Abstract
Trimethylamine oxide (TMAO) originates from trimethylamine (TMA), which is oxidized in the liver by hepatic flavin-containing monooxygenases (FMO3). TMA is produced by its dietary precursors such as choline, carnitine, and phosphatidylcholine by gut microbiota. TMAO attracts attention, identified as a novel and independent risk factor for promoting obesity, atherosclerosis and cardiovascular disease (CVD), chronic kidney disease (CKD), insulin tolerance, and colon cancer. Probiotics have been considered as live microorganisms, providing benefits to their host when they are given in sufficient quantities and administered continuously. The objective of this study is to suggest a method to select potential probiotic strains to reduce the serum concentration of TMAO in mice fed with choline. In this work, we chose three lactobacilli with strong adherence capability, and fed multistrain formula (MF) to the mice challenged with choline. On days 7, 14, and day 28, it was found that the MF-containing L. amylovorus LAM1345, Lpb. plantarum LP1145, and Lim. fermentum LF33 showed a significant reduction in serum TMAO and TMA levels. For the single strains, LP1145 reduced TMAO on days 14 and 28, and strain LAM1345 reduced TMAO significantly on days 7 and day 14. For strain LF1143 from strain LF33, it showed no significant effect on TMAO and TMA. Thus, MF showed the best effect, which may be due to the additive and synergetic effect and the contribution of strain LP1145 and LAM1345. Finally, for the LAM1345 and LP1145 strains, we used molecular identification and typing methods to assure that these two strains are unique strains. The methods used for LAM 1345 were leader peptidase A (lepA) gene analysis and phylogenetic analysis, while for strain LP 1145and other strains of Lpb. plantarum subsp. plantarum sequences were compared using the whole-genome multilocus sequence typing (wgMLST) method.
Keywords:
TMAO; cardiovascular disease; choline-fed mice; lactobacilli; Lpb. plantarum; L. amylovorus; WgMLST 1. Introduction
Cardiovascular disease (CVD) is a prominent cause of death and mortality in the world, particularly in affluent countries [1,2]. The human gut microbiota contains over 1000 different gut microbiomes, which play significant roles in metabolism, minerals and vitamins bioactivation, immunological function, and digestion [3]. Food components such as choline, phosphatidylcholine, and L-carnitine can be metabolized by some gut bacteria and used as growth factors [4,5]. The gut microbiome and the extrinsic variables that cause disease have gotten much attention in recent years. The gut microbiota and its metabolites have been implicated in host physiology and metabolic disorders in numerous studies [3,6]. Trimethylamine-N-oxide (TMAO) is a chemical produced by gut microbial metabolism from choline, betaine, and carnitine. The host’s liver hepatic flavin-containing monooxygenase 3 (FMO 3) converts trimethylamine (TMA) to TMAO [7]. High plasma TMAO levels can be caused by gut dysbiosis. Studies have suggested that high serum TMAO levels have been linked to an increased risk of atherosclerosis [5], chronic kidney disease, cancer, and non-alcoholic fatty liver disease in the recent decade [5,8]. Therefore, lowering the serum TMAO level could be a potential target to prevent CVD and other diseases. Several therapeutic strategies have been investigated to target the reduction in TMAO levels, which includes the use of some analogs or the inhibition of TMA precursors. In these studies, serum TMAO levels were decreased by antibiotics and some natural or synthetic molecules such as allicin, 3,3-dimethyl-1-butanol (DMB), resveratrol (RSV), by remodeling the gut microbiota or inhibiting the activity of TMA lyase of TMA-producing bacteria [9]. Another proposed therapeutic approach is the use of probiotics potentially effective in altering the microbiota composition. Numerous studies suggested that the use of probiotics, single strain, or multistrain could influence the intestinal microbiome composition but failed to reduce serum TMAO levels [10,11,12,13].
The aim of this study is to find a strategy for the selection of potential probiotic strains that are able to reduce the TMAO level in vivo. In this study, we selected some lactobacilli with basic probiotic properties and used them for in vivo study in mice supplemented with choline. Additionally, to assure that our selected strains are unique strains, we also used genetic identification and phylogenetic methods to characterize our selected strains.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
Bacterial strains were collected from the Bioresource Collection and Research Center (BCRC, Hsinchu, Taiwan), the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Germany) and the American Type Culture Collection (ATCC, Manassas, VA, USA). These bacteria stain numbers and sources, and growth conditions are shown in Table 1.

Table 1.
List of bacterial strains used in the present study.
Stock cultures were maintained in 50% glycerol broth at −80 °C. Cells were grown at 37 °C for 18–24 h for DNA preparation, bacteria plate counting, and serial dilutions. Bacteria strains were incubated anaerobically at 37 °C using an anaerobic jar and anaeropack (MGC, Tokyo, Japan). Nutrient broth (NB) and tryptic soy broth (TSB) were purchased from DifcoTM (Becton, Dickinson and Company, Sparks, MD, USA) and Acumedia (Neogen, Lansing, MI, USA), respectively. The de Man, Rogosa, and Sharpe (MRS) agar medium was purchased from DifcoTM (USA). Agar (Acumedia, Neogen, Lansing, MI, USA) was mixed with NB/TSB/MRS at a final concentration of 1.5% (w/v). Ten-fold serial dilutions of culture were prepared, 0.1 mL spotted on agar plates (1.5%), and colony-forming units per mL (CFU/mL) were counted.
2.2. Screening of Lactic Acid Bacteria (LAB) Strains and Their Antagonistic Effect against TMA-Producing Bacteria
2.2.1. Resistance to Artificial Gastric and Intestinal Fluids
Simulated gastric digestion was tested essentially as described in Zarate et al. 2000 [14]. Simulated gastric juice prepared with the following composition: KCl 7 mmol L−1; NaCl, 125 mmol L−1; NaHCO3, 45 mmol L−1 and pepsin, 3 g L−1, HCl used to adjust the final pH 2 and 2.5 and with NaOH to pH 7. In general, 1 mL of cell suspension containing approximately 108–109 CFU mL−1 of LAB was transferred into 9 mL of simulated gastric juice with different pH 2, pH 2.5, and pH 7. The mixture was incubated at 37 °C for 0, 1.5, and 3 h. After incubation, viable LAB cells counts were determined by plating serial dilutions. Simulated intestinal fluid (SIF) was prepared using 0.1% (wt/v) pancreatin (Sigma) and 0.15% (w/v) Oxgall bile salts (Sigma) in water, and pH (pH 8.0) was adjusted with 5 mol l−1 NaOH. After 180 min of gastric digestion, cells were centrifuged (3000× g, 5 min), washed with PBS, suspended in SIF were incubated. Viable bacterial counts were taken at 0, 1.5, and 3 h by plating serial dilutions on MRS agar.
2.2.2. Antimicrobial Activity of LAB Strains
The agar diffusion method was used to study the antibacterial activity of LAB strains. LAB strains were cultured individually in an MRS broth medium in anaerobic jars and incubated for 20 h at 37 °C. Bacterial cells were removed by centrifugation (5000× g, 5 min), and the supernatants were recovered. For each TMA bacteria strains, bacterial cells were cultivated on agar medium. Then, 100 μL of the spent culture supernatant (SCS) (108 CFU/mL) of LAB was added, and the cultures were incubated at 37 °C for 14 h. The diameter of the inhibition zone around the wells was measured. LAB strains with inhibition zones < 11, 11–16, 17–22, and > 23 mm were classified as strains of no –; mild ++; strong ++; and very strong +++ inhibition, respectively [15].
2.2.3. Adhesion to Caco-2 Cells for LAB Strains and TMA-Producing Strains
The conditions described by Gopal et al. [16] and Tsai et al. [17] for the adhesion study of LAB cells were followed. Caco-2 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, Basel, Switzerland) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% non-essential amino acids (NEAA, Invitrogen) at 37 °C in a humidified incubator (5% CO2). Caco-2 cells were transferred (5 × 104 cells mL−1) to a 24-well multidish containing medium without antibiotics. The mixture was stored at 37 °C in a 5% CO2, 95% air atmosphere until a monolayer was formed in each well. Before the bacteria was added, the Caco-2 monolayer was washed twice with sterile PBS. The bacterial monocultures were grown overnight (18 h) and washed twice with sterile PBS. Bacteria cell suspension (1 × 108 CFUmL−1) was added to a 24-well multidish and then incubated for 2 h. After incubation, it is washed three times with sterile PBS and fixed with formalin and stained with gram stain. The adhesion number of LAB cells was counted using Gopal et al. method [16]. The bacteria number was calculated in ten random microscopic fields.
2.3. Mouse Models
C57BL/6J female mice (5 weeks old) were purchased from National Laboratory Animal Center (NLAC), Taipei, Taiwan. The animal experiment was approved by the Institutional Animal Care and Use Committee (IACUC) of Hungkuang University (HK-10605). All mice were maintained at a controlled temperature of 23 ± 2 °C and relative humidity of 50–70% with a 12 h light/dark period. Each mouse was housed individually in cages and had free access to water and food ad libitum. All mice feed was purchased from Harlan Tekland (Madison, WI, USA). All mice were fed with a normal diet for 1-week adaptation (Table 2).

Table 2.
Choline-deficient diet. After the adaptation period, mice were fed either a choline-deficient diet or a normal diet with 1% choline chloride. The experiment was carried for 2–4 weeks, and during the experimental period, the blood of the mice was sampled from the eyes on 0, 7, 14, and/or 28 days.
2.4. Probiotics Treatment
Lactobacilli, i.e., Lpb. plantarum LP1145, L. amylovorus LAM 1345 and Lim. fermentum LF33 were cultured individually in MRS broth at 37 °C. Bacterial strains were centrifuged and washed with PBS. Every day, 0.2 mL (109 CFU/mL) of probiotic strains, i.e., multistrain formula (MF) or single strain, were given to each mouse in each treatment group (n = 7) by oral gavage until the end of the experiment. The probiotic mixture MF is the mixture of the selected strains in an equal volume of LAB solution, each containing 109 CFU/mL. Blood samples were collected and allowed to clot at 4 °C, and serum samples were prepared by centrifugation (3000× g at 4 °C for 10 min). The serum aliquots were filtered and stored at −80 °C until analysis using UPLC-MS/MS.
2.5. UPLC-MS/MS Analysis
Derivatization of TMA and TMAO was carried out using ethyl bromoacetate according to the published methods with minor modifications [18,19,20,21,22,23,24,25,26,27,28,29,30]. Sample preparation and UPLC-MS/MS analysis conditions were previously published by Ramireddy et al. [19]. Briefly, 1 μL of samples were injected, and analysis was performed on Waters ACQUITY UPLC system with Xevo TQ MS (mass spectrometry), operated in positive ion electrospray ionization (ESI+) mode using MassLynx 4.1 SCN810 software. The precursor product ion pairs operated in multiple reaction monitoring (MRM) mode were: m/z146→118, m/z146.1→59 for TMA and d9TMA, m/z 76→59, m/z 76→42 for TMAO and d9TMAO, respectively.
2.6. Counting Lactobacilli Population in Cecum Digesta of Mice
On day 28, the cecum digesta taken from the mice of the MF group and LP1145 group after sacrifice was homogenized in PBS and centrifuged (1000× g). The upper phase was collected, followed by centrifugation. The pellet was then collected and used as intact bacterium cell suspension for DNA isolation using the EZNA stool DNA kit (Omega Bio-tek, Norcross, GA) following the manufacturer’s instructions. The extracted DNA samples were stored at −20 °C until use.
2.7. Quantification of Lactobacilli Populations by qPCR
To quantify the abundance of lactobacilli in mice intestines, real-time quantitative PCR (qPCR) was carried out using an ABI 7500 Fast system. Total DNA isolated from the mice cecum samples of control, MF, and LP1145 mice groups on day 28 after sacrifice was used for the detection of total lactobacilli counts. The Lactobacillus genus-specific primers were used, the primer sequences and PCR conditions are shown in Table 3. The final volume 20 μL of the real-time PCR mixture made up of 6.8 μL nuclease-free water, 10 μL SYBR premix, 0.4 μL ROX reference dye (KAPA Biosystems, Woburn, MA, USA), 2 μL of cDNA, and 0.4 μL (0.25 mM) each of the forward and reverse primers. For quantification of total lactobacilli, a standard curve was generated using a 10-fold dilution series standard ranging from 108 to 102 CFU/mL with real-time PCR. The bacterial cell count was calculated from a standard curve. All reactions were carried out in duplicate.

Table 3.
Primers and PCR conditions used in this study.
2.8. Identification of Lactobacillus Amylovorus Strain LAM 1345
To identify that strain LAM1345 is a unique strain, two tests were conducted, including biochemical method and genotypic identification. The preliminary tests were conducted for strain LAM1345, including gram staining, morphological observation, motility, catalase reaction, growth conditions (aerobic or anaerobic), whether endospores formed, and acid as well as bile tolerance. Gram staining and catalase by MRS agar (Difco lactobacilli MRS agar) were used to identify LAM1345 as LAB.
2.8.1. Genotypic Characterization Method
DNA Extraction
Extraction of genomic DNA from the culture of LAM 1345 was performed using a Genomic DNA Mini Kit (Geneaid Biotech, GB100/GB300) according to the manufacturer’s protocol. The integrity, purity, and concentration of extracted DNA were confirmed absorbance at 260 nm by using a UV spectrophotometer (Eppendorf, Hamburg, Germany). The PCR primers and conditions are shown in Table 3. Each reaction amplification mixture (20 μL final volume) contained 10 ng of genomic DNA 1 μL, 10 μM of each primer 5 μL, 10 μL of 2 × Taq master mix (Ampliqon), dd water was added up to a 20 μL volume. The PCR product was analyzed by agarose gel electrophoresis (1.5% w/v) and visualized by staining with ethidium bromide. The PCR amplified product sent for 16S rDNA gene sequencing (Genomics BioSci & Tech Co., Ltd., Taipei Taiwan), obtained sequences were aligned using BLAST (http://www.ncbi.nlm.nih.gov/blast (accesses on 8 December 2020) [20]. Phylogenetic analysis was performed using clustalW [21].
2.8.2. Leader Peptidase A (lep A) Gene Analysis
The LepA gene PCR primers and conditions are shown in Table 3. The PCR products were purified and sent to a sequencing company (Genomics Biosci and Tec Co. Taipei, Taiwan) to perform sequence analysis. The sequences were then compared with sequences of other strains using BLAST and clustalW.
2.8.3. Phylogenetic Analysis
The neighbor-joining method was used to create a phylogenetic tree. To determine the stability of our phylogenetic tree, the percentage of replicate trees in which the associated taxa clustered together are shown next to the branches (1000 replicates) [22]. The Kimura 2-parameter method [23] was used to calculate the evolutionary distances. Codon positions 1st + 2nd + 3rd + noncoding were considered in the study, which included 17 nucleotide sequences. Gaps and missing data were removed from all positions. The total number of places in the final data set was 995. MEGA7 was used to perform evolutionary analysis.
2.9. Lpb. plantarum LP1145 Identification
2.9.1. Genomic DNA Extraction, Sequencing, and Annotation
Genomic DNA for long reads using Oxford Nanopore Technologies (ONT) and short reads Illumina sequencing were extracted using QIAGEN Genomic-tip 20/G Kit. All extracted genomic DNA quality was determined using QuantiFluor® dsDNA System. Sequencing was performed using ONT GridION for longer raw reads and Illumina Mini-Seq for short reads to generate a paired-end library of 2 × 310 bp for a large number of high accuracy short reads. ONT raw data obtained from the sequencer were decoded and demultiplexed on a sequencer by built-in software. The Illumina raw reads were quality trimmed (Phred Q score below 20) and de novo assembled using the CLC Genomics Workbench V.10., with default parameters. The validated ONT and Illumina sequences were assembled using the software SPAdes v3.13.0. Open reading frames (ORFs) were predicted using GLIMMER [24]. The rRNA and tRNA genes were annotated using RNAmmer and tRNAscan-SE, respectively. The identified coding regions were annotated by screening against the NCBI NT database using NCBI ncbi-blast 2.2.28+, while the translated sequences were searched against the NCBI NR database using DIAMOND with default parameters [25]. Average nucleotide identity (ANI) calculations were performed using the OrthoANIu tool EZBioCloud (https://www.ezbiocloud.net/tools/ani accessed on 11 December 2021) and digital DNA-DNA hybridization (dDDH) values were performed using the genome-to-genome distance calculation (GGDC) website (http://tygs.dsmz.de accessed on 11 December 2021). The ANI and dDDH values are shown in Table 4.

Table 4.
Pairwise comparison of average nucleotide identity (ANI) and digital DNA-DNA (dDDH) hybridization values between Lpb. plantarum and the closely related type strains.
2.9.2. Identification of Genes Related to Antimicrobial Resistance (AMR) and Virulence Factors, Biogenic Amine Producing Genes
BLASTp was used to find the AMR genes in the Comprehensive Antibiotic Resistance Database (CARD), NCBI AMRFinderPlus, ResFinder, and ARG-ANNOT (BLASTp, 70%coverage, 60% identity) [26]. To identify the virulence genes, BLAST searched against the Virulence Factors Database (VFDB), MvirDB, CGE Virulence Finder, CGE Pathogen Finder, and PAIDB [27]. The cutoffs were as follows: e-value (1.0e−20), coverage (70%) and identity (60%).
Microbial biogenic amine-related genes of tyrosine decarboxylase, histidine decarboxylase, ornithine decarboxylase, agmatine deiminase, and lysine decarboxylase were searched by BLASTn. The BLAST results showing a cutoffs e-value of 1.0e−20, identity (60%), and coverage >70% were considered.
2.9.3. Whole-Genome Multilocus Sequence Typing (WgMLST)
The whole genome of Lpb. plantarum LP1145 was sequenced. The National Center for Biotechnology Information (NCBI) bacterial genome database was used to retrieve 51 public genome sequences of Lpb. plantarum strains (Table S1). The wgMLST analysis was performed using the cano-wgMLST _BacCompare web-based tool [28] as described by Huang et al. [29], to compare the genome sequences on the Linux platform.
The whole-genome sequences were compared with the constructed pan-genome allele database (PGAdb) using BLASTN V2.2.30 cano-wgMLST _BacCompare tool consists inbuilt process, Prokka pipeline, used for contig annotations, Roary V3.10.2 pipeline, for building PGAdb, PHYLIP v3.6 program [30] to construct a genetic relatedness tree.
2.10. Lim. fermentum LF1143 Identification
To confirm strain of LF1143 is a unique strain, in original Lim. fermentum sample (LF33), which was identified by the use of API 50 CHL kit, showed to be Lim. fermentum I and Lim. fermentum II, each counted about 42%–48%, respectively (data not shown). To identify the strain of LF1143, bacterial cells in the dd water-diluted solution of Lim. fermentum LF33 was cultured on MRS agar plate at 37 °C for 24 hrs.; then, single colonies were picked for further culture in MRS medium for bacteria cell collection as well as DNA preparation. The PCR conditions and primers specific for Lim. fermentum and L. reuteri strains are shown in Table 3.
