Paeonol Disrupts the Integrity of Aspergillus flavus Cell Walls via Releasing Surface Proteins, Inhibiting the Biosynthesis of β-1,3-Glucan and Promoting the Degradation of Chitin, and an Identification of Cell Surface Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Fungal Strain and Culture Conditions
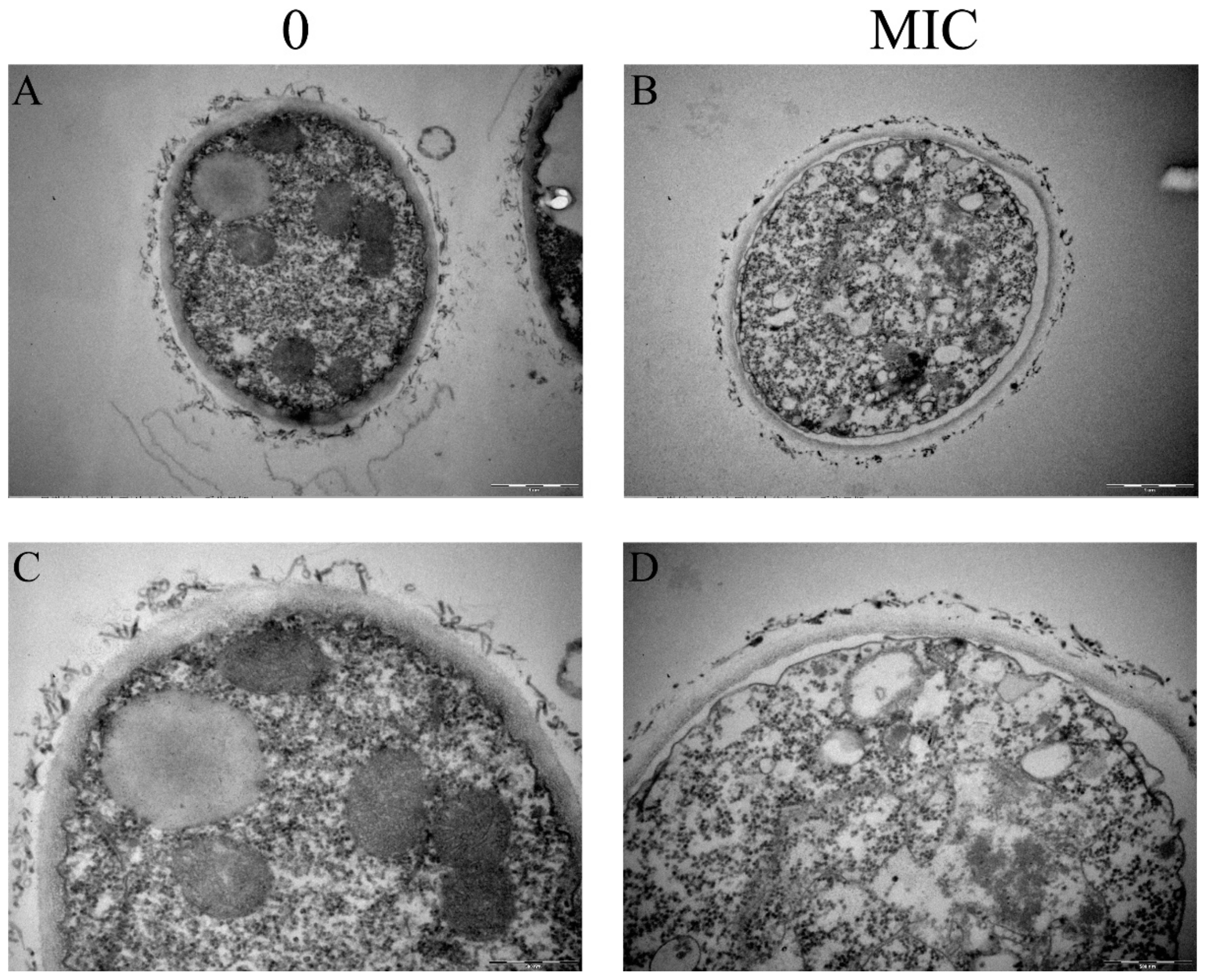
2.3. Transmission Electron Microscopy
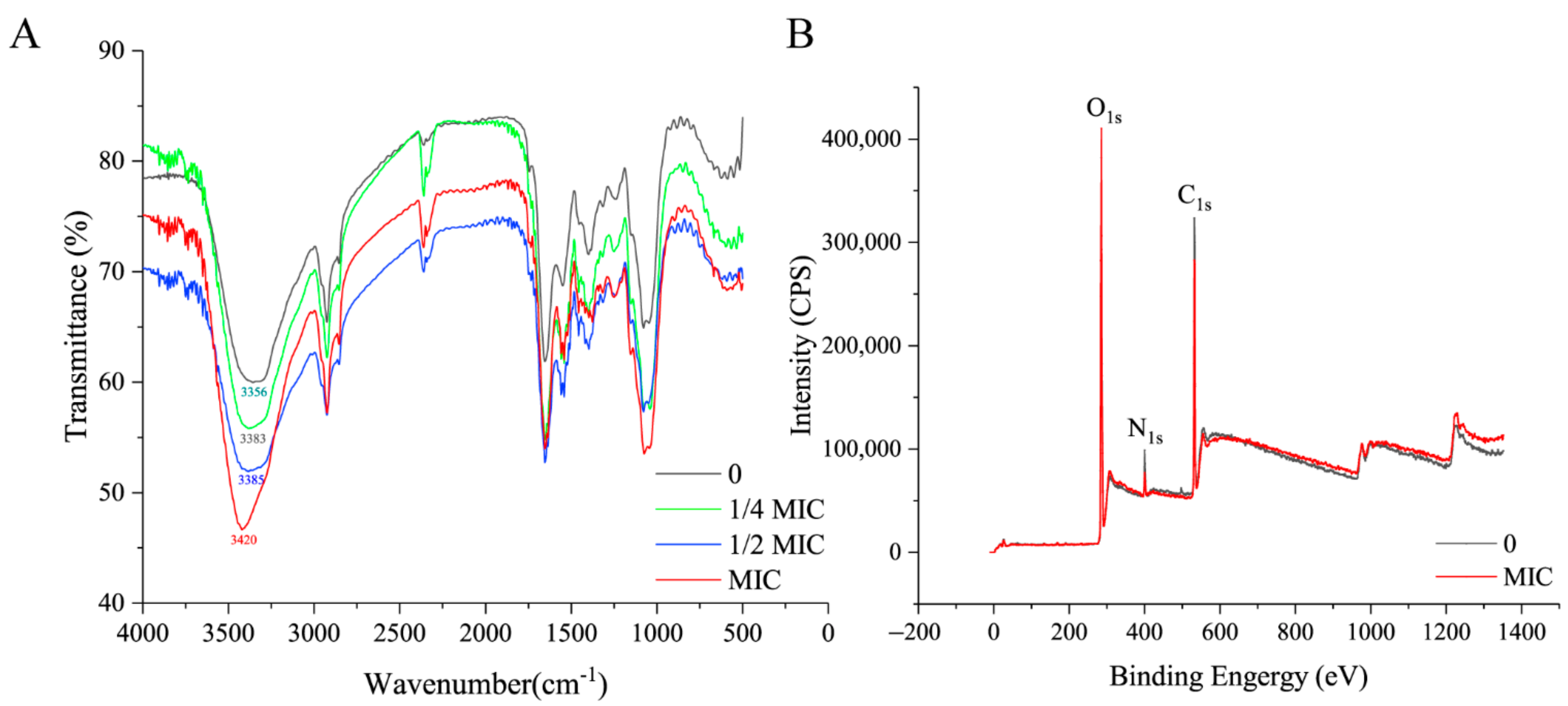
2.4. FT-IR Assay
2.5. XPS Assay
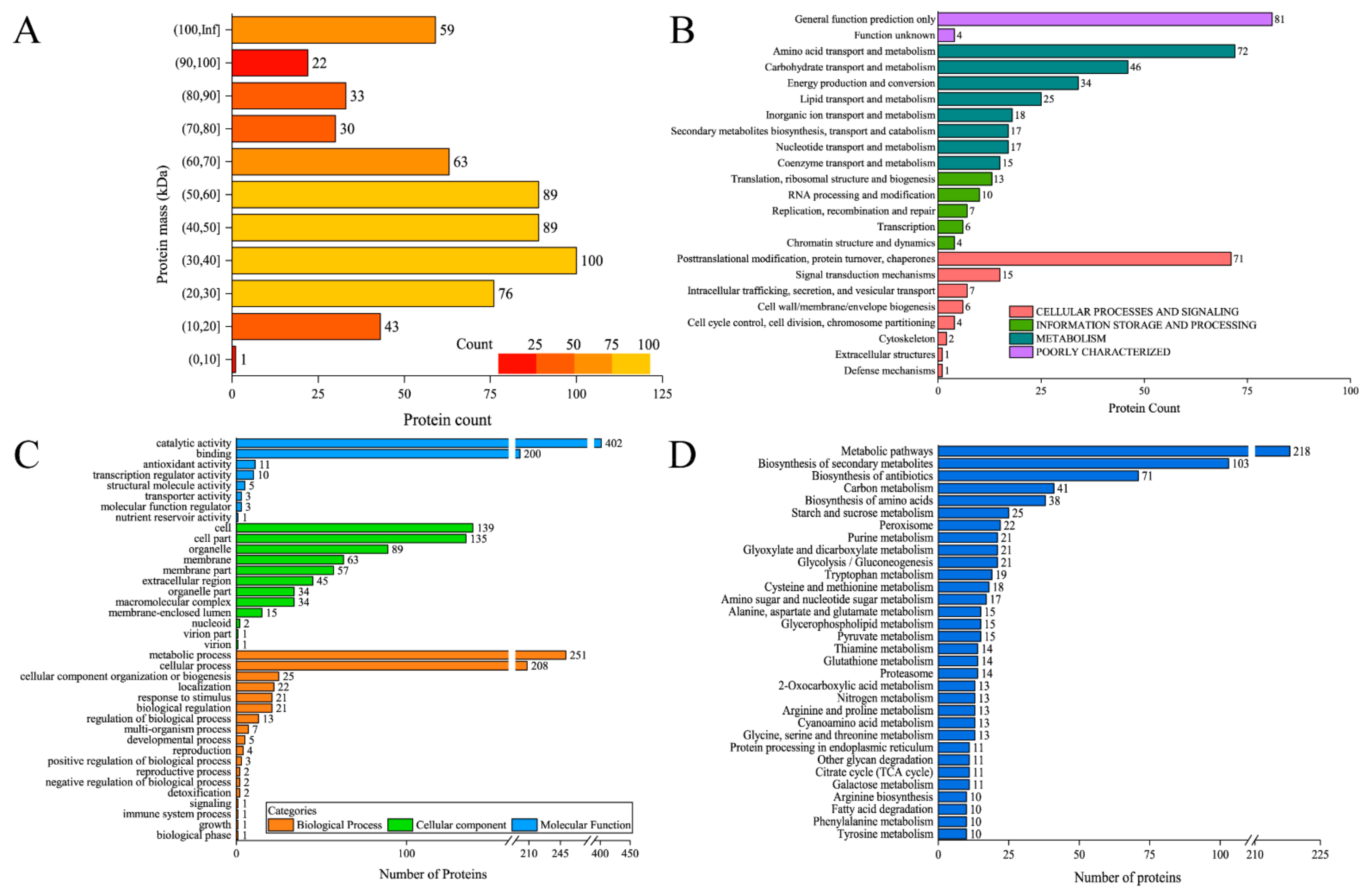
2.6. Proteomic Analysis of Cell Surface Proteins
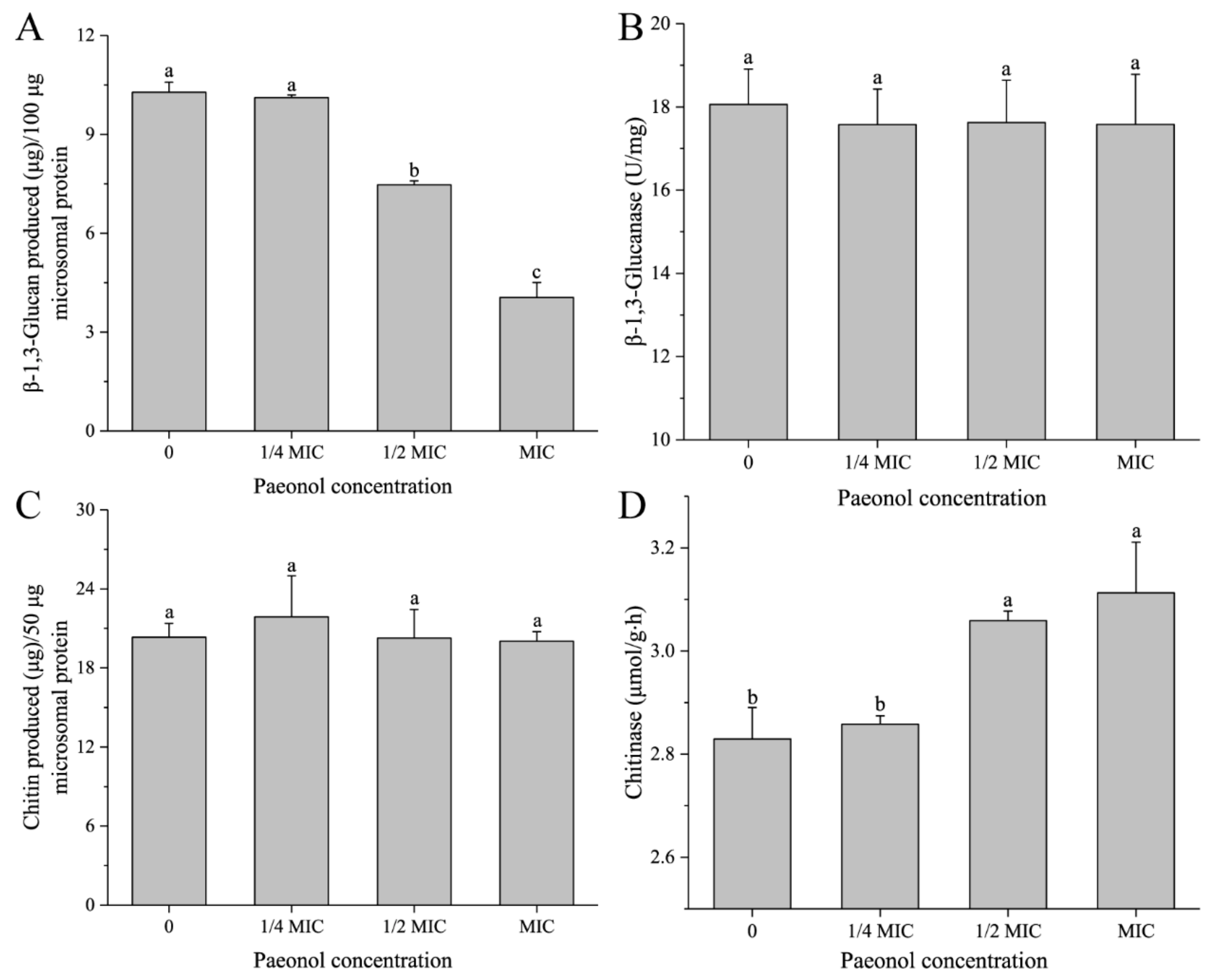
2.7. Determination of the Activity of β-1,3-Glucan Synthase
2.8. Determination of the Activity of Chitin Synthase
2.9. Determination of the Activities of β-1,3-Glucanase and Chitinase
2.10. Real-Time Quantitative Reverse Transcription PCR (QRT-PCR)
2.11. Antifungal Effect of Paeonol on Peanut Butter
2.12. Statistical Analysis
3. Results and Discussion
3.1. Paeonol Destroyed Both the Outer and Inner Layer of Cell Walls
3.2. Paeonol Changed OH Groups, Released Surface Proteins, and Increased Lipid Content
3.3. Identification of the Composition of Surface Proteins
3.4. Paeonol Regulated the Activities of Enzymes Participating in β-1,3-Glucan and Chitin Metabolism
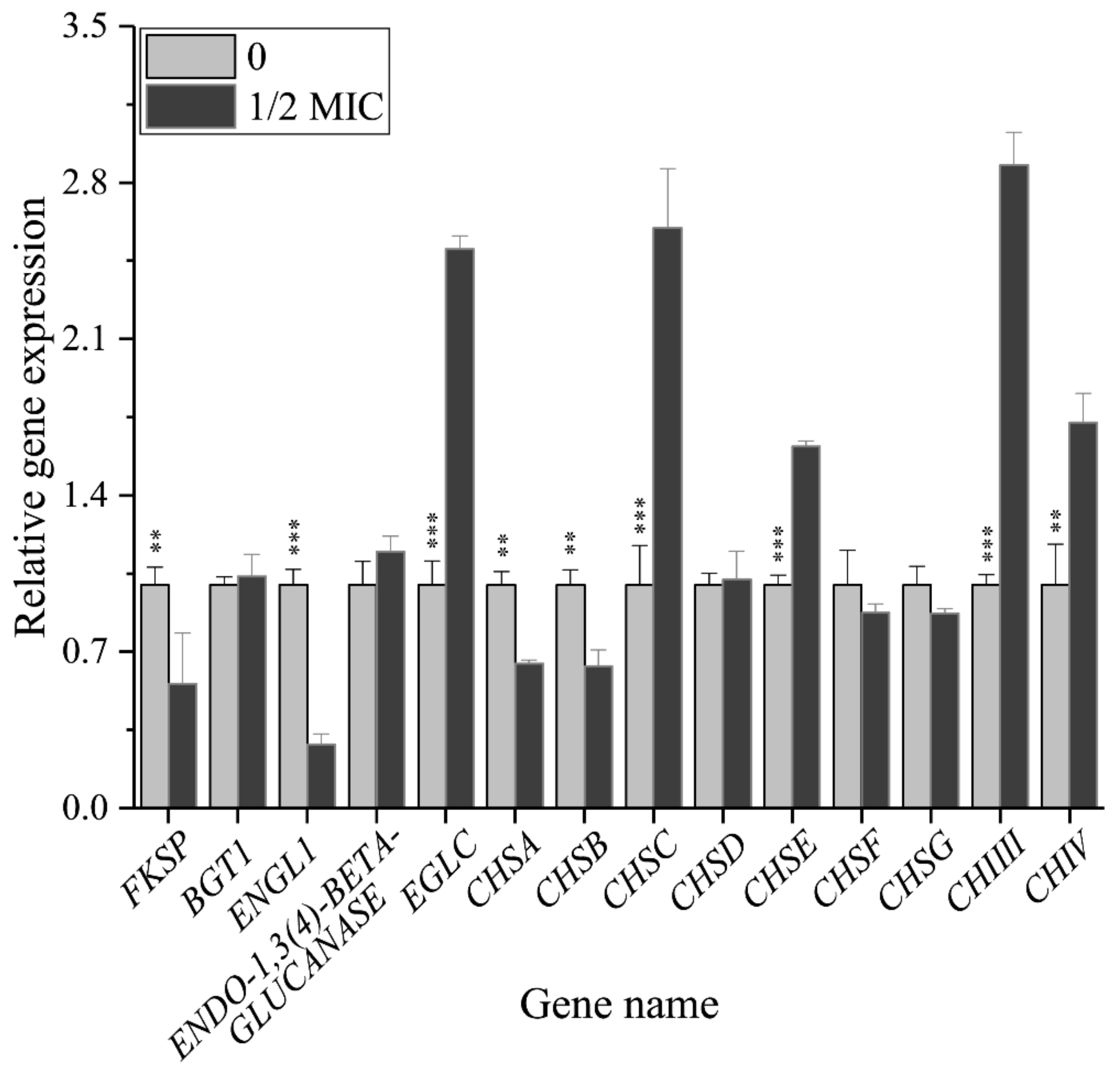
3.5. Paeonol Regulated the Relative Expressions of Genes Encoding the Enzymes Participating in Β-1,3-Glucan and Chitin Metabolism
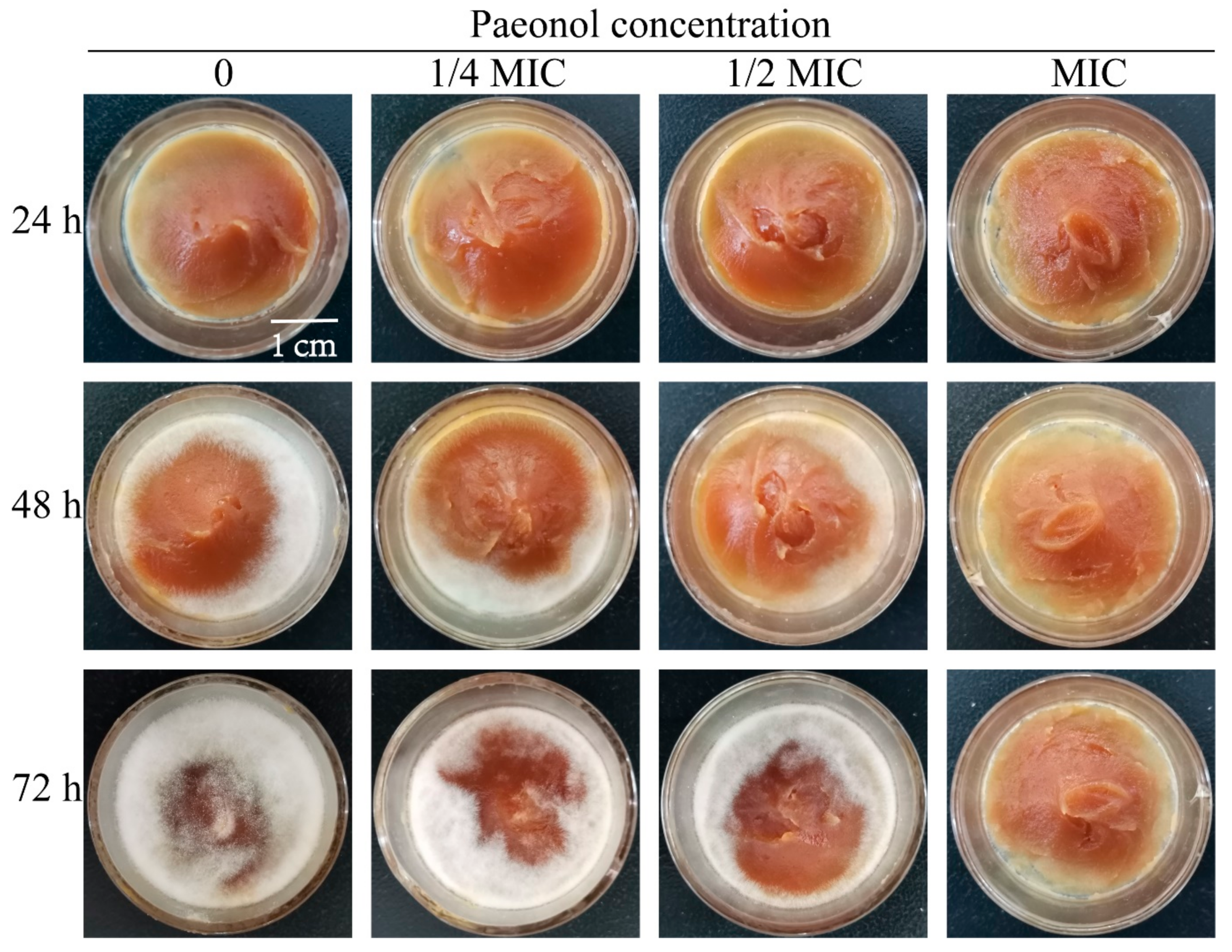
