Impact of Plasma-Activated Water Treatment on Quality and Shelf-Life of Fresh Spinach Leaves Evaluated by Comprehensive Metabolomic Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Raw Materials
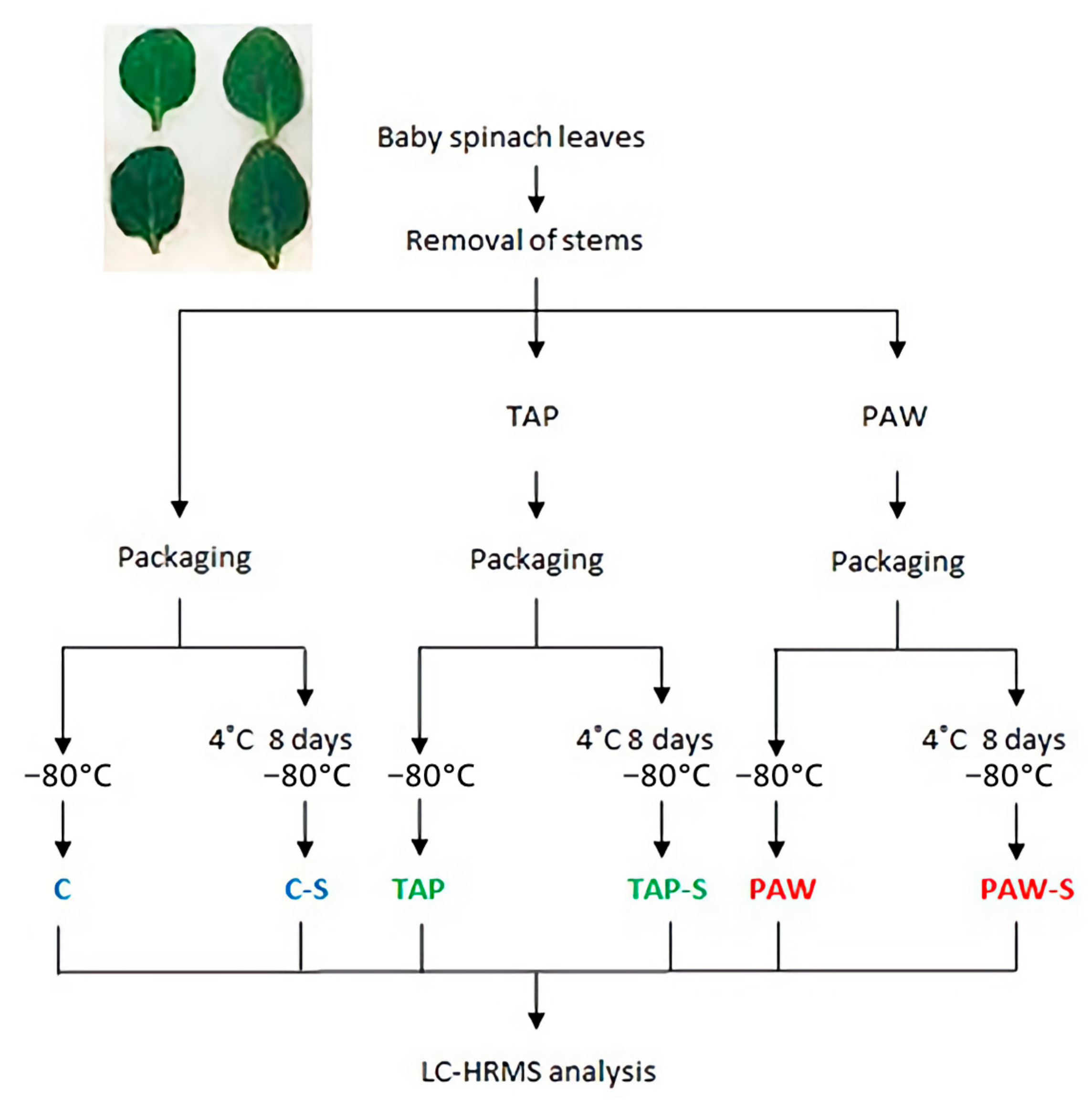
2.3. Sample Treatments and Experimental Design
2.4. PAW Generation and Characterisation
2.5. Untargeted Metabolomics Analysis by Liquid Chromatography High-Resolution Mass Spectrometry (LC-HRMS)
2.5.1. Sample Extraction
2.5.2. Untargeted LC-HRMS and LC-HRMS/MS
- (1)
- Reverse-phase (RP) chromatography using a Hypersil GOLD aQ (Thermo Fisher Scientific, San Jose, CA, USA; 100 × 2.1, 1.9 µm) column was performed by eluting the column with a mobile phase consisting of water + 0.1% formic acid (FA) (A) and MeOH + 0.1% FA (B). The elution proceeded isocratically at a constant flow rate of 0.4 mL/min for 0.5 min with 100% A, followed by linear gradient elution to 95% B in 4.5 min, which was retained until 12.5 min. After flushing the column with 95% B for 2 min, it was returned to the starting conditions and equilibrated for 2.5 min.
- (2)
- Hydrophilic interaction chromatography (HILIC) was performed using a zwitterionic SeQuant ZIC-pHILIC column (Merck, Darmstadt, Germany; 150 × 4.6 mm, 5 µm). The column was eluted with a mobile phase consisting of 20 mM ammonium carbonate (A, pH 8.3) and MeCN (B). Elution proceeded isocratically at a constant flow rate of 0.3 mL/min for 1 min with 80% B, followed by linear gradient elution to 20% B in 29 min. Subsequently, the column was flushed with 8% B for 5 min, and then returned to the starting conditions and equilibrated for 9 min.
2.5.3. Processing and QC of Metabolomics Data
2.6. Statistical Analysis
2.7. Annotation
2.7.1. Level of Identification
2.7.2. MS Peaks to Pathway Analysis
3. Results
3.1. Quality Assessment of LC-HRMS Datasets
3.2. Comparison of Processing Methods at Baseline (D1)
3.3. Comparison of Processing Methods after 8 Days of Refrigerated Storage (D8)
3.4. Comparison of Stored (D8) to Baseline (D1) Samples for Each Processing Method
3.5. Storage- and Processing-Induced Changes in Metabolic Pathways
4. Discussion
4.1. Storage Period as a Dominant Factor Determining Metabolite Profiles in Spinach Leaves
4.2. Characteristic Changes in the Metabolite Profiles of Stored Spinach Leaves
4.3. PAW Treatment Sustains Anti-Senescent Mechanisms in Detached Spinach Leaves
4.4. Enhancement of Carbohydrate Pathways by PAW Treatment May Have a Protective Effect
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Disclaimer
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lisiewska, Z.; Kmiecik, W.; Gębczyński, P.; Sobczyńska, L. Amino acid profile of raw and as-eaten products of spinach (Spinacia oleracea L.). Food Chem. 2011, 126, 460–465. [Google Scholar] [CrossRef]
- Roberts, J.L.; Moreