar] [CrossRef] [PubMed]
- Rajaram, S.R.; Braun, H.J. Wheat yield potential. In International Symposium on Wheat Yield Potential: Challenges to International Wheat Breeding; Reynolds, M., Pietragalla, P.J., Braun, H.J., Eds.; CIMMYT: Mexico City, México, 2008; pp. 103–107. [Google Scholar]
- Fenn, D.; Lukow, O.M.; Bushuk, W.; Depauw, R.M. Milling and baking quality of 1BL/1RS translocation wheats. I. Effects of genotype and environment. Cereal Chem. 1994, 71, 189–195. [Google Scholar]
- Liu, L.; He, H.Z.; Yan, Y.; Xia, X.C.; Peña, R.J. Allelic variations at the Glu-1 and Glu-3 loci, presence of the 1B.1R translocation, and their effects on mixographic properties in Chinese bread wheats. Euphytica 2005, 142, 197–204. [Google Scholar] [CrossRef]
- Aguiriano, E.; Ruiz, M.; Fité, R.; Carrillo, J.M. Genetic variation for glutenin and gliadins associated with quality in durum wheat (Triticum turgidum L. ssp. turgidum) landraces from Spain. Span. J. Agric. Res. 2008, 6, 599–609. [Google Scholar] [CrossRef]
- Gupta, R.B.; Paul, J.G.; Cornish, B.B.; Palmer, G.A.; Bekes, F.; Rathjen, A.J. Allelic variation at glutenin subunit and gliadin loci, Glu-1, Glu-3 and Gli-1, of common wheats. I. Its additive and interaction effects on dough properties. J. Cereal Sci. 1994, 19, 9–17. [Google Scholar] [CrossRef]
- Ito, M.; Fushie, S.; Maruyama-Funatsuki, W.; Ikeda, T.M.; Nishio, Z.; Nagasawa, K.; Tabiki, T.; Yamauchi, H. Effect of allelic variation in three glutenin loci on dough properties and breadmaking qualities of winter wheat. Breed Sci. 2011, 61, 281–287. [Google Scholar] [CrossRef][Green Version]
- Krystkowiak, K.; Langner, M.; Adamski, T.; Salmanowicz, B.P.; Kaczmarek, Z.; Krajewski, P.; Surma, M. Interactions between Glu-1 and Glu-3 loci and associations of selected molecular markers with quality traits in winter wheat (Triticum aestivum L.) DH lines. J. Appl. Genet. 2017, 58, 37–48. [Google Scholar] [CrossRef]
- He, Z.H.; Liu, L.; Xia, X.C.; Liu, J.J.; Peña, R.J. Composition of HMW and LMW Glutenin Subunits and Their Effects on Dough Properties, Pan Bread, and Noodle Quality of Chinese Bread Wheats. Cereal Chem. 2005, 82, 345–350. [Google Scholar] [CrossRef]
- Nieto-Taladriz, M.T.; Perretant, M.R.; Rousset, M. Effect of gliadins and HMW and LMW subunits of glutenin on dough properties in the F6 recombinant inbred lines from a bread wheat cross. Theor. Appl. Genet. 1994, 88, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Laidig, F.; Piepho, H.P.; Rentel, D.; Drobek, T.; Meyer, U.; Huesken, A. Breeding progress, environmental variation and correlation of winter wheat yield and quality traits in german official variety trials and on-farm during 1983–2014. Theor. Appl. Genet. 2017, 130, 223–245. [Google Scholar] [CrossRef] [PubMed]