3.6. Paeonol Can Effectively Control the Pathogenicity of A. flavus on Peanut Butter
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rudramurthy, S.M.; Paul, R.A.; Chakrabarti, A.; Mouton, J.W.; Meis, J.F. Invasive aspergillosis by Aspergillus flavus: Epidemiology, diagnosis, antifungal resistance, and management. J. Fungi 2019, 5, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Z.; Zhang, Y.; Yu, W.; Zhang, J.; Wang, J.; Wan, F.; Kim, Y.; Liu, Y.; Kou, X. Recent advances in aflatoxin B1 detection based on nanotechnology and nanomaterials—A review. Anal. Chim. Acta 2019, 1069, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Klingelhöfer, D.; Zhu, Y.; Braun, M.; Bendels, M.H.K.; Brüggmann, D.; Groneberg, D.A. Aflatoxin—publication analysis of a global health threat. Food Control 2018, 89, 280–290. [Google Scholar] [CrossRef]
- Yu, J.; Cleveland, T.E.; Bennett, J.W. Aspergillus flavus genomics: Gateway to human and animal health, food safety, and crop resistance to diseases. Rev. Iberoam. Micol. 2005, 22, 194–202. [Google Scholar] [CrossRef]
- Lima, S.L.; Colombo, A.L.; de Almeida Júnior, J.N. Fungal cell wall: Emerging antifungals and drug resistance. Front. Microbiol. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Gow, N.A.R.; Latgé, J.P.; Munro, C.A. The fungal cell wall: Structure, biosynthesis, and function. Microbiol. Spectr. 2017, 5, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic Fungal Cell Wall Architecture in Stress Adaptation and Immune Evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef]
- Liu, W.; Yuan, L.; Wang, S.Z. Recent progress in the discovery of antifungal agents targeting the cell wall. J. Med. Chem. 2020, 63, 12429–12459. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, X.M.; Xie, Y.L.; Zhong, Y. o-Vanillin, a promising antifungal agent, inhibits Aspergillus flavus by disrupting the integrity of cell walls and cell membranes. Appl. Microbiol. Biot. 2021, 105, 5147–5158. [Google Scholar] [CrossRef]
- Xie, X.; Lipke, P.N. On the evolution of fungal and yeast cell walls. Yeast 2010, 27, 479–488. [Google Scholar] [CrossRef] [Green Version]
- Latgé, J.P. Tasting the fungal cell wall. Cell. Microbiol. 2010, 12, 863–872. [Google Scholar] [CrossRef]
- Gravelat, F.N.; Beauvais, A.; Liu, H.; Lee, M.J.; Snarr, B.D.; Chen, D.; Xu, W.; Kravtsov, I.; Hoareau, C.M.Q.; Vanier, G.; et al. Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal beta-glucan from the immune system. PLoS Pathog. 2013, 9, e1003575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocio, G.R.; Haroldo, C.O.; Rivera, J.; Nuria, T.C. The fungal cell wall: Candida, Cryptococcus, and Aspergillus Species. Front. Microbiol. 2019, 10, 2993. [Google Scholar]
- Briard, B.; Muszkieta, L.; Latge, J.P.; Fontaine, T. Galactosaminogalactan of Aspergillus fumigatus, a bioactive fungal polymer. Mycologia 2016, 108, 572–580. [Google Scholar] [CrossRef] [Green Version]