2.11. Statistical Analysis
The findings were analyzed using a one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test to establish statistical significance. Statistical significance was considered as p < 0.05. TMAO and TMA data from 7 mice in each experiment (control and probiotic groups) were taken for statistical analysis. Data were expressed as the mean ± standard deviation (SD) unless otherwise stated.
3. Results
3.1. Acid and Bile Tolerance among Lactobacilli
Based on the methods for the assay of the acid and bile resistance of the LAB strains, all the 6 LAB strains, i.e., Lpb. plantarum LP1145, L. amylovorus LAM1345, Lim. fermentum LF33 and other three strains, i.e., L. gasseri (BCRC 14619), L. salivarius (BCRC 14789), Lpb. plantarum (LPL07) were acid-tolerant at pH ≥ 2.5 and less tolerant at pH 2.0. In addition, LP 1145, LAM 1345, and LF33 strains are tolerant to bile. Only the strain of L. gasseri was less tolerant.
3.2. Growth Inhibition of TMA Bacteria by LAB Supernatants
Antimicrobial activity levels of LAB strains against TMA-producing bacteria are shown in Table 5. We randomly selected nine of the TMA bacteria strains and studied their inhibition by LAB strains. Probiotic strains used in this study were able to inhibit the growth of some of the TMA-producing bacteria, such as Providencia alcalifaciens; Escherichia fergusonii; Proteus mirabilis; Klebsiella Pneumoniae; and Providencia rustigianii, with inhibition zones greater than or equal to 15 mm. However, most of the LAB strains showed little or no antimicrobial activity against some of the TMA bacteria strains, such as Clostridium sporogenes; Escherichia coli; Clostridium tetani; and Providencia rettgeri strains, shown in Table 5.

Table 5.
Inhibition of TMA-producing bacteria by lactic acid bacteria.
3.3. Adherence of TMA Bacteria and Lactobacilli to Caco-2 Cells
We examined the adherence capacity of probiotic and TMA-producing strains to the human intestinal cell line Caco-2 (Table 6). All the strains examined were able to adhere to Caco-2 cells with variable levels. The adhered number of the TMA-producing strains showed poor adherence, i.e., less than 20 bacterial count per cell (Table 6). Probiotic strains showed variable ability to adhere to human Caco-2 cells, from 17.7 to > 200 CFU per Caco-2 cell. For strains of LAM 1345, LP1145, and Lim. fermentum LF33, which contains Lim. fermentum I and II as assayed with API 50 CHL kit, all these strains showed high adherence, i.e., > 200 CFU per Caco-2 cell. The probiotic strains exhibited stronger adherence to Caco-2 cells, and they may inhibit the adherence of TMA-producing strains by competition and exclusion.

Table 6.
Adhesion of TMA and LAB bacteria to Caco2 cells.
3.4. Probiotic Strains Reducing Serum TMAO Levels in Mice Challenged with Choline
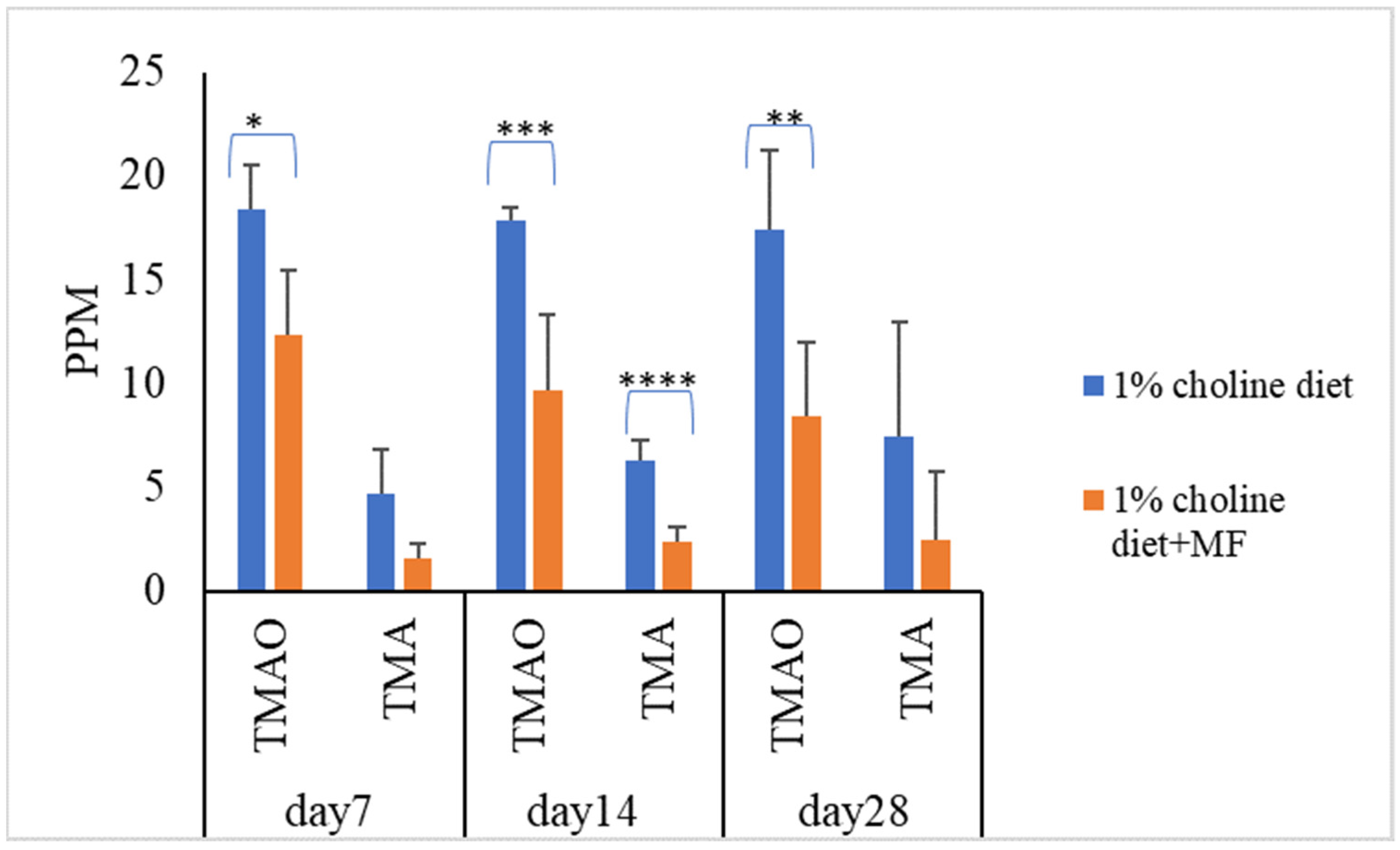
To know the effect of probiotic strains on reducing serum TMAO levels in vivo, mice were divided into five groups (n = 7 in each experiment group). Mice in the control group were fed with a 1% choline diet, and mice in the other four groups were fed with 1% choline diet plus LAB multistrain formula (MF) or single LAB strains, i.e., LP1145, LAM 1345, LF1143 for 7, 14, and/or 28 days. After feeding, on day 7, day 14, and/or day 28, serum TMAO levels were measured. For MF group Lim. fermentum LF33 (contains type I and type II) and for single strain Lim. fermentum I -LF1143 was used for analysis. Results showed that probiotics in the MF group significantly reduced serum TMAO and TMA levels on days 7, 14, and 28 (Figure 1).
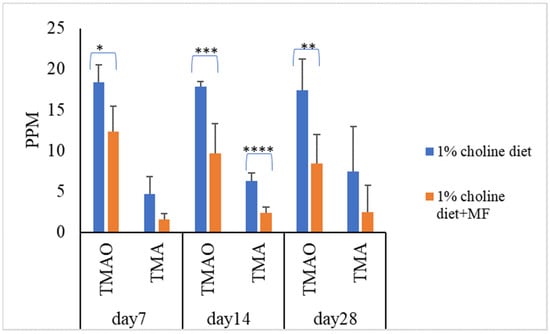
Figure 1.
Probiotic multistrain formula decreases the plasma TMAO and TMA levels in C57BL/6J mice. Experimental conditions were as described in Methods. Data are shown as mean values ± SD (n = 7). A p-value of <0.05 was considered to be statistically significant, * p <0.05, ** p < 0.01, *** p <0.001, **** p < 0.0001.
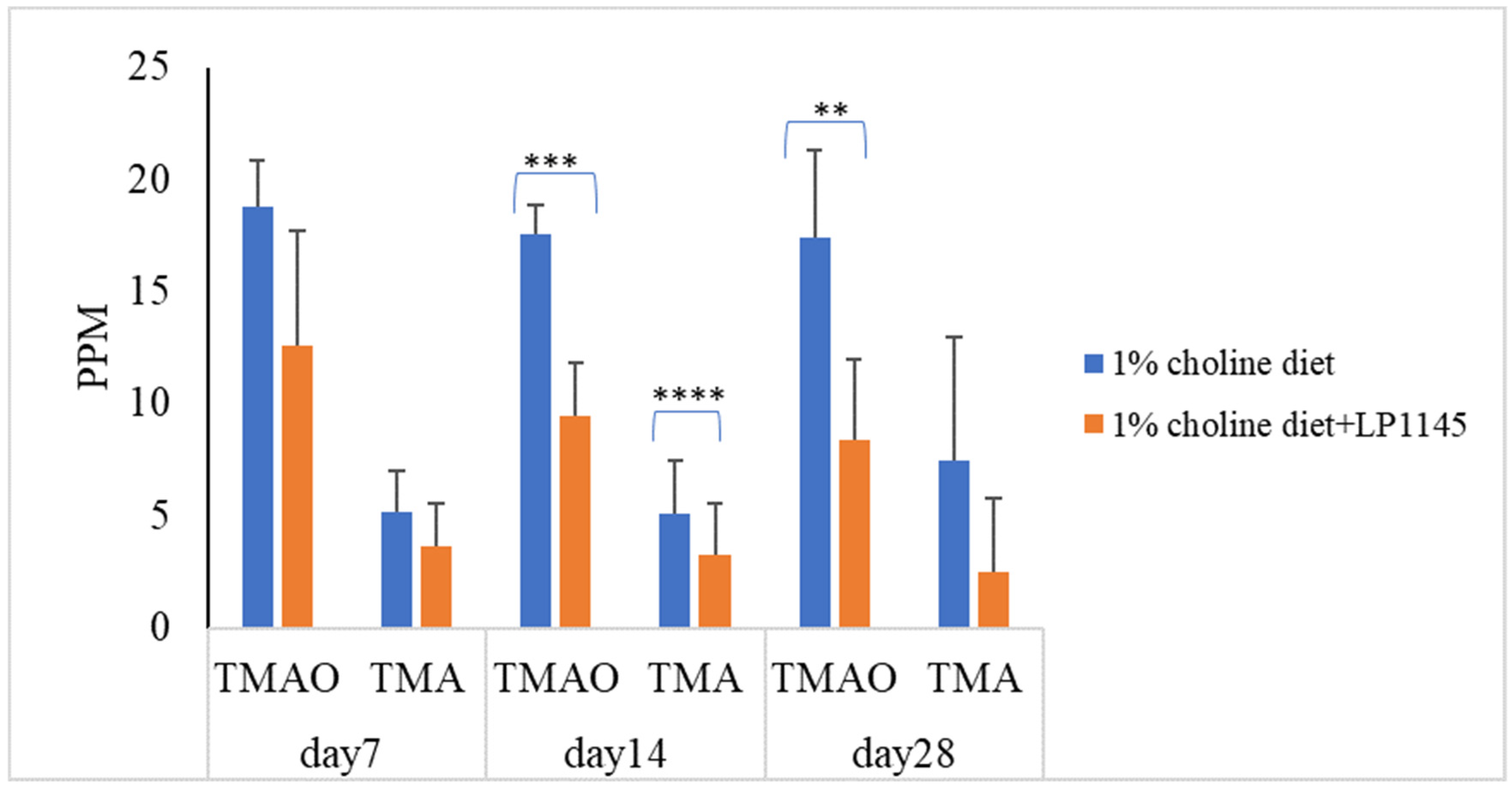
Further, to find the roles played by the individual strains of LP1145, LAM 1345, and LF1143 on the serum levels of TMAO and TMA, attempts were made. The results showed that LP 1145 significantly reduced the serum level of TMAO on days 14 and 28 and TMA level on day 14 when compared with the result from the control group (Figure 2).
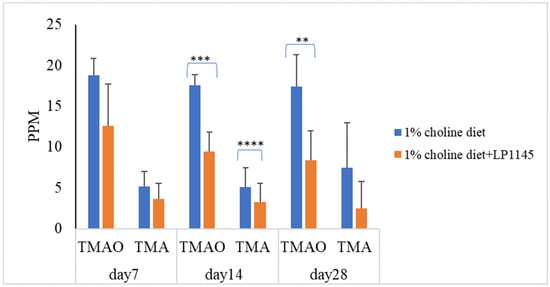
Figure 2.
Probiotic strain Lpb. plantarum decreases the plasma TMAO and TMA levels in C57BL/6J mice. Experimental conditions were as described in Methods. Data are expressed as mean values ± SD (n = 7). ** p < 0.01, *** p <0.001, **** p < 0.0001.
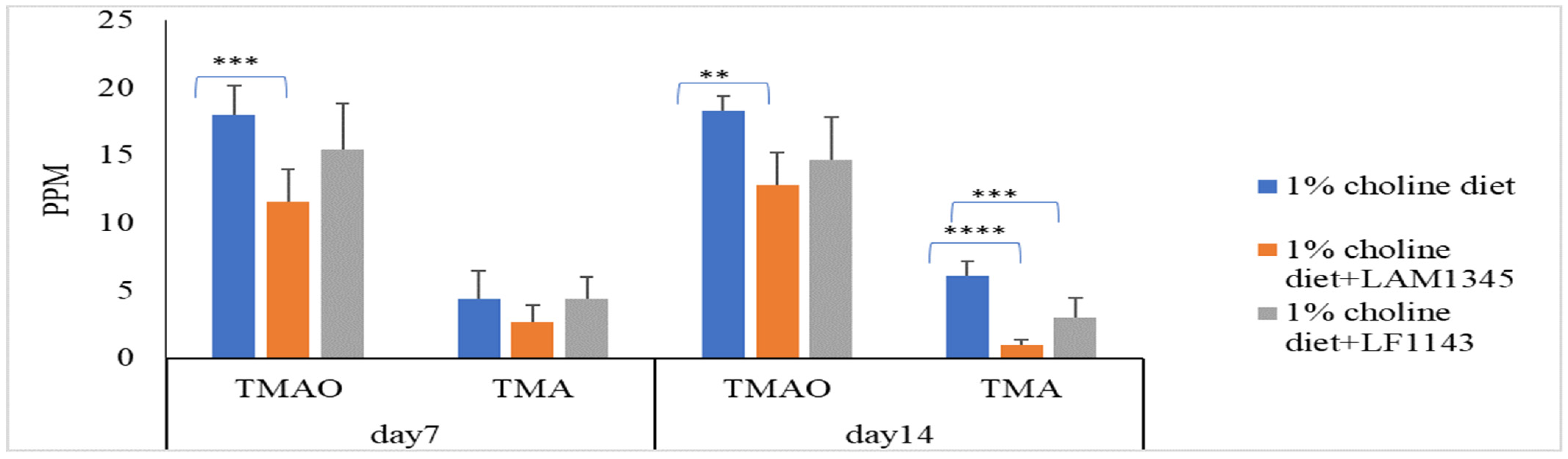
As for the effect of LAM1345 and LF1143 on TMAO and TMA levels in mice serum, LAM1345 showed a significant reduction effect on serum TMAO and TMA levels on day 7 and day 14 when compared to those from the control group (Figure 3). However, a significant reduction effect on serum TMAO levels was not observed for strain LF1143. Thus, the significant reduction in TMAO in mice serum by the MF group measured on days 7, 14, and 28 was mainly due to the contribution of strain LP1145 and LAM1345. In addition, for strains of LAM1345 and LF1143, since the results of that of TMAO reduction on day 14 were not improved as compared with that obtained on day 7, for these two strains, an animal study did not proceed to day 28.
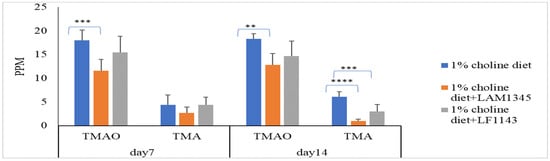
Figure 3.
Effect of L. amylovorusLAM1345 and Lim. fermentum LF1145 on serum TMAO and TMA levels. Experimental conditions were as described in Methods. Values are represented as mean values ± SD (n = 7) ** p < 0.01, *** p <0.001, **** p < 0.0001.
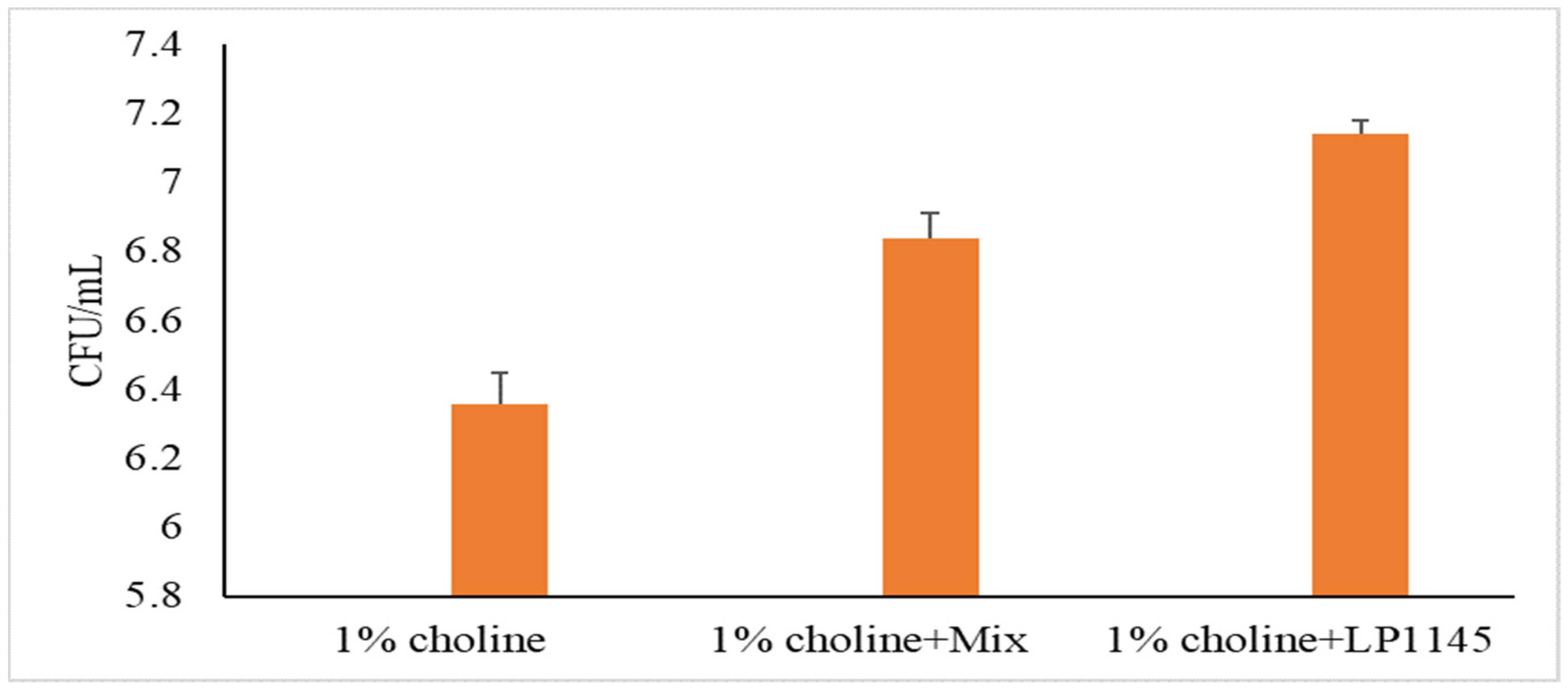
For MF and Lpb. plantarum groups, we also measured the intestinal total LAB counts after sacrificing the mice on day 28. A significant increase in lactobacilli population was observed as compared to that of the control group (Figure 4).
