Genomic and Metabolic Features of an Unexpectedly Predominant, Thermophilic, Assistant Starter Microorganism, Thermus thermophilus, in Chinese Inner Mongolian Cheese
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chinese Inner Mongolian Cheese Samples and Food-Borne Pathogens Detection
2.2. High-Throughput Sequencing Based on the Bacterial 16S rRNA Genes of Cheese Samples
2.3. Strain Isolation and Genome DNA Extraction
2.4. Genome Sequencing and Assembly
2.5. Genome Component Prediction
2.6. Genome Annotation
2.7. Verification of the Milk Fermentation Ability and Characteristics of T. thermophilus N-1
3. Results
3.1. Relative Abundance of T. thermophilus in the Bacterial Community of Chinese Inner Mongolian Cheese
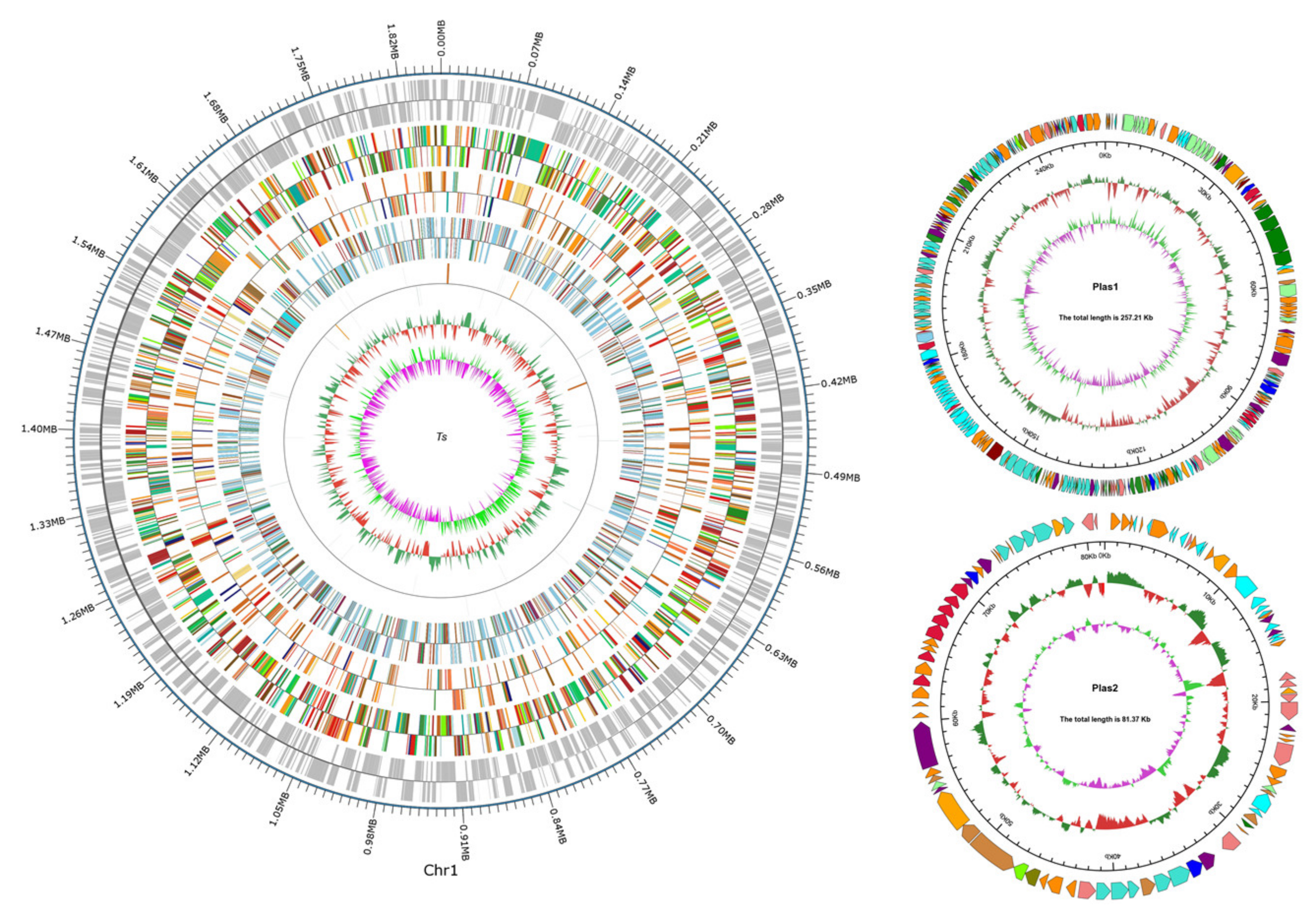
3.2. General Genome Features of T. thermophilus N-1
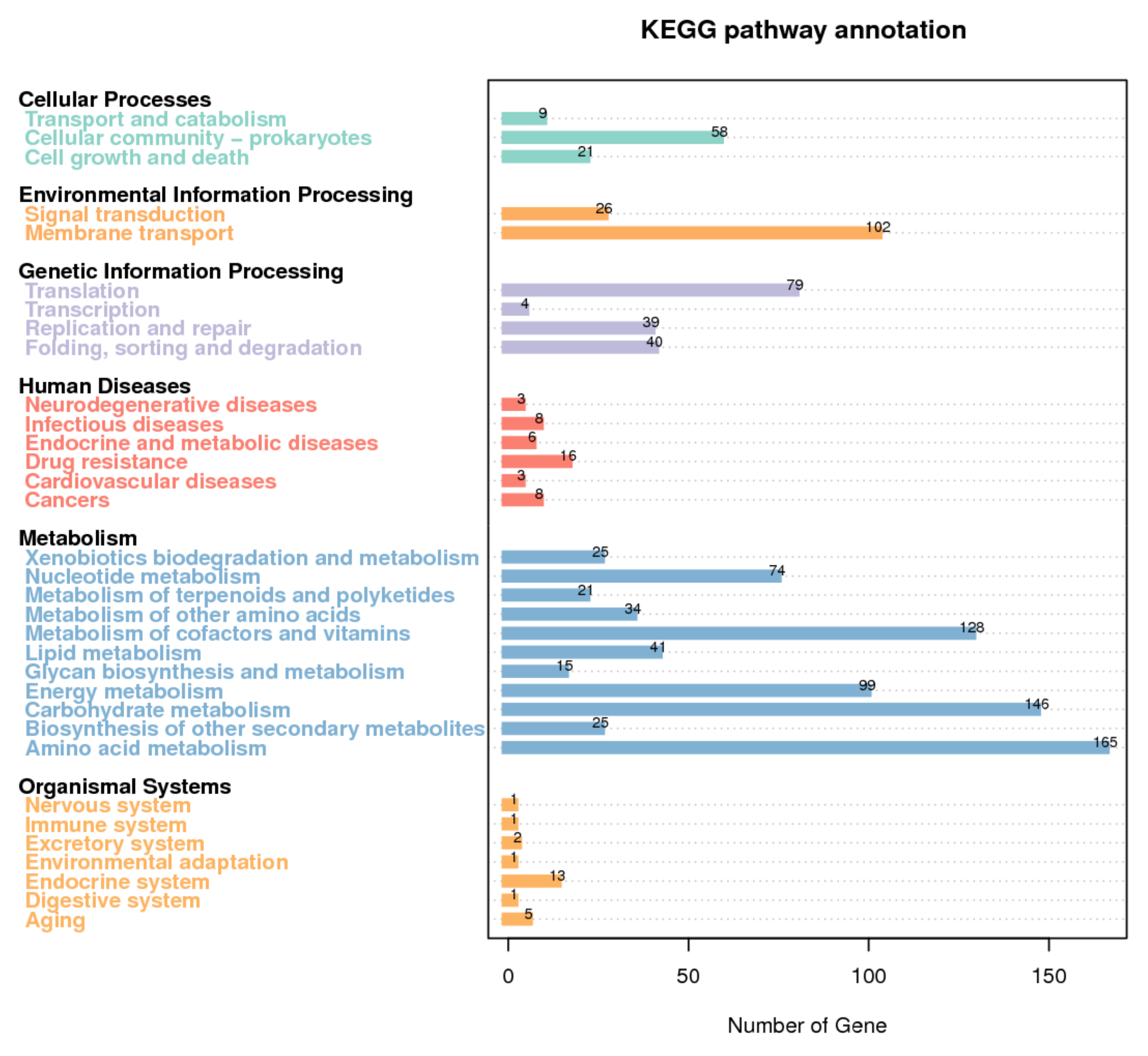
3.3. Functional Annotation of T. thermophilus N-1
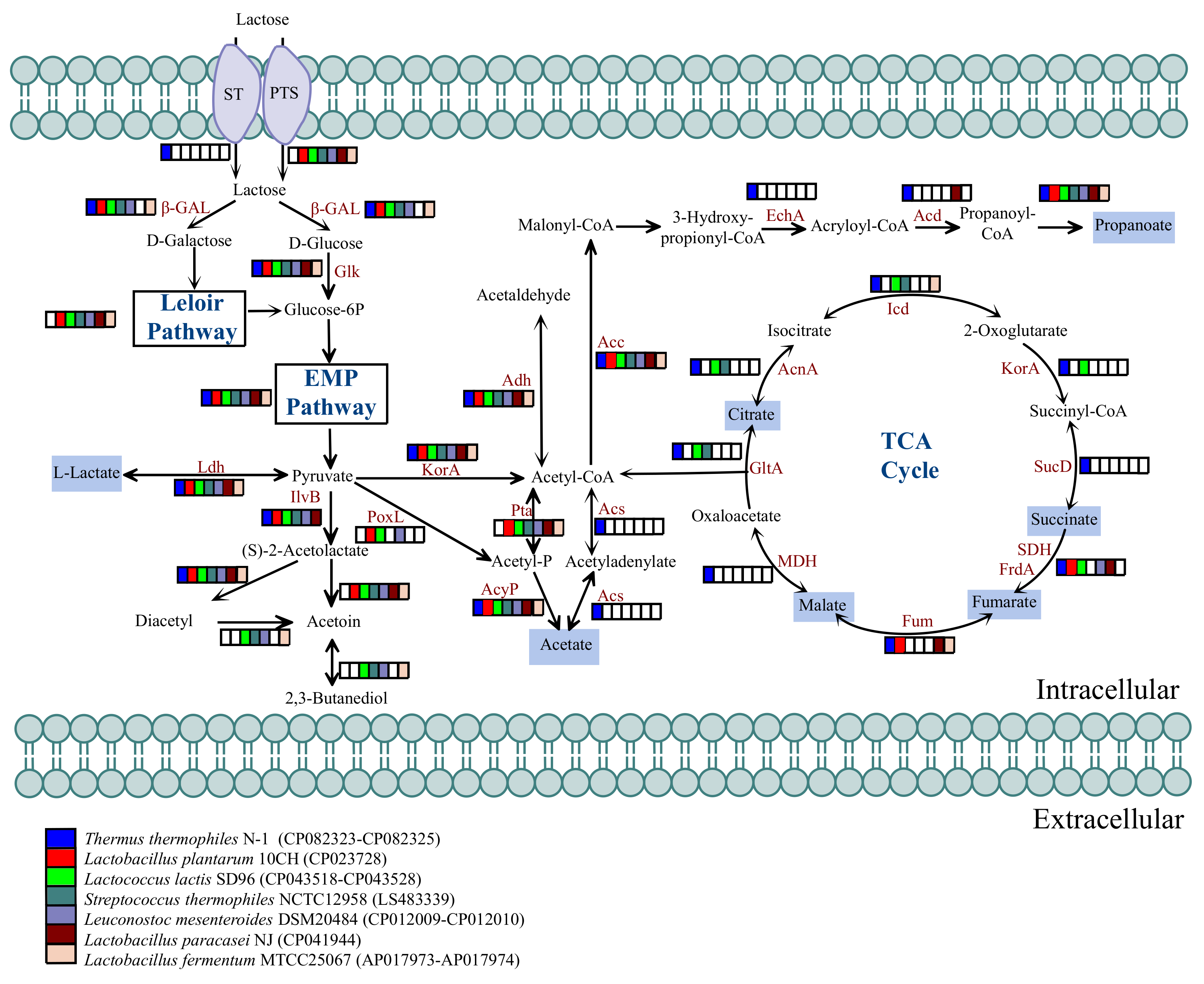
3.4. Distribution of Enzymes Associated with Lactose Metabolism
3.5. Distribution of Enzymes Associated with Organic Acid Generation
3.6. Validation of T. thermophilus N-1 in Milk during Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khattab, A.R.; Guirguis, H.A.; Tawfik, S.M.; Farag, M.A. Cheese ripening: A review on modern technologies towards flavor enhancement, process acceleration and improved quality assessment. Trends Food Sci. Technol. 2019, 88, 343–360. [Google Scholar] [CrossRef]
- Gao, M.L.; Hou, H.M.; Teng, X.X.; Zhu, Y.L.; Hao, H.S.; Zhang, G.L. Microbial diversity in raw milk and traditional fermented dairy products (Hurood cheese and Jueke) from Inner Mongolia, China. Genet. Mol. Res. 2017, 16, 1–3. [Google Scholar] [CrossRef]