- OuYang, Q.L.; Duan, X.F.; Li, L.; Tao, N.G. Cinnamaldehyde Exerts Its Antifungal Activity by Disrupting the Cell Wall Integrity of Geotrichum citri-aurantii. Front. Microbiol. 2019, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhu, X.M.; Xie, Y.L.; Ren, S.L. 2-Hydroxy-4-methoxybenzaldehyde inhibits the growth of Aspergillus flavus via damaging cell wall, cell membrane, manipulating respiration thus creating a promising antifungal effect on corn kernels. Int. J. Food Sci. Technol. 2020, 56, 178–184. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.; Zhu, X.M.; Xie, Y.L. Antifungal efficacy of paeonol on Aspergillus flavus and its mode of action on cell walls and cell membranes. LWT 2021, 149, 111985. [Google Scholar] [CrossRef]
- Lee, K.K.; Maccallum, D.M.; Jacobsen, M.D.; Walker, L.A.; Odds, F.C.; Gow, N.A.; Munro, C.A. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 2012, 56, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, P.M.; Tupe, S.G.; Deshpande, M.V. Chitin synthase inhibitors as antifungal agents. Mini Rev. Med. Chem. 2013, 13, 222–236. [Google Scholar] [PubMed]
- Hu, L.L.; Ban, F.F.; Li, H.B.; Qian, P.P.; Shen, Q.S.; Zhao, Y.Y.; Mo, H.Z.; Zhou, X. Thymol induces conidial apoptosis in Aspergillus flavus via stimulating K+ eruption. J. Agric. Food Chem. 2018, 66, 8530–8536. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.H.J.; Neucere, J.N.; Cleveland, T.E. Surface proteins of two aflatoxin-producing isolates of Aspergillus flavus and Aspergillus parasiticus mycelia. 2. HPLC mapping by gel-permeation, ion-exchange, and reverse-phase chromatography. J. Agri. Food Chem. 1992, 40, 1613–1616. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Selvakumar, D.; Miyamoto, M.; Furuichi, Y.; Komiyama, T. Inhibition of fungal β-1,3-glucan synthase and cell growth by HM-1 killer toxin single-chain anti-idiotypic antibodies. Antimicrob. Agents Chemother. 2006, 50, 3090–3097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belewa, V.; Baijnath, H.; Frost, C.; Somai, B.M. Tulbaghia violacea Harv. plant extract affects cell wall synthesis in Aspergillus flavus. J. Appl. Microbiol. 2017, 122, 921–931. [Google Scholar]
- Magellan, H.; Boccara, M.; Drujon, T.; Soulie, M.C.; Guillou, C.; Dubois, J.; Becker, H.F. Discovery of two new inhibitors of Botrytis cinerea chitin synthase by a chemical library screening. Bioorgan. Med. Chem. 2013, 21, 4997–5003. [Google Scholar] [CrossRef]
- Zeng, R.; Zhang, A.; Chen, J.; Fu, Y. Postharvest quality and physiological responses of clove bud extract dip on ‘Newhall’ navel orange. Sci. Hortic. 2012, 138, 253–258. [Google Scholar] [CrossRef]
- Hua, K.C.; Feng, J.T.; Yang, X.G.; Wang, F.; Zhang, H.; Yang, L.; Zhang, H.R.; Xu, M.Y.; Li, J.K.; Qiao, R.Q.; et al. Assessment of the defatting efficacy of mechanical and chemical treatment for allograft cancellous bone and its effects on biomechanics properties of bone. Orthop. Surg. 2020, 12, 617–630. [Google Scholar] [CrossRef]
- Yang, N.; Qiu, F.; Zhu, F.; Qi, L. Therapeutic potential of zinc oxide-loaded syringic acid against in vitro and in vivo model of lung cancer. Int. J. Nanomed. 2020, 15, 8249–8260. [Google Scholar] [CrossRef]
- Wang, Y.; Feng, K.W.; Yang, H.H.; Yuan, Y.H.; Yue, T.L. Antifungal mechanism of cinnamaldehyde and citral combination against Penicillium expansum based on FT-IR fingerprint, plasma membrane, oxidative stress and volatile profile. RSC Adv. 2018, 8, 5806–5815. [Google Scholar] [CrossRef] [Green Version]
- Serrano, L.; Egües, I.; Alriols, M.G.; Llano, P.R.; Labidi, J. Miscanthus sinensis fractionation by different reagents. Chem. Eng. J. 2010, 156, 49–55. [Google Scholar] [CrossRef]
- Dague, E.; Delcorte, A.; Latgé, J.P.; Dufrêne, Y.F. Combined use of atomic force microscopy, X-ray photoelectron spectroscopy, and secondary ion mass spectrometry for cell surface analysis. Langmuir 2008, 24, 2955–2959. [Google Scholar] [CrossRef] [PubMed]
- Beauvais, A.; Latgé, J.P. Special issue: Fungal cell wall. J. Fungi 2018, 4, 91. [Google Scholar] [CrossRef] [Green Version]
- Latgé, J.P.; Beauvais, A.; Chamilos, G. The cell wall of the human fungal pathogen Aspergillus fumigatus: Biosynthesis, organization, immune response, and virulence. Annu. Rev. Microbiol. 2017, 71, 99–116. [Google Scholar] [CrossRef] [PubMed]
- Mouyna, I.; Hartl, L.; Latgé, J.P. β-1,3-glucan modifying enzymes in Aspergillus fumigatus. Front. Microbiol. 2013, 4, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, W.; Li, C.; Liang, J.; Sun, S. The Aspergillus fumigatus β-1,3-glucanosyltransferase Gel7 plays a compensatory role in maintaining cell wall integrity under stress conditions. Glycobiology 2014, 24, 418–427. [Google Scholar] [CrossRef] [Green Version]