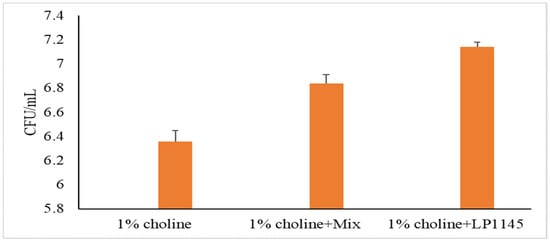
Figure 4.
Total lactobacilli counts in mice cecum. Experimental conditions were as described in Methods. Values are shown in mean value ± SD (n = 7).
From the above study, we found two probiotic strains were able to reduce the TMAO and TMA level in the serum of mice challenged with choline., i.e., strain L. amylovorus LAM1345 and Lpb. plantarum LP1145. To assure that these strains are unique strains, we attempted to use molecular identification methods to distinguish these strains from other strains of the same species. In addition, to distinguish the strain of Lim. fermentum I from strain Lim. fermentum II, as described earlier, PCR primers specific for Lim. fermentum and L. reuteri were used to identify these strains.
3.5. Lactobacillus Amylovorus Identification
3.5.1. Identification of Bacteria through Bacteriological and Biochemical Tests
The isolate was morphologically similar to Lactobacillus spp. and was found Gram-positive, catalase-negative, no motility, grown anaerobically, no endospore formation. The strain showed both acid and bile tolerance characteristics. The LAM 1345 strain is highly similar to L. acidophilus. If we use a biochemical method, such as API50, it may be detected as L. acidophilus strain. Thus, we performed genomic analysis to identify this strain.
3.5.2. Sequence Analysis and Phylogenetic Tree
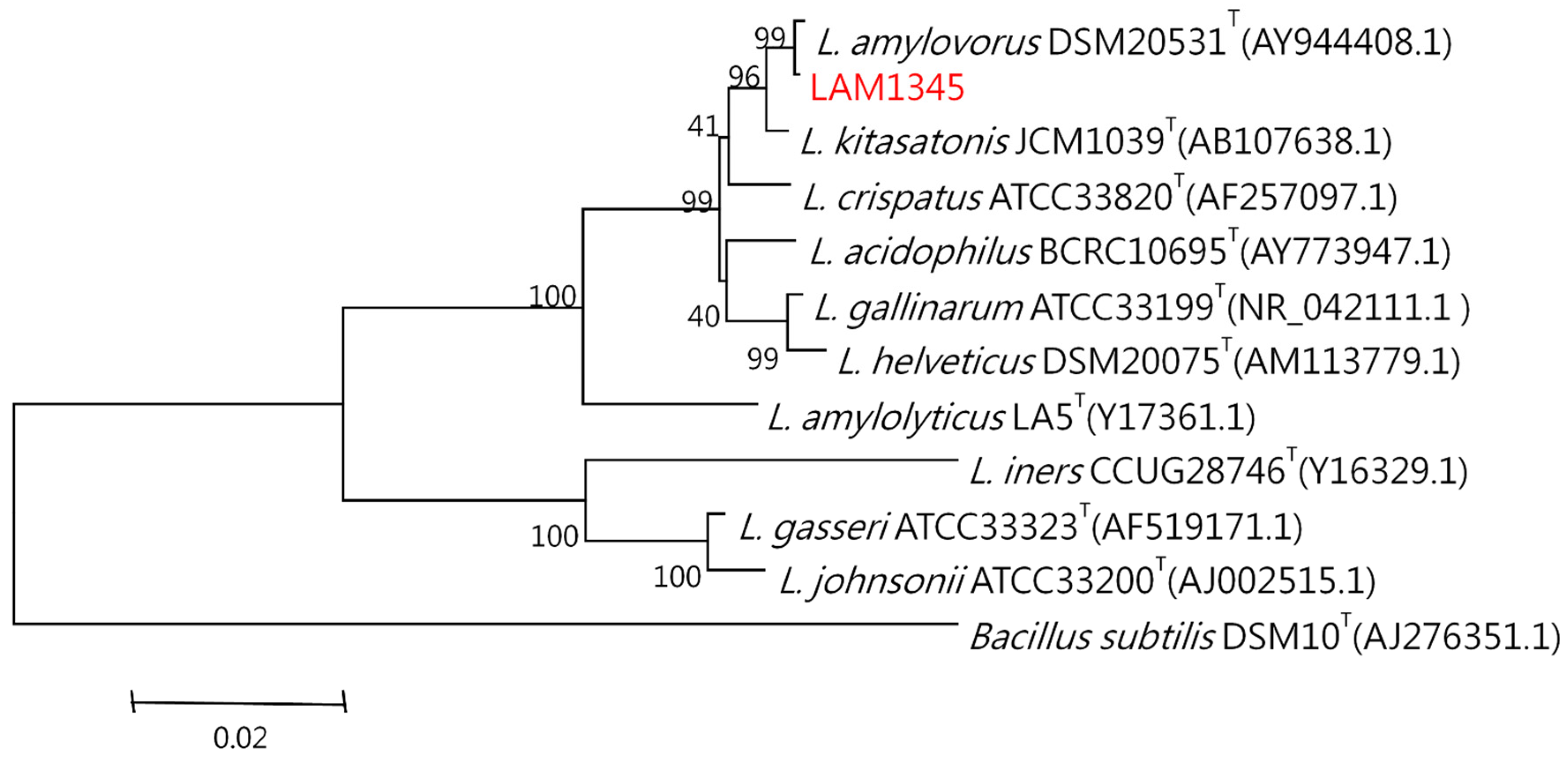
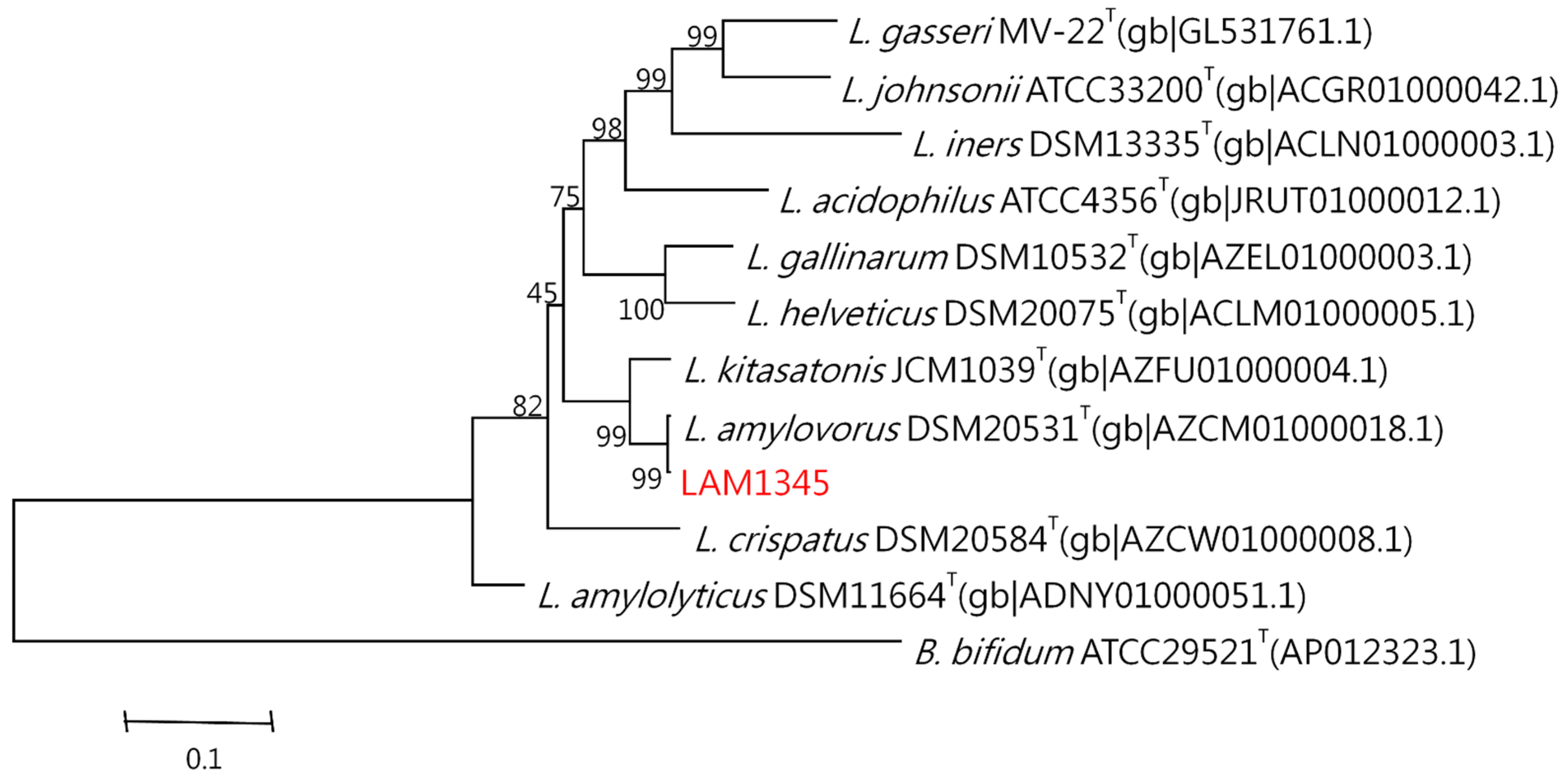
The genomic DNA was isolated according to the previously described method. The 16S rDNA was amplified at approximately 1500 bp, and the product was sequenced. In addition, the LepA gene of LAM1345 was amplified, and approximately 1163 bp of the DNA sequence was determined (unpublished data). DNA sequencing results were analyzed with NCBI-BLAST. MSA was analyzed using clustalW. The phylogenetic tree was depicted in Figure 5 and Figure 6, showing the genetic distances of various lactobacilli. The strain LAM 1345′ of 16s rDNA gene sequence was aligned with neighboring-type strains, and taxonomic relationships were determined. Highest degrees of 16s rDNA sequence identity with L. amylovorus DSM 20,531 T (accession number: AY944408.1, 99%) (Figure 5). Phylogenetic and homology analysis revealed that isolated strain LAM1345 could be L. amylovorus. Additionally, a phylogenetic tree to determine the relationship of the LepA gene inLAM1345. The strain LAM1345 showed the closest phylogenetic relationship with L. amylovorus DSM 20531 (accession number: AZCM01000018.1; 99%) (Figure 6).