- Eichi, V.R.; Okamoto, M.; Garnett, T.; Eckermann, P.; Darrier, B.; Riboni, M.; Langridge, P. Strengths and Weaknesses of National Variety Trial Data for Multi-Environment Analysis: A Case Study on Grain Yield and Protein Content. Agronomy 2020, 10, 753. [Google Scholar] [CrossRef]
- Achilli, A.L.; Roncallo, P.F.; Echenique, V. Genetic gains in grain yield and agronomic traits of Argentinian durum wheat from 1934 to 2015. CERZOS-CONICET, Bahía Blanca, Buenos Aires. Argentina 2021. manuscript being submitted. [Google Scholar]
- Mefleh, M.; Conte, P.; Fadda, C.; Giunta, F.; Piga, A.; Hassoun, G.; Motzo, R. From ancient to old and modern durum wheat varieties: Interaction among cultivar traits, management, and technological quality. J. Sci. Food Agric. 2019, 99, 2059–2067. [Google Scholar] [CrossRef]
- Kibite, S.; Evans, L.E. Causes of negative correlations between grain yield and grain protein concentration in common wheat. Euphytica 1984, 33, 801–810. [Google Scholar] [CrossRef]
- Taghouti, M.; Nsarellah, N.; Rhrib, K.; Benbrahim, N.; Amallah, L.; Rochdi, A. Evolution from durum wheat landraces to recent improved varieties in Morocco in terms of productivity increase to the detriment of grain quality. Rev. Mar. Sci. Agron. Vét. 2017, 5, 351–358. [Google Scholar]
- Pronin, D.; Börner, A.; Weber, H.; Scherf, K.A. Wheat (Triticum aestivum L.) Breeding from 1891 to 2010 Contributed to Increasing Yield and Glutenin Contents but Decreasing Protein and Gliadin Contents. J. Agric. Food Chem. 2020, 68, 13247–13256. [Google Scholar] [CrossRef]
- Roncallo, P.F.; Larsen, A.O.; Achilli, A.L.; Ammar, K.; Huerta Espino, J.; Gonzalez, L.; Campos, P.; Dreisigacker, S.; Beker, M.; Echenique, V. Effect of major genes and loci identified by association mapping on yield, plant height and head date in durum wheat. In Proceedings of the RAFV Conference 202, XXXIII Argentinian Meeting of Plant Physiology, Santa Fe, Argentina, 13–17 September 2021; Available online: https://rafv2020.wixsite.com/santa-fe (accessed on 2 September 2021).
- Cubadda, R.; Carcea, M.; Pasqui, L.A. Suitability of the gluten index method for assessing gluten strength in durum wheat and semolina. Cereal Foods World 1992, 37, 866–869. [Google Scholar]
- Dexter, J.E.; Matsuo, R.R.; Kosmolak, F.G.; Leisle, D.; Marchylo, B.A. The suitability of the SDS-sedimentation test for assessing gluten strength in durum wheat. Can. J. Plant Sci. 1980, 60, 427. [Google Scholar] [CrossRef]
- Dick, J.W.; Quick, J.S. A modified screening test for the rapid estimation of gluten strength in early-generation durum wheat breeding lines. Cereal Chem. 1983, 60, 315. [Google Scholar]
- Clarke, F.R.; Clarke, J.M.; Ames, N.A.; Knox, R.E.; Ross, J.R. Gluten index compared with SDS sedimentation volume for early generation selection for gluten strength in durum wheat. Can. J. Plant Sci. 2010, 90, 1–11. [Google Scholar] [CrossRef]


| Argentina (85) | Chile (26) | CIMMYT (10) | France (21) | Italy (33) | USA (4) | WANA (17) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus/Banding Pattern | Allele | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % |
| HMW glutenin subunits | |||||||||||||||||
| Glu-A1 (2) | |||||||||||||||||
| 2 * | b | 1 | 0.5 | 1 | 1.2 | ||||||||||||
| null | c | 195 | 99.5 | 84 | 98.8 | 26 | 100.0 | 10 | 100.0 | 21 | 100.0 | 33 | 100.0 | 4 | 100.0 | 17 | 100.0 |
| Glu-B1 (10) | |||||||||||||||||
| 7 + 8 | b | 74 | 37.8 | 28 | 32.9 | 18 | 69.2 | 5 | 50.0 | 4 | 19.0 | 12 | 39.4 | 1 | 25.0 | 6 | 35.3 |
| 6 + 8 | d | 52 | 26.5 | 35 | 41.2 | 6 | 23.1 | 1 | 10.0 | 4 | 19.0 | 1 | 3.0 | 3 | 75.0 | 2 | 11.8 |
| 20x + 20y | e | 50 | 25.5 | 16 | 18.8 | 2 | 7.7 | 2 | 20.0 | 10 | 47.6 | 12 | 33.3 | 8 | 47.1 | ||
| 13 + 16 | f | 5 | 2.6 | 1 | 1.2 | 1 | 4.8 | 3 | 9.1 | ||||||||
| 7 + 15 | z | 7 | 3.6 | 3 | 3.5 | 2 | 20.0 | 2 | 9.5 | ||||||||
| 6 | an | 1 | 0.5 | 1 | 1.2 | ||||||||||||
| 7 + 22 | ch | 4 | 2.0 | 1 | 1.2 | 3 | 9.1 | ||||||||||
| 14 + 22 * | new (cr) | 1 | 0.5 | 1 | 3.0 | ||||||||||||
| 6 * + 15 * | new (cs) | 1 | 0.5 | 1 | 5.9 | ||||||||||||
| 6 + 20y | new (ct) | 1 | 0.5 | 1 | 3.0 | ||||||||||||
| LMW glutenin subunits | |||||||||||||||||
| Glu-A3 (9) | |||||||||||||||||
| 6 | a | 113 | 57.7 | 50 | 58.8 | 17 | 65.4 | 8 | 80.0 | 7 | 33.3 | 25 | 75.8 | 6 | 35.3 | ||
| 5 | b | 7 | 3.6 | 1 | 3.8 | 2 | 6.1 | 1 | 25.0 | 3 | 17.6 | ||||||
| 6 + 10 | c | 52 | 26.5 | 23 | 27.1 | 7 | 26.9 | 2 | 20.0 | 9 | 42.9 | 1 | 3.0 | 2 | 50.0 | 8 | 47.1 |
| 6 + 11 | d | 4 | 2.0 | 1 | 1.2 | 1 | 3.8 | 1 | 4.8 | 1 | 3.0 | ||||||
| 11 | e | 3 | 1.5 | 2 | 9.5 | 1 | 3.0 | ||||||||||
| null | h | 12 | 6.1 | 8 | 9.4 | 1 | 4.8 | 3 | 9.1 | ||||||||
| 6.1 + 10 | ax | 3 | 1.5 | 3 | 3.5 | ||||||||||||
| 5 + 10 | 1 | 0.5 | 1 | 25.0 | |||||||||||||
| 10 + 11 | 1 | 0.5 | 1 | 4.8 | |||||||||||||
| Glu-B3 (8) | |||||||||||||||||
| 2 + 4 + 15 + 19 | a | 176 | 89.8 | 80 | 94.1 | 24 | 92.3 | 10 | 100.0 | 19 | 90.5 | 29 | 87.9 | 2 | 50.0 | 12 | 70.6 |
| 8 + 9 + 13 + 16 | b | 8 | 4.1 | 1 | 3.8 | 2 | 6.1 | 2 | 50.0 | 3 | 17.6 | ||||||
| 2 + 4 + 15 + 16 | g | 4 | 2.0 | 1 | 1.2 | 1 | 3.0 | 2 | 11.8 | ||||||||
| 1 + 3 + 14 + 18 | h | 1 | 0.5 | 1 | 3.0 | ||||||||||||
| 2 + 4 + 15 + 18 | ax | 2 | 1.0 | 2 | 9.5 | ||||||||||||
| 2 + 4 + 16 | ag | 1 | 0.5 | 1 | 1.2 | ||||||||||||
| 2 + 4 + 8 + 9 + 15 + 19 | new (aw) | 3 | 1.5 | 3 | 3.5 | ||||||||||||
| Null | j | 1 | 0.5 | 1 | 3.8 | ||||||||||||
| Glu-B2 (3) | |||||||||||||||||
| 12 | a | 183 | 93.4 | 81 | 95.3 | 25 | 96.2 | 10 | 100.0 | 20 | 95.2 | 29 | 87.9 | 2 | 50.0 | 16 | 94.1 |
| null | b | 11 | 5.6 | 4 | 4.7 | 1 | 3.8 | 1 | 4.8 | 3 | 9.1 | 2 | 50.0 | ||||
| 12 * | c | 2 | 1.0 | 1 | 3.0 | 1 | 5.9 | ||||||||||
| LMW-GS Model | |||||||||||||||||
| 1 | 8 | 4.1 | 1 | 3.8 | 2 | 6.1 | 2 | 50.0 | 3 | 17.6 | |||||||
| 2 | 187 | 95.4 | 85 | 100.0 | 24 | 92.3 | 10 | 100.0 | 21 | 100.0 | 31 | 93.9 | 2 | 50.0 | 14 | 82.4 | |
| none | 1 | 0.5 | 1 | 3.8 | |||||||||||||
| 196 | 100 | 85 | 100.0 | 26 | 100.0 | 10 | 100.0 | 21 | 100.0 | 33 | 100.0 | 4 | 100.0 | 17 | 100.0 | ||
| Loci | ||||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype | Glu-A1 | Glu-B3 | Glu-B2 | Glu-B1 | Glu-A3 | N | Frequency (%) | Quality Traits Assessed 1 |
| Hap_1 | Ax2 * | 2 + 4 + 15 + 19 | 12 | 6 + 8 | 6 | 1 | 0.5 | yes |
| Hap_2 | null | 1 + 3 + 14 + 18 | 12 * | 14 + 22 * | null | 1 | 0.5 | yes |
| Hap_3 | null | 2 + 4 + 15 + 16 | 12 | 20x + 20y | 6 + 10 | 1 | 0.5 | yes |
| Hap_4 | null | 2 + 4 + 15 + 16 | 12 | 6 * + 15 * | 6 | 1 | 0.5 | yes |
| Hap_5 | null | 2 + 4 + 15 + 16 | 12 | 20x + 20y | 6 | 1 | 0.5 | yes |
| Hap_6 | null | 2 + 4 + 15 + 16 | null | 6 + 8 | 6 | 1 | 0.5 | no |
| Hap_7 | null | 2 + 4 + 15 + 18 | 12 | 20x + 20y | 11 | 1 | 0.5 | yes |
| Hap_8 | null | 2 + 4 + 15 + 18 | 12 | 7 + 15 | null | 1 | 0.5 | yes |
| Hap_9 | null | 2 + 4 + 15 + 19 | 12 | 13 + 16 | 6 | 2 | 1.0 | yes |
| Hap_10 | null | 2 + 4 + 15 + 19 | 12 | 13 + 16 | 6 + 10 | 1 | 0.5 | yes |
| Hap_11 | null | 2 + 4 + 15 + 19 | 12 | 20x + 20y | 6 | 25 | 12.8 | yes |
| Hap_12 | null | 2 + 4 + 15 + 19 | 12 | 20x + 20y | 6.1 + 10 | 3 | 1.5 | yes |
| Hap_13 | null | 2 + 4 + 15 + 19 | 12 | 20x + 20y | 6 + 10 | 11 | 5.6 | yes |
| Hap_14 | null | 2 + 4 + 15 + 19 | 12 | 20x + 20y | null | 3 | 1.5 | yes |
| Hap_15 | null | 2 + 4 + 15 + 19 | 12 | 6 + 20y | 6 | 1 | 0.5 | no |
| Hap_16 | null | 2 + 4 + 15 + 19 | 12 | 6 + 8 | 6 | 25 | 12.8 | yes |
| Hap_17 | null | 2 + 4 + 15 + 19 | 12 | 6 + 8 | 6 + 10 | 20 | 10.2 | yes |
| Hap_18 | null | 2 + 4 + 15 + 19 | 12 | 7 + 15 | 11 | 1 | 0.5 | yes |
| Hap_19 | null | 2 + 4 + 15 + 19 | 12 | 7 + 15 | 6 | 5 | 2.6 | yes |
| Hap_20 | null | 2 + 4 + 15 + 19 | 12 | 7 + 22 | 6 | 2 | 1.0 | yes |
| Hap_21 | null | 2 + 4 + 15 + 19 | 12 | 7 + 22 | null | 2 | 1.0 | yes |
| Hap_22 | null | 2 + 4 + 15 + 19 | 12 | 7 + 8 | 6 | 44 | 22.4 | yes |
| Hap_23 | null | 2 + 4 + 15 + 19 | 12 | 7 + 8 | 6 + 10 | 18 | 9.2 | yes |
| Hap_24 | null | 2 + 4 + 15 + 19 | 12 | 7 + 8 | 6 + 11 | 4 | 2.0 | yes |
| Hap_25 | null | 2 + 4 + 15 + 19 | 12 | 7 + 8 | null | 2 | 1.0 | no |
| Hap_26 | null | 2 + 4 + 15 + 19 | null | 13 + 16 | 11 | 1 | 0.5 | yes |
| Hap_27 | null | 2 + 4 + 15 + 19 | null | 20x + 20y | 6 | 1 | 0.5 | yes |
| Hap_28 | null | 2 + 4 + 15 + 19 | null | 20x + 20y | 6 + 10 | 1 | 0.5 | yes |
| Hap_29 | null | 2 + 4 + 15 + 19 | null | 20x + 20y | null | 1 | 0.5 | yes |
| Hap_30 | null | 2 + 4 + 15 + 19 | null | 6 + 8 | 6 | 1 | 0.5 | no |
| Hap_31 | null | 2 + 4 + 15 + 19 | null | 7 + 8 | 10 + 11 | 1 | 0.5 | yes |
| Hap_32 | null | 2 + 4 + 16 | 12 | 13 + 16 | 6 | 1 | 0.5 | no |
| Hap_33 | null | 2 + 4 + 8 + 9 + 15 + 19 | 12 | 6 | 6 | 1 | 0.5 | no |
| Hap_34 | null | 2 + 4 + 8 + 9 + 15 + 19 | 12 | 6 + 8 | null | 2 | 1.0 | no |
| Hap_35 | null | 8 + 9 + 13 + 16 | 12 | 20x + 20y | 5 | 1 | 0.5 | yes |
| Hap_36 | null | 8 + 9 + 13 + 16 | 12 | 7 + 8 | 5 | 3 | 1.5 | yes |
| Hap_37 | null | 8 + 9 + 13 + 16 | 12 * | 7 + 8 | 5 | 1 | 0.5 | yes |
| Hap_38 | null | 8 + 9 + 13 + 16 | null | 20x + 20y | 5 | 1 | 0.5 | yes |
| Hap_39 | null | 8 + 9 + 13 + 16 | null | 6 + 8 | 5 | 1 | 0.5 | yes |
| Hap_40 | null | 8 + 9 + 13 + 16 | null | 6 + 8 | 5 + 10 | 1 | 0.5 | yes |
| Hap_41 | null | null | null | 7 + 8 | 6 | 1 | 0.5 | yes |
| Total (N or %) | 2 | 8 | 3 | 10 | 9 | 196 | 100.0 | |
| 2011 | GPC 1 | SDSS Test | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus/Banding Pattern | Allele | N | % | CA 2011 | BW 2011 | PS 2011 | LSmean | CA 2011 | BW 2011 | PS 2011 | LSmean |
| HMW glutenin subunits | |||||||||||
| Glu-A1 (2) | |||||||||||
| 2 * | b | 1 | 0.8 | 13.92 a | 12.29 a | 11.5 a | 12.58 a | 84.50 a | 86.66 a | 68.51 a | 80.17 a |
| null | c | 131 | 99.2 | 14.62 a | 13.39 a | 11.95 a | 13.32 a | 62.11 a | 66.51 a | 54.16 a | 60.92 a |
| Glu-B1 (10) | |||||||||||
| 7 + 8 | b | 42 | 31.8 | 14.49 a | 13.28 a | 11.93 a | 13.23 a | 67.6 b | 71.18 b | 61.94 b | 66.34 b |
| 6 + 8 | d | 33 | 25.0 | 14.81 a | 13.32 ab | 11.85 a | 13.32 a | 71.97 b | 77.19 b | 62.58 b | 70.63 b |
| 20x + 20y | e | 43 | 32.6 | 14.59 a | 13.43 ab | 11.96 a | 13.33 a | 48.69 ab | 52.66 ab | 38.87 ab | 46.71 ab |
| 13 + 16 | f | 4 | 3.0 | 14.48 a | 13.45 ab | 11.78 a | 13.23 a | 61.0 b | 74.37 b | 54.24 ab | 63.34 b |
| 7 + 15 | z | 4 | 3.0 | 14.57 a | 13.49 ab | 12.15 a | 13.41 a | 73.13 b | 77.85 b | 69.14 b | 73.38 b |
| 6 | an | na | na | na | na | na | na | na | na | na | na |
| 7 + 22 | ch | 4 | 3.0 | 14.54 a | 13.84 ab | 12.35 a | 13.59 a | 69.0 b | 71.62 b | 60.43 b | 66.92 b |
| 14 + 22 * | new (cr) | 1 | 0.8 | 14.99 a | 14.78 b | 13.18 a | 14.3 a | 24.5 a | 19.61 a | 20.6 a | 21.83 a |
| 6 * + 15 * | new (cs) | 1 | 0.8 | 14.99 a | 14.17 ab | 12.47 a | 13.9 a | 80.5 b | 88.57 b | 70.82 b | 79.83 b |
| 6 + 20y | new (ct) | na | na | na | na | na | na | na | na | na | na |
| LMW glutenin subunits | |||||||||||
| Glu-A3 (9) | |||||||||||
| 6 | a | 74 | 56.1 | 14.65 a | 13.41 ab | 11.99 a | 13.35 a | 64.72 b | 68.53 b | 56.55 ab | 63.28 b |
| 5 | b | 6 | 4.5 | 14.84 a | 13.85 ab | 12.35 a | 13.67 a | 22.0 a | 22.36 a | 18.25 a | 20.94 a |
| 6 + 10 | c | 37 | 28.0 | 14.46 a | 13.21 ab | 11.8 a | 13.15 a | 64.31 b | 70.92 b | 56.27 ab | 63.83 b |
| 6 + 11 | d | 1 | 0.8 | 15.09 a | 13.25 ab | 11.94 a | 13.4 a | 56.5 ab | 75.98 b | 52.84 ab | 61.67 b |
| 11 | e | 3 | 2.3 | 14.6 a | 13.23 ab | 11.88 a | 13.24 a | 69.8 b | 71.37 b | 52.37 ab | 64.61 b |
| null | h | 7 | 5.3 | 14.94 a | 14.14 b | 12.4 a | 13.84 a | 57.0 ab | 60.11 ab | 48.49 ab | 55.0 ab |
| 6.1 + 10 | ax | 3 | 2.3 | 14.09 a | 12.64 a | 11.0 a | 12.57 a | 59.2 ab | 57.3 ab | 54.02 ab | 56.83 ab |
| 5 + 10 | na | na | na | na | na | na | na | na | na | na | |
| 10 + 11 | 1 | 0.8 | 14.34 a | 13.01 ab | 11.9 a | 13.12 a | 78.0 b | 86.87 b | 75.79 b | 80.17 b | |
| Glu-B3 (8) | |||||||||||
| 2 + 4 + 15 + 19 | a | 120 | 90.9 | 14.59 a | 13.34 a | 11.9 a | 13.28 a | 64.9 c | 69.6 b | 56.79 a | 63.78 b |
| 8 + 9 + 13 + 16 | b | 6 | 4.5 | 14.84 a | 13.85 ab | 12.35 a | 13.67 a | 22.0 a | 22.36 a | 18.25 a | 20.94 a |
| 2 + 4 + 15 + 16 | g | 3 | 2.3 | 14.41 a | 13.69 ab | 12.47 a | 13.53 a | 51.8 abc | 52.75 ab | 45.42 a | 49.78 ab |
| 1 + 3 + 14 + 18 | h | 1 | 0.8 | 14.99 a | 14.78 b | 13.18 a | 14.3 a | 24.5 ab | 15.5 a | 20.6 a | 21.83 a |
| 2 + 4 + 15 + 18 | ax | 2 | 1.5 | 15.34 a | 13.54 a | 12.35 a | 13.75 a | 58.3 bc | 67.07 b | 41.03 a | 55.25 ab |
| 2 + 4 + 16 | ag | na | na | na | na | na | na | na | na | na | na |
| 2 + 4 + 8 + 9 + 15 + 19 | new (aw) | na | na | na | na | na | na | na | na | na | na |
| Null | j | na | na | na | na | na | na | na | na | na | na |
| Glu-B2 (3) | |||||||||||
| 12 | a | 125 | 94.7 | 14.60 a | 13.37 a | 11.92 a | 13.3 a | 63.76 b | 68.35 b | 55.48 b | 62.52 b |
| null | b | 5 | 3.8 | 14.86 a | 13.51 a | 12.23 a | 13.55 a | 42.8 ab | 44.22 ab | 39.5 ab | 42.23 ab |
| 12 * | c | 2 | 1.5 | 14.47 a | 14.08 a | 12.84 a | 13.78 a | 18.75 a | 16.93 a | 15.59 a | 17.17 b |
| LMW-GS Model | |||||||||||
| 1 | 6 | 4.5 | 14.6 a | 13.85 b | 12.35 a | 13.67 a | 22.0 a | 22.4 a | 18.3 a | 20.9 a | |
| 2 | 126 | 95.5 | 14.84 a | 13.36 a | 11.93 a | 13.3 a | 64.2 b | 68.8 b | 56.0 b | 60.98 b | |
| none | na | na | na | na | na | na | na | na | na | na | |
| 2014/17 | GPC 1 | SDSS Test | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus/Banding Pattern | Allele | N | % | CA 2014 | BW 2014 | PS 2014 | BW 2017 | LSmean | CA 2014 | BW 2014 | PS 2014 | LSmean |
| HMW glutenin subunits | ||||||||||||
| Glu-A1 (2) | ||||||||||||