- Mouyna, I.; Aimanianda, V.; Hartl, L.; Prevost, M.C.; Sismeiro, O.; Dillies, M.A.; Jagla, B.; Legendre, R.; Coppee, J.Y.; Latgé, J.P. GH16 and GH81 family β-(1,3)-glucanases in Aspergillus fumigatus are essential for conidial cell wall morphogenesis. Cell. Microbiol. 2016, 18, 1285–1293. [Google Scholar] [CrossRef] [Green Version]
- Gastebois, A.; Clavaud, C.; Aimanianda, V.; Latgé, J.P. Aspergillus fumigatus: Cell wall polysaccharides, their biosynthesis and organization. Future Microbiol. 2009, 4, 583–595. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Yuan, K.; Zhou, Q.; Lu, C.; Du, L.; Liu, F. Mechanism of antifungal activity of Perilla frutescens essential oil against Aspergillus flavus by transcriptomic analysis. Food Control 2021, 123, 107703–107733. [Google Scholar] [CrossRef]
- Shreaz, S.; Wani, W.A.; Behbehani, J.M.; Raja, V.; Irshad, M.; Karched, M.; Ali, I.; Siddiqi, W.A.; Hun, L.T. Cinnamaldehyde and its derivatives, a novel class of antifungal agents. Fitoterapia 2016, 112, 116–131. [Google Scholar] [CrossRef] [PubMed]

| %C | %N | %O | N/C | O/C | N/O | %CPr | %CPs | %CLp | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 72.57 | 5.57 | 21.86 | 0.08 | 0.30 | 0.25 | 19.96 | 18.45 | 34.15 |
| MIC | 77.72 | 3.21 | 19.07 | 0.04 | 0.25 | 0.17 | 11.51 | 18.40 | 47.81 |
| Pathway | Protein Name |
|---|---|
| Other glycan degradation | XP_041140530.1;XP_041141235.1;XP_041141593.1;XP_041142346.1;XP_041144446.1;XP_041145896.1;XP_041147338.1;XP_041147528.1;XP_041148927.1;XP_041151327.1;XP_041151574.1 |