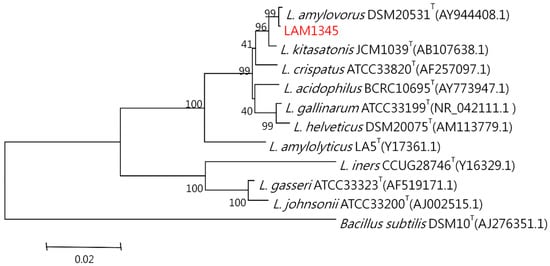
Figure 5.
Phylogenetic tree based on the 16S rRNA gene sequence showing the phylogenetic relationships between the LAM1345 strain and related lactobacilli species.
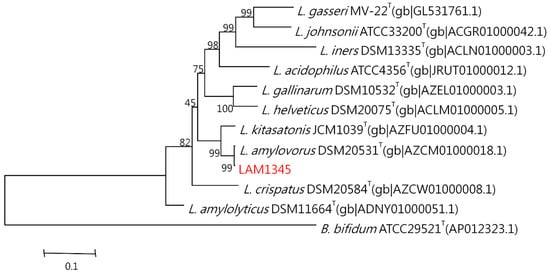
Figure 6.
Phylogenetic tree of the lepA gene of strain LAM1345 strain and related lactobacilli species.
3.6. Lpb. plantarum LP1145 Identification
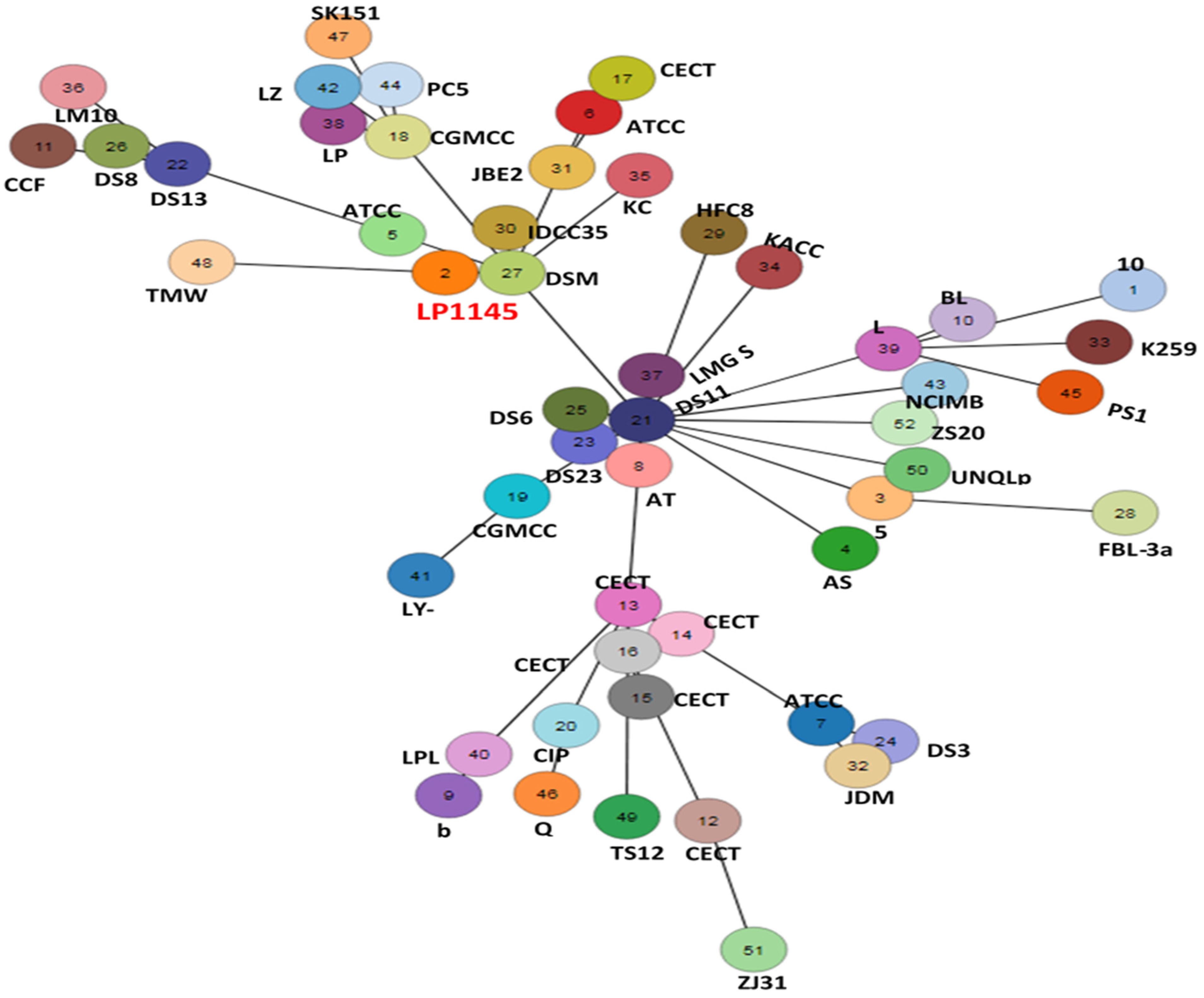
The cano-wgMLST_BacCompare analysis platform was used to compare the genome sequences for strain typing of 52 Lpb. plantarum strains. The Lpb. plantarum PGAdb contained 10,273 genes, of which 2223 (21.6%) genes belonged to the core genome, 4713 (45.9%) belonged to the accessory genome, and 3337 (32.5%) were unique genes.
The 52 strains were assigned to different sequence types using the wgMLST analysis based on allele profiles of the 2223 core genes (Figure 7). The phylogenetic tree revealed that the strain LP1145 was most closely related to the DSM 20174T (Figure 8) and showed 11 loci differences, including 35 SNPs, 3 insertion/deletions (Table S2).
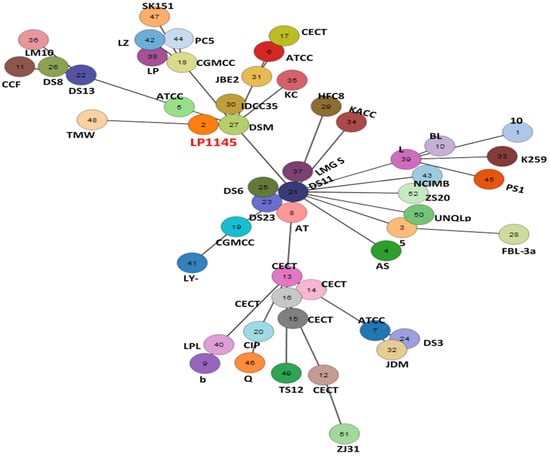
Figure 7.
The allele-based minimum spanning tree constructed with cgMLST profiles for the 52 Lactiplantibacillus plantarum subsp. plantarum strains on the basis of a comparison of 2233 core genes. Each circle represents a different sequence type.

Figure 8.
The allele-based UPGMA tree constructed with cgMLST profiles for the 52 Lactiplantibacillus plantarum subsp. plantarum strains on the basis of a comparison of 2233 differentiated core genes.