| 2 * | b | 1 | 0.6 | 13.2 a | 10.61 a | 11.54 a | 10.51 a | 11.64 a | 72.08 a | 68.99 a | 59.08 a | 66.84 a |
| null | c | 161 | 94.7 | 12.98 a | 12.2 a | 12.37 a | 11.14 a | 12.16 a | 58.81 a | 66.38 a | 66.28 a | 63.83 a |
| Glu-B1 (10) | ||||||||||||
| 7 + 8 | b | 65 | 38.2 | 12.49 ab | 12.31 a | 12.31 a | 10.93 a | 12.0 a | 61.99 b | 73.03 b | 71.16 b | 68.69 b |
| 6 + 8 | d | 44 | 25.9 | 13.68 ab | 11.94 a | 12.34 a | 11.23 ab | 12.29 a | 64.82 b | 66.39 ab | 67.48 b | 66.31 b |
| 20x + 20y | e | 47 | 27.6 | 13.08 ab | 12.26 a | 12.41 a | 11.3 ab | 12.27 a | 48.12 ab | 55.19 ab | 56.46 ab | 53.27 ab |
| 13 + 16 | f | 4 | 2.4 | 11.73 ab | 11.6 a | 11.98 a | 11.23 ab | 11.59 a | 59.34 ab | 77.87 b | 78.02 b | 71.61 b |
| 7 + 15 | z | 4 | 2.4 | 12.32 ab | 11.81 a | 12.42 a | 10.94 a | 11.9 a | 61.59 b | 69.14 b | 78.82 b | 69.63 b |
| 6 | an | na | na | |||||||||
| 7 + 22 | ch | 4 | 2.4 | 14.04 ab | 12.58 a | 12.33 a | 11.26 ab | 12.48 ab | 74.9 b | 80.61 b | 72.01 b | 76.15 b |
| 14 + 22 * | new (cr) | 1 | 0.6 | 15.38 b | 12.5 a | 14.82 b | 12.7 b | 14.02 b | 18.41 a | 21.97 a | 21.28 a | 20.31 a |
| 6 * + 15 * | new (cs) | 1 | 0.6 | 11.43 a | 13.31 a | 13.67 b | 11.82 ab | 12.35 a | 68.67 b | 87.02 b | 72.0 b | 75.48 b |
| 6 + 20y | new (ct) | na | na | |||||||||
| LMW glutenin subunits | ||||||||||||
| Glu-A3 (9) | ||||||||||||
| 6 | a | 98 | 57.6 | 12.98 a | 12.23 a | 12.39 ab | 11.11 ab | 12.07 a | 61.92 abc | 69.18 bc | 70.11 b | 67.10 c |
| 5 | b | 6 | 3.5 | 13.06 a | 13.12 a | 13.05 ab | 11.69 ab | 12.77 a | 18.72 a | 19.9 a | 23.6 a | 20.66 a |
| 6 + 10 | c | 48 | 28.2 | 12.89 a | 12.08 a | 12.23 ab | 11.08 ab | 12.07 a | 58.11 abc | 68.68 bc | 64.69 ab | 63.81 bc |
| 6 + 11 | d | 2 | 1.2 | 13.80 a | 11.97 a | 12.44 ab | 10.73 ab | 12.24 a | 67.58 bc | 73.09 c | 60.76 ab | 67.15 c |
| 11 | e | 3 | 1.8 | 11.92 a | 11.77 a | 12.03 ab | 11.42 ab | 11.78 a | 61.25 abc | 67.0 abc | 72.51 b | 66.55 c |
| null | h | 8 | 4.7 | 13.35 a | 12.15 a | 12.65 ab | 11.44 ab | 12.43 a | 57.36 abc | 64.48 abc | 64.27 ab | 62.03 bc |
| 6.1 + 10 | ax | 3 | 1.8 | 13.86 a | 11.29 a | 11.15 a | 10.8 ab | 11.72 a | 51.01 abc | 42.33 abc | 58.57 ab | 50.7 abc |
| 5 + 10 | 1 | 0.6 | 14.24 a | 11.99 a | 11.96 ab | 12.3 ab | 12.64 a | 31.45 ab | 21.96 ab | 23.78 a | 25.97 ab | |
| 10 + 11 | 1 | 0.6 | 11.79 a | 12.33 a | 13.29 b | 10.42 a | 11.69 a | 78.4 c | 80.56 c | 90.74 b | 83.22 c | |
| Glu-B3 (8) | ||||||||||||
| 2 + 4 + 15 + 19 | a | 156 | 117.3 | 12.99 a | 12.15 a | 12.32 a | 11.10 ab | 12.13 a | 61.17 ab | 68.92 bc | 68.42 b | 66.18 b |
| 8 + 9 + 13 + 16 | b | 7 | 5.3 | 13.23 a | 12.96 a | 12.98 ab | 11.78 ab | 12.75 ab | 20.54 a | 20.20 a | 23.63 a | 21.42 a |