| Various types of N-glycan biosynthesis | XP_041141235.1;XP_041142920.1;XP_041147528.1 |
| N-Glycan biosynthesis | XP_041142920.1;XP_041146481.1;XP_041146483.1 |
| Glycosaminoglycan degradation | XP_041141235.1;XP_041147528.1;XP_041151178.1 |
| Aflatoxin biosynthesis | XP_041143468.1;XP_041147095.1;XP_041147899.1 |
| Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | XP_041148932.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Zhao, Y.; Xie, Y. Paeonol Disrupts the Integrity of Aspergillus flavus Cell Walls via Releasing Surface Proteins, Inhibiting the Biosynthesis of β-1,3-Glucan and Promoting the Degradation of Chitin, and an Identification of Cell Surface Proteins. Foods 2021, 10, 2951. https://doi.org/10.3390/foods10122951
Li Q, Zhao Y, Xie Y. Paeonol Disrupts the Integrity of Aspergillus flavus Cell Walls via Releasing Surface Proteins, Inhibiting the Biosynthesis of β-1,3-Glucan and Promoting the Degradation of Chitin, and an Identification of Cell Surface Proteins. Foods. 2021; 10(12):2951. https://doi.org/10.3390/foods10122951
Chicago/Turabian StyleLi, Qian, Ying Zhao, and Yanli Xie. 2021. "Paeonol Disrupts the Integrity of Aspergillus flavus Cell Walls via Releasing Surface Proteins, Inhibiting the Biosynthesis of β-1,3-Glucan and Promoting the Degradation of Chitin, and an Identification of Cell Surface Proteins" Foods 10, no. 12: 2951. https://doi.org/10.3390/foods10122951