- Al-Khusaibi, M. Arab Traditional Foods: Preparation, Processing and Nutrition BT—Traditional Foods: History, Preparation, Processing and Safety; Al-Khusaibi, M., Al-Habsi, N., Shafiur Rahman, M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 9–35. ISBN 978-3-030-24620-4. [Google Scholar]
- Davidson, P.M.; Critzer, F.M. Interventions to Inhibit or Inactivate Bacterial Pathogens in Foods BT—Microbial Food Safety: An Introduction; Oyarzabal, O.A., Backert, S., Eds.; Springer: New York, NY, USA, 2012; pp. 189–202. ISBN 978-1-4614-1177-2. [Google Scholar]
- Aldrete-Tapia, A.; Escobar-Ramírez, M.C.; Tamplin, M.L.; Hernández-Iturriaga, M. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef]
- Dugat-Bony, E.; Garnier, L.; Denonfoux, J.; Ferreira, S.; Sarthou, A.S.; Bonnarme, P.; Irlinger, F. Highlighting the microbial diversity of 12 French cheese varieties. Int. J. Food Microbiol. 2016, 238, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Joung, J.Y.; Choi, Y.S.; Kim, Y.; Oh, N.S. Characterisation of microbial diversity and chemical properties of Cheddar cheese prepared from heat-treated milk. Int. Dairy J. 2016, 63, 92–98. [Google Scholar] [CrossRef]
- Wei, W.; Hu, X.; Hou, Z.; Wang, Y.; Zhu, L. Microbial community structure and diversity in different types of non-bovine milk. Curr. Opin. Food Sci. 2021, 40, 51–57. [Google Scholar] [CrossRef]
- Issa, A.T.; Tahergorabi, R. Chapter 22—Milk Bacteria and Gastrointestinal Tract: Microbial Composition of Milk. In Dietary Interventions in Gastrointestinal Diseases; Watson, R.R., Preedy, V.R.B.T.-B.F., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 265–275. ISBN 978-0-12-814468-8. [Google Scholar]
- Park, A.-R.; Oh, D.-K. Galacto-oligosaccharide production using microbial β-galactosidase: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 85, 1279–1286. [Google Scholar] [CrossRef]
- Pantazaki, A.A.; Papaneophytou, C.P.; Pritsa, A.G.; Liakopoulou-Kyriakides, M.; Kyriakidis, D.A. Production of polyhydroxyalkanoates from whey by Thermus thermophilus HB8. Process Biochem. 2009, 44, 847–853. [Google Scholar] [CrossRef]
- Thybo, C.D.; Lillevang, S.K.; Skibsted, L.H.; Ahrné, L. Calcium balance during direct acidification of milk for Mozzarella cheese production. LWT 2020, 131, 109677. [Google Scholar] [CrossRef]
- Li, J.; Huang, Q.; Zheng, X.; Ge, Z.; Lin, K.; Zhang, D.; Chen, Y.; Wang, B.; Shi, X. Investigation of the Lactic Acid Bacteria in Kazak Cheese and Their Contributions to Cheese Fermentation. Front. Microbiol. 2020, 11, 228. [Google Scholar] [CrossRef]
- Akalin, A.S.; Gönç, S.; Akbaş, Y. Variation in Organic Acids Content during Ripening of Pickled White Cheese. J. Dairy Sci. 2002, 85, 1670–1676. [Google Scholar] [CrossRef] [Green Version]
- Bluma, A.; Ciprovica, I. Diversity of lactic acid bacteria in raw milk. Res. Rural Dev. 2015, 1, 157–161. [Google Scholar]
- Xiao, F.; Zhu, J.-J.; Zhang, H.-M.; Ya, M.; Yong, F. Inner Mongolian traditional dairy product processing technology and hygiene quality analysis. China Diary Ind. 2016, 44, 35–42. [Google Scholar]
- D’amico, D.J.; Groves, E.; Donnelly, C.W. Low Incidence of Foodborne Pathogens of Concern in Raw Milk Utilized for Farmstead Cheese Production. J. Food Prot. 2008, 71, 1580–1589. [Google Scholar] [CrossRef]
- Nalepa, B.; Markiewicz, L.H. PCR-DGGE markers for qualitative profiling of microbiota in raw milk and ripened cheeses. LWT 2017, 84, 168–174. [Google Scholar] [CrossRef]
- Holbrook, R.; Anderson, J.M.; Baird-Parker, A.C. The Performance of a Stable Version of Baird-Parker’s Medium for Isolating Staphylococcus aureus. J. Appl. Bacteriol. 1969, 32, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Leong, W.; Geier, R.; Engstrom, S.; Ingham, S.; Ingham, B.; Smukowski, M. Growth of Listeria monocytogenes, Salmonella spp., Escherichia coli O157:H7, and Staphylococcus aureus on Cheese during Extended Storage at 25 °C. J. Food Prot. 2014, 77, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zeng, C.; Yang, S.; Hou, Z.; Wang, Y.; Hu, X.; Senoo, K.; Wei, W. Diversity and specificity of the bacterial community in Chinese horse milk cheese. Microbiologyopen 2020, 9, e1066. [Google Scholar] [CrossRef] [PubMed]
- Reiner, J.; Pisani, L.; Qiao, W.; Singh, R.; Yang, Y.; Shi, L.; Khan, W.A.; Sebra, R.; Cohen, N.; Babu, A. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet–Biedl Syndrome 9 (BBS9) deletion. Npj Genom. Med. 2018, 3, 3. [Google Scholar] [CrossRef]
- Ardui, S.; Ameur, A.; Vermeesch, J.R.; Hestand, M.S. Single molecule real-time (SMRT) sequencing comes of age: Applications and utilities for medical diagnostics. Nucleic Acids Res. 2018, 46, 2159–2168. [Google Scholar] [CrossRef] [Green Version]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.; Bridges, S.; Magbanua, Z.V.; Peterson, D.G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 2008, 36, 2284–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Moxon, S.; Marshall, M.; Khanna, A.; Eddy, S.R.; Bateman, A. Rfam: Annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005, 33, 121–124. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Lu, Z.; Wang, S.; Jing-Yan Wang, J.; Gao, X. CMsearch: Simultaneous exploration of protein sequence space and structure space improves not only protein homology detection but also protein structure prediction. Bioinformatics 2016, 32, i332–i340. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.; Wan, I.; Jones, S.J.; Brinkman, F.S.L. IslandPath: Aiding detection of genomic islands in prokaryotes. Bioinformatics 2003, 19, 418–420. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Jaroszewski, L.; Godzik, A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 2002, 18, 77–82. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Zeppa, G.; Conterno, L.; Gerbi, V. Determination of organic acids, sugars, diacetyl, and acetoin in cheese by high-performance liquid chromatography. J. Agric. Food Chem. 2001, 49, 2722–2726. [Google Scholar] [CrossRef] [PubMed]
- Irene, G.-R.; Paula, G.; Borja, S.; Miguel, G.; Abelardo, M.; Rute, N.A. Catabolism of glucose and lactose in Bifidobacterium animalis subsp. lactis, studied by 13C nuclear magnetic resonance. Appl. Environ. Microbiol. 2013, 79, 7628–7638. [Google Scholar] [CrossRef] [Green Version]
- Comas, I.; González-Candelas, F.; Zúñiga, M. Unraveling the evolutionary history of the phosphoryl-transfer chain of the phosphoenolpyruvate:phosphotransferase system through phylogenetic analyses and genome context. BMC Evol. Biol. 2008, 8, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salminen, S.; Wright, A.V.; Lahtinen, S.; Ouwehand, A.C. Lactic Acid Bacteria: Microbiological and Functional Aspects; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Fox, P.F.; Wallace, J.M. Formation of flavor compounds in cheese. Adv. Appl. Microbiol. 1997, 45, 17–86. [Google Scholar]
- Sablé, S.; Cottenceau, G. Current knowledge of soft cheeses flavor and related compounds. J. Agric. Food Chem. 1999, 47, 4825–4836. [Google Scholar] [CrossRef]
- Singh, T.K.; Drake, M.A.; Cadwallader, K.R. Flavor of cheddar cheese: A chemical and sensory perspective. Compr. Rev. Food Sci. Food Saf. 2003, 2, 166–189. [Google Scholar] [CrossRef]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.-Y.; Wei, Y.-N.; Lin, L.-C.; Chen, S.-J.; Feng, P.-P.; Xiao, D.-G.; Lin, X.; Zhang, C.-Y. Increasing yield of 2,3,5,6-Tetramethylpyrazine in baijiu through Saccharomyces cerevisiae metabolic engineering. Front. Microbiol. 2020, 11, 2946. [Google Scholar] [CrossRef]
- Guinee, T.P. Salting and the role of salt in cheese. Int. J. Dairy Technol. 2004, 57, 99–109. [Google Scholar] [CrossRef]
- Marteinsson, V.T. Isolation and characterization of Thermus thermophilus Gy1211 from a deep-sea hydrothermal vent. Extremophiles 1999, 3, 247–251. [Google Scholar] [CrossRef]
- Farahnaky, A.; Mousavi, S.H.; Nasiri, M. Role of salt in Iranian ultrafiltered Feta cheese: Some textural and physicochemical changes during ripening. Int. J. Dairy Technol. 2013, 66, 359–365. [Google Scholar] [CrossRef]
- Drider, D.; Bekal, S.; Prévost, H. Genetic organization and expression of citrate permease in lactic acid bacteria. Genet. Mol. Res. 2004, 3, 273–281. [Google Scholar]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef] [Green Version]
- Pirttijärvi, T.S.M.; Wahlström, G.; Rainey, F.A.; Saris, P.E.J.; Salkinoja-Salonen, M.S. Inhibition of bacilli in industrial starches by nisin. J. Ind. Microbiol. Biotechnol. 2001, 26, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W. Use of starter cultures in fermentation on a household scale. Food Control 1997, 8, 241–258. [Google Scholar] [CrossRef]
- Park, M.K.; Kim, Y.-S. Comparative metabolic expressions of fermented soybeans according to different microbial starters. Food Chem. 2020, 305, 125461. [Google Scholar] [CrossRef] [PubMed]
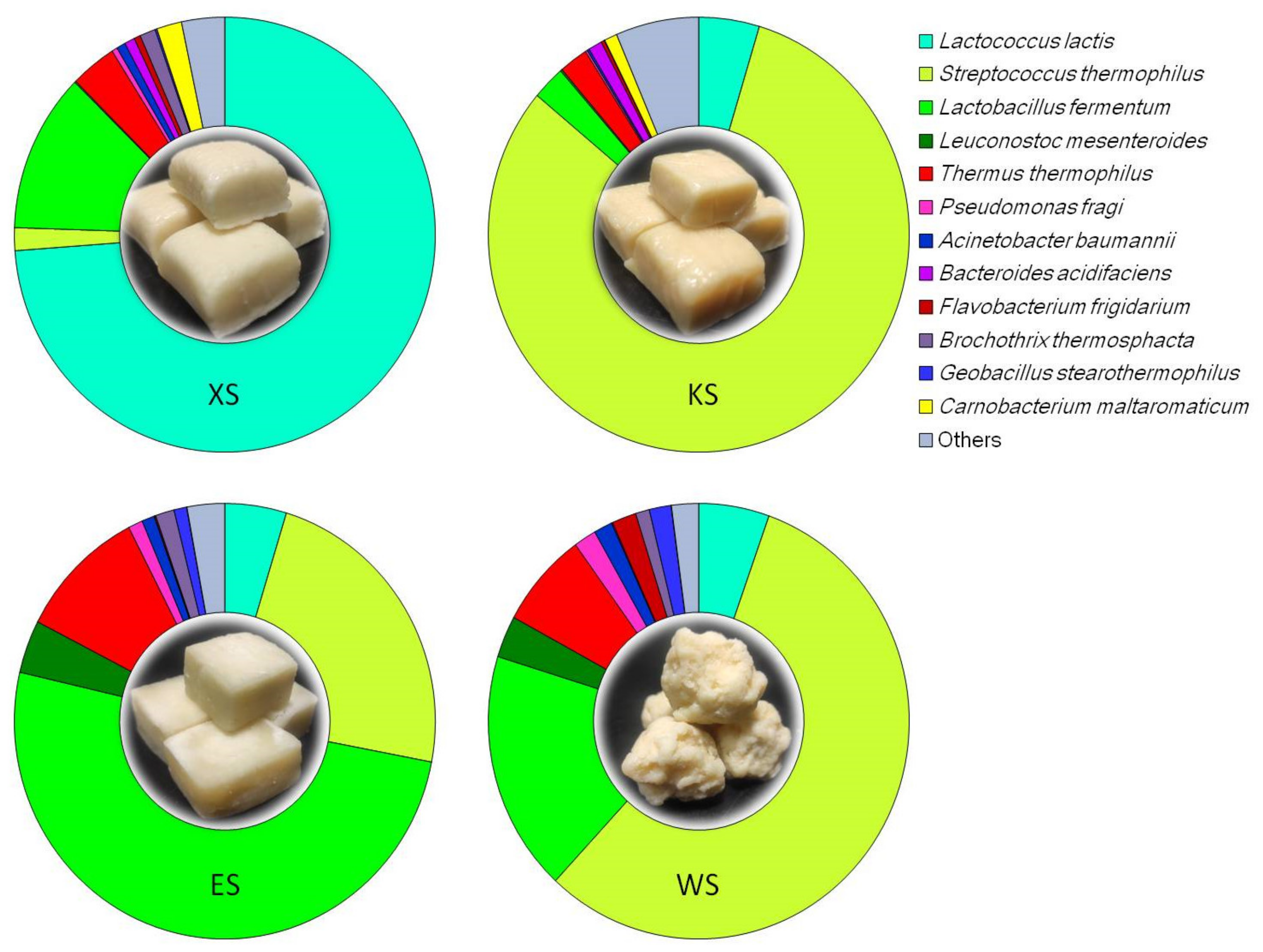

| Sample Name | Raw Reads | Clean Reads | OTU Number | Shannon Index | Simpson Index | Chao1 | ACE | Library Coverage |