3.7. Lim. fermentum Identification
To ensure that the strains we used were unique strains, we also tried to identify the strains in Lim. fermentum LF33 sample, if we use the API 50 CHL biochemical kit, two strains are identified as Lim fermentum, i.e., Lim. fermentum type I 48.5% and Lim. fermentum type II 42.8%. It was found that when we use specific PCR methods, it was found that in addition to strain LF1143, part of the strains are L. reuteri strains (initially Lim. fermentum II known as L. reuteri) (data not shown). We did not pursue molecular identification of these strains because serum TMAO levels were not considerably lowered by LF1143.
4. Discussion
Gut microbiota has received considerable attention because of its potential involvement in reducing the risk of cardiovascular disease (CVD) and other diseases. There is growing interest in research on the role of probiotics in the prevention and treatment of CVD and multiple diseases. The use of probiotics could be a safer and potentially more effective strategy to change the composition of the bacteria. A potential strain identification method is necessary for evaluating their probiotic properties. In this study, we found two novel LAB strains with potential probiotic properties and the ability to reduce the serum TMAO level in choline-challenged mice. According to the Food and Agriculture Organization and the World Health Organization (FAO/WHO), the requirement for probiotic strains are survival in the gastrointestinal tract, able to adhere to epithelial cells and cell lines, antimicrobial activity against potentially harmful microorganisms, ability to inhibit pathogen adherence to intestinal cell surfaces, and activity of the bile salt hydrolase [35]. In this study, the probiotic characteristics, including acid bile tolerance, antimicrobial activity, and adhesion capacity, were evaluated. The most significant properties of probiotics are their ability to survive in the stomach and intestine. The acid condition in the stomach and bile salt condition in the duodenum has been identified as the two most significant barriers to LAB survival in the host’s GI tract [36]. The probiotic strains used in this study showed enough tolerance to low pH and bile salts. Antimicrobial activity against TMA-producing bacteria was studied. These TMA production strains were able to produce TMA at different levels [19]. Most of the probiotic strains showed antimicrobial activity (Table 5). The capacity of probiotics to survive and adhere to the intestine is the key factor of probiotics. It not only allows probiotics to live longer in the GI tract but also enhances their interaction with the host. Probiotic strains we selected as potential strains have been found not only able to adhere to Caco-2 cells (Table 6) but also to stimulate the production of TNF-a by macrophage RAW264.7 (data not shown). Strain-specific cell-surface components play major roles in the adhesion ability of lactobacilli to the intestine mucosa. For L. amylovorus and Lpb. plantarum species, both have been shown able to increase their adherence to human epithelial cells due to their surface layer protein [37,38]. These surface layer proteins regulate the immune system by mediating bacterial adherence to the gut mucosa [39].
Probiotics are bacterial strains that lack the genes required to convert choline and carnitine to TMA [40]. Varied bacteria strains have different effects on choline metabolism and TMAO levels. Different bacteria strains can influence choline metabolism and TMAO levels in different ways. Recent studies have revealed that several bacteria strains such as Enterobacter aerogenes ZDY01, Lactiplantibacillus plantarum subsp. plantarum ZDY04, and Lacticaseibacillus paracasei can reduce TMAO and TMA levels by modifying the gut microbiota [41,42,43]. In contrast, in human clinical trials, supplementation of single or multistrain probiotics serum TMAO levels was not reduced [10,11,13,44,45]. New potential probiotic strains are needed to reduce TMAO levels in vivo and in humans. In this study, we have isolated new strains L. amylovorus LAM1345 and Lpb. plantarum LP1145 with probiotic properties. Reports have shown that Lim. fermentum and L. amylovorus are probiotics able to change body adiposity and gut microbiota in healthy people [28]. Our LAM1345, as well as LP1145, were able to reduce the TMAO level in serum and thus may be the potential probiotics able to lower the risk of CVD and other diseases.
The L. amylovorus strain has bile salt hydrolase (BSH) activity, which allows the deconjugation of bile salts; bile salt-activated signaling pathways, which are linked to metabolic disorders such as atherosclerosis, type 2 diabetes, obesity, and non-alcoholic steatohepatitis [46,47]. Study shows that daily consumption of L. amylovorus improves pre-obesity states and affects the gut microbial population [46]. However, no research was found on the effect of L. amylovorus on TMAO levels. This is the first study that we are aware of that focuses on TMAO level reduction by L. amylovorus strain.
Furthermore, we also use the high-resolution molecular methods to identify that our strain of LAM1345 is a novel strain. We tested physiological and biochemical properties. In general, rRNA and rDNA are the potential targets for the identification and phylogenetic analysis of these bacteria because of their ubiquity and their resistance to evolutionary changes. In this study, we sequenced 16srDNA and compared it with the NCBI database, and found that the identified strain belongs to L. amylovorus. Recently a unique elongation factor 4 (EF4), also known as LepA, was discovered in bacteria. LepA is one of the most conserved proteins found in all bacteria [48]. Sequencing and phylogenetic analysis showed that our isolate belongs to L. amylovorus with 99% similarity (Figure 6).
Lpb. plantarum plays an important role in the pharmaceutical industry and in the medical field without side effects [49]. A recent study showed Lpb. plantarum ZDY04 reduces the serum TMAO levels in mice [42]. However, how this strain can work in humans is unknown. In another study, supplementation of Lpb. plantarum 299v did not change the serum TMAO concentrations in men with coronary artery disease [45]. Finding a new potential probiotic strain to prevent CVD is necessary. In this study, we identified a new Lpb. plantarum LP1145 strain with probiotic properties, which significantly reduces the serum TMAO levels. Since Lpb. Plantarum ZDY04 has been published, although we do not have the ZDY04 strain for comparison. Comparative genomic analysis performed by the wgMLST method indicated that our LP1145 is a novel strain and closely related to the standard strain DSM20174T (Figure 7 and Figure 8).
Our probiotic strain mixture, i.e., multistrain formula (MF), showed a significant reduction in serum TMAO levels from day 7 to day 28. For single strains, we selected L. amylovorus, Lpb. Plantarum, and Lim. fermentum type I (strain LF1143) rather than type II strain, i.e., L. reuteri, for effect on TMAO reduction study because all these three probiotic strains are effective in lowering the serum TMAO levels of mice. However, strain LF1143H may not be strong enough to reduce the serum TMAO level since Lim. fermentum has been shown to be a probiotic capable of modifying body adiposity and intestinal bacteria in healthy people. It may help to prevent CVD, cancer and diabetes, etc. [50]. These diseases have reported relatedness to high serum levels of TMAO. In addition, the multistrain formula has been reported to have additive and synergistic effects, and that may serve additional roles, such as improving strain colonization and adhesion [51]. Thus, the multistrain formula (MF), which contains the probiotic strains were able to reduce the serum level of TMAO. Finally, while all the investigations were carried out in mice, comparable outcomes could be expected in humans if the right conditions for human subjects and clinical trials are used. Currently, we are trying to use these LAB strains for a human clinical study.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/foods10122931/s1, Table S1: Genomic characteristics of Lactiplantibacillus plantarum subsp. plantarum strains. Table S2: Discriminated loci between Lactiplantibacillus plantarum subsp. plantarum LP1145 and DSM 20174T.