| 2 + 4 + 15 + 16 | g | 3 | 2.3 | 11.96 a | 12.32 a | 12.26 a | 11.32 ab | 11.95 a | 49 ab | 67.52 abc | 58.82 ab | 58.59 ab |
| 1 + 3 + 14 + 18 | h | 1 | 0.8 | 15.38 b | 12.5 a | 14.82 b | 12.70 b | 14.02 b | 18.41 a | 21.97 ab | 21.88 a | 20.31 a |
| 2 + 4 + 15 + 18 | ax | 2 | 1.5 | 10.92 a | 12.54 a | 12.81 ab | 10.88 ab | 11.96 a | 45.83 ab | 44.0 abc | 70.91 b | 53.48 ab |
| 2 + 4 + 16 | ag | na | na | |||||||||
| 2 + 4 + 8 + 9 + 15 + 19 | new (aw) | na | na | |||||||||
| Null | j | 1 | 0.8 | 14.61 ab | 10.97 a | 12.11 a | 10.60 a | 12.06 a | 66.5 b | 82.04 c | 81.69 b | 76.94 b |
| Glu-B2 (3) | ||||||||||||
| 12 | a | 159 | 93.5 | 12.96 a | 12.19 a | 12.33 a | 11.11 a | 12.14 a | 60.06 b | 68.02 b | 67.54 b | 65.22 b |
| null | b | 9 | 5.3 | 13.16 a | 11.96 a | 12.44 a | 11.36 a | 12.25 a | 48.00 b | 48.89 b | 54.30 b | 50.31 b |
| 12 * | c | 2 | 1.2 | 14.08 a | 12.88 a | 14.48 b | 12.07 a | 13.43 b | 14.65 a | 16.43 a | 16.66 a | 15.52 a |
| LMW-GS Model | ||||||||||||
| 1 | 7 | 4.1 | 13.23 a | 12.96 b | 12.98 a | 11.78 a | 12.74 a | 20.54 a | 20.20 a | 23.63 a | 21.42 a | |
| 2 | 162 | 95.3 | 12.96 a | 12.16 ab | 12.34 a | 11.11 a | 12.14 a | 60.5 b | 68.3 b | 67.98 b | 65.6 b | |
| none | 1 | 0.588 | 14.61 a | 10.88 a | 12.11 a | 10.60 a | 12.06 a | 66.5 b | 82.04 b | 81.68 b | 76.94 b | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roncallo, P.F.; Guzmán, C.; Larsen, A.O.; Achilli, A.L.; Dreisigacker, S.; Molfese, E.; Astiz, V.; Echenique, V. Allelic Variation at Glutenin Loci (Glu-1, Glu-2 and Glu-3) in a Worldwide Durum Wheat Collection and Its Effect on Quality Attributes. Foods 2021, 10, 2845. https://doi.org/10.3390/foods10112845
Roncallo PF, Guzmán C, Larsen AO, Achilli AL, Dreisigacker S, Molfese E, Astiz V, Echenique V. Allelic Variation at Glutenin Loci (Glu-1, Glu-2 and Glu-3) in a Worldwide Durum Wheat Collection and Its Effect on Quality Attributes. Foods. 2021; 10(11):2845. https://doi.org/10.3390/foods10112845
Chicago/Turabian StyleRoncallo, Pablo F., Carlos Guzmán, Adelina O. Larsen, Ana L. Achilli, Susanne Dreisigacker, Elena Molfese, Valentina Astiz, and Viviana Echenique. 2021. "Allelic Variation at Glutenin Loci (Glu-1, Glu-2 and Glu-3) in a Worldwide Durum Wheat Collection and Its Effect on Quality Attributes" Foods 10, no. 11: 2845. https://doi.org/10.3390/foods10112845
APA StyleRoncallo, P. F., Guzmán, C., Larsen, A. O., Achilli, A. L., Dreisigacker, S., Molfese, E., Astiz, V., & Echenique, V. (2021). Allelic Variation at Glutenin Loci (Glu-1, Glu-2 and Glu-3) in a Worldwide Durum Wheat Collection and Its Effect on Quality Attributes. Foods, 10(11), 2845. https://doi.org/10.3390/foods10112845