|---|---|---|---|---|---|---|---|---|
| XS | 80076 | 72864 | 646 | 2.335 | 0.464 | 614.919 | 625.801 | 0.998 |
| KS | 73117 | 69204 | 403 | 1.103 | 0.235 | 398.943 | 409.096 | 0.998 |
| ES | 88637 | 80066 | 389 | 3.753 | 0.817 | 351.643 | 354.244 | 0.999 |
| WS | 87213 | 80137 | 413 | 3.884 | 0.816 | 390.818 | 400.815 | 0.999 |
| Features | Chromosome | Plasmid 1 | Plasmid 2 |
|---|---|---|---|
| Genome topology | Circular | Circular | Circular |
| Genome size (bp) | 1,855,628 | 257,206 | 81,373 |
| G+C content (mol%) | 69.72 | 69.33 | 67.95 |
| Open reading frames | 2328 | - | - |
| tRNA genes | 48 | - | - |
| rRNA genes | 6 | - | - |
| sRNA genes | 0 | - | - |
| GenBank accession number | CP082323 | CP082324 | CP082325 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Hou, Z.; Hu, X.; Liu, X.; Dai, T.; Wang, X.; Zeng, C.; Wang, Y.; Wang, C.; Yang, S.; et al. Genomic and Metabolic Features of an Unexpectedly Predominant, Thermophilic, Assistant Starter Microorganism, Thermus thermophilus, in Chinese Inner Mongolian Cheese. Foods 2021, 10, 2962. https://doi.org/10.3390/foods10122962
Zhu L, Hou Z, Hu X, Liu X, Dai T, Wang X, Zeng C, Wang Y, Wang C, Yang S, et al. Genomic and Metabolic Features of an Unexpectedly Predominant, Thermophilic, Assistant Starter Microorganism, Thermus thermophilus, in Chinese Inner Mongolian Cheese. Foods. 2021; 10(12):2962. https://doi.org/10.3390/foods10122962
Chicago/Turabian StyleZhu, Lin, Zhaozhi Hou, Xinyu Hu, Xu Liu, Tian Dai, Xinyu Wang, Chunlin Zeng, Yuan Wang, Caizheng Wang, Shujing Yang, and et al. 2021. "Genomic and Metabolic Features of an Unexpectedly Predominant, Thermophilic, Assistant Starter Microorganism, Thermus thermophilus, in Chinese Inner Mongolian Cheese" Foods 10, no. 12: 2962. https://doi.org/10.3390/foods10122962