Author Contributions
Conceptualization, H.-Y.T. and C.-Y.H.; methodology, C.-C.C. (Chien-Chi Chen), L.R., S.-R.W. and J.-S.L.; software, L.R., C.-H.H. and C.-C.C. (Chien-Chi Chen); validation, H.-Y.T.; formal analysis, Y.-C.C., C.-H.H. and C.-C.C. (Chih-Chieh Chenn); investigation, L.R., S.-R.W., S.-H.C. and C.-C.C. (Chih-Chieh Chenn); data curation, L.R., S.-R.W. and H.-Y.T.; writing original draft preparation, L.R. and S.-L.Y.; writing review and editing, H.-Y.T.; supervision H.-Y.T., S.-L.Y. and C.-Y.H.; funding acquisition, H.-Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by the Ministry of Science and Technologygrant no. MOST-104-2320-B-241-002-MY2, MOST 103-2313-B241-001, and NSC 102-2632-B-241-001-MY3-3.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We would like to give our deep thanks to MOST for supporting this work. In addition, we would like to thank SynBio Tech Inc., Kaohsiung, Taiwan, and BCRC, Hsinchu, Taiwan, for the assistance of molecular identification work.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Dahlof, B. Cardiovascular disease risk factors: Epidemiology and risk assessment. Am. J. Cardiol. 2010, 105, 3A–9A. [Google Scholar] [CrossRef]
- Lloyd-Jones, D.M. Cardiovascular risk prediction: Basic concepts, current status, and future directions. Circulation 2010, 121, 1768–1777. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, K.; Wang, X.; Pang, Y.; Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell 2021, 12, 360–373. [Google Scholar] [CrossRef]
- Bennett, B.J.; de Aguiar Vallim, T.Q.; Wang, Z.; Shih, D.M.; Meng, Y.; Gregory, J.; Allayee, H.; Lee, R.; Graham, M.; Crooke, R.; et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013, 17, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Coutinho-Wolino, K.S.; de F Cardozo, L.F.M.; Cardozo, L.; de Oliveira Leal, V.; Mafra, D.; Stockler-Pinto, M.B. Can diet modulate trimethylamine N-oxide (TMAO) production? What do we know so far? Eur. J. Nutr. 2021, 60, 3567–3584. [Google Scholar] [CrossRef]
- Iglesias-Carres, L.; Hughes, M.D.; Steele, C.N.; Ponder, M.A.; Davy, K.P.; Neilson, A.P. Use of dietary phytochemicals for inhibition of trimethylamine N-oxide formation. J. Nutr. Biochem. 2021, 91, 108600. [Google Scholar] [CrossRef]
- Tripolt, N.J.; Leber, B.; Triebl, A.; Kofeler, H.; Stadlbauer, V.; Sourij, H. Effect of Lactobacillus casei Shirota supplementation on trimethylamine-N-oxide levels in patients with metabolic syndrome: An open-label, randomized study. Atherosclerosis 2015, 242, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Boutagy, N.E.; Neilson, A.P.; Osterberg, K.L.; Smithson, A.T.; Englund, T.R.; Davy, B.M.; Hulver, M.W.; Davy, K.P. Probiotic supplementation and trimethylamine-N-oxide production following a high-fat diet. Obesity 2015, 23, 2357–2363. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, Z.; Lv, Y.; Tong, L.; Liu, T.; Yi, H.; Zhou, X.; Yu, Z.; Tian, X.; Cui, Q.; et al. Reduction of intestinal trimethylamine by probiotics ameliorated lipid metabolic disorders associated with atherosclerosis. Nutrition 2020, 79–80, 110941. [Google Scholar] [CrossRef]
- Chen, S.; Jiang, P.P.; Yu, D.; Liao, G.C.; Wu, S.L.; Fang, A.P.; Chen, P.Y.; Wang, X.Y.; Luo, Y.; Long, J.A.; et al. Effects of probiotic supplementation on serum trimethylamine-N-oxide level and gut microbiota composition in young males: A double-blinded randomized controlled trial. Eur. J. Nutr. 2021, 60, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Zarate, G.; Chaia, A.P.; Gonzalez, S.; Oliver, G. Viability and beta-galactosidase activity of dairy propionibacteria subjected to digestion by artificial gastric and intestinal fluids. J. Food Prot. 2000, 63, 1214–1221. [Google Scholar] [CrossRef] [PubMed]
- Rammelsberg, M.; Radler, F. Antibacterial polypeptides of Lactobacillus species. J. Appl. Bacteriol. 1990, 69, 177–184. [Google Scholar] [CrossRef]
- Gopal, P.K.; Prasad, J.; Smart, J.; Gill, H.S. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int. J. Food Microbiol. 2001, 67, 207–216. [Google Scholar] [CrossRef]
- Tsai, C.C.; Huang, L.F.; Lin, C.C.; Tsen, H.Y. Antagonistic activity against Helicobacter pylori infection in vitro by a strain of Enterococcus faecium TM39. Int. J. Food Microbiol. 2004, 96, 1–12. [Google Scholar] [CrossRef]
- Johnson, D.W. A flow injection electrospray ionization tandem mass spectrometric method for the simultaneous measurement of trimethylamine and trimethylamine N-oxide in urine. J. Mass Spectrom. 2008, 43, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Ramireddy, L.; Tsen, H.-Y.; Chiang, Y.-C.; Hung, C.Y.; Chen, F.-C.; Yen, H.-T. The gene expression and bioinformatic analysis of choline trimethylamine-lyase (CutC) and its activating enzyme (CutD) for gut microbes and comparison with their TMA production levels. Curr. Res. Microb. Sci. 2021, 2, 100043. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Park, Y.K.; Kim, J.F. PAIDB v2.0: Exploration and analysis of pathogenicity and resistance islands. Nucleic Acids Res. 2015, 43, D624–D630. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Lin, J.W.; Chen, C.C. cano-wgMLST_BacCompare: A Bacterial Genome Analysis Platform for Epidemiological Investigation and Comparative Genomic Analysis. Front. Microbiol. 2019, 10, 1687. [Google Scholar] [CrossRef]
- Huang, C.H.; Chen, C.C.; Chiu, S.H.; Liou, J.S.; Lin, Y.C.; Lin, J.S.; Huang, L.; Watanabe, K. Development of a high-resolution single-nucleotide polymorphism strain-typing assay using whole genome–based analyses for the Lactobacillus acidophilus probiotic strain. Microorganisms 2020, 8, 1445. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.J. Use of Tuf Gene Sequences for the Quantititative and Qualitative Assay of Lactic Acid Bacteria and Comparison of Tuf with 16SrRNA Gene for the Phylogenetic Analysis of Bifidobacterium. Ph.D. Thesis, National Chung Hsing University, Taichung, Taiwan, 2009. [Google Scholar]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Dickson, E.M.; Riggio, M.P.; Macpherson, L. A novel species-specific PCR assay for identifying Lactobacillus fermentum. J. Med. Microbiol. 2005, 54, 299–303. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, Y.; Kato, N.; Liu, C.; Matsumiya, Y.; Kato, H.; Watanabe, K. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 2000, 187, 167–173. [Google Scholar] [CrossRef]
- FAO/WHO. Guidelines for Evaluation of Probiotics in Food; Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food London, ON, Canada, 30 April and 1 May 2002; FAO: Rome, Italy; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- Hsu, T.C.; Yi, P.J.; Lee, T.Y.; Liu, J.R. Probiotic characteristics and zearalenone-removal ability of a Bacillus licheniformis strain. PLoS ONE 2018, 13, e0194866. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M.; Zhao, J.; Xia, Y.; Lai PF, H.; Ai, L. A surface protein from Lactobacillus plantarum increases the adhesion of Lactobacillus strains to human epithelial cells. Front. Microbiol. 2018, 9, 2858. [Google Scholar] [CrossRef] [PubMed]
- Hynonen, U.; Kant, R.; Lahteinen, T.; Pietila, T.E.; Beganovic, J.; Smidt, H.; Uroic, K.; Avall-Jaaskelainen, S.; Palva, A. Functional characterization of probiotic surface layer protein-carrying Lactobacillus amylovorus strains. BMC Microbiol. 2014, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Hynonen, U.; Palva, A. Lactobacillus surface layer proteins: Structure, function and applications. Appl. Microbiol. Biotechnol. 2013, 97, 5225–5243. [Google Scholar] [CrossRef]
- Zeisel, S.H.; Warrier, M. Trimethylamine N-Oxide, the Microbiome, and Heart and Kidney Disease. Annu. Rev. Nutr. 2017, 37, 157–181. [Google Scholar] [CrossRef]
- Qiu, L.; Tao, X.; Xiong, H.; Yu, J.; Wei, H. Lactobacillus plantarum ZDY04 exhibits a strain-specific property of lowering TMAO via the modulation of gut microbiota in mice. Food Funct. 2018, 9, 4299–4309. [Google Scholar] [CrossRef]
- Qiu, L.; Yang, D.; Tao, X.; Yu, J.; Xiong, H.; Wei, H. Enterobacter aerogenes ZDY01 Attenuates Choline-Induced Trimethylamine N-Oxide Levels by Remodeling Gut Microbiota in Mice. J. Microbiol. Biotechnol. 2017, 27, 1491–1499. [Google Scholar] [CrossRef]
- Hsu, C.N.; Hou, C.Y.; Chan, J.Y.H.; Lee, C.T.; Tain, Y.L. Hypertension Programmed by Perinatal High-Fat Diet: Effect of Maternal Gut Microbiota-Targeted Therapy. Nutrients 2019, 11, 2908. [Google Scholar] [CrossRef]
- Malik, M.; Suboc, T.M.; Tyagi, S.; Salzman, N.; Wang, J.; Ying, R.; Tanner, M.J.; Kakarla, M.; Baker, J.E.; Widlansky, M.E. Lactobacillus plantarum 299v Supplementation Improves Vascular Endothelial Function and Reduces Inflammatory Biomarkers in Men with Stable Coronary Artery Disease. Circ. Res. 2018, 123, 1091–1102. [Google Scholar] [CrossRef]
- Montrucchio, C.; De Nicolo, A.; D’Ettorre, G.; D’Ascenzo, F.; Lazzaro, A.; Tettoni, M.; D’Avolio, A.; Bonora, S.; Celani, L.; Di Perri, G.; et al. Serum Trimethylamine-N-oxide Concentrations in People Living with HIV and the Effect of Probiotic Supplementation. Int. J. Antimicrob Agents 2020, 55, 105908. [Google Scholar] [CrossRef]
- Grill, J.P.; Cayuela, C.; Antoine, J.M.; Schneider, F. Isolation and characterization of a Lactobacillus amylovorus mutant depleted in conjugated bile salt hydrolase activity: Relation between activity and bile salt resistance. J. Appl. Microbiol. 2000, 89, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Polacek, N.; Vesper, O.; Staub, E.; Einfeldt, E.; Wilson, D.N.; Nierhaus, K.H. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 2006, 127, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Arasu, M.V.; Al-Dhabi, N.A.; Ilavenil, S.; Choi, K.C.; Srigopalram, S. In vitro importance of probiotic Lactobacillus plantarum related to medical field. Saudi J. Biol. Sci. 2016, 23, S6–S10. [Google Scholar] [CrossRef] [PubMed]
- Omar, J.M.; Chan, Y.-M.; Jones, M.L.; Prakash, S.; Jones, P.J.H. Lactobacillus fermentum and Lactobacillus amylovorus as probiotics alter body adiposity and gut microflora in healthy persons. J. Funct. Foods 2013, 5, 116–123. [Google Scholar] [CrossRef]
- Timmerman, H.M.; Koning, C.J.; Mulder, L.; Rombouts, F.M.; Beynen, A.C. Monostrain, multistrain and multispecies probiotics- a acomparison of functionality and efficacy. Int. J. Food Microbiol. 2004, 96, 219–233. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).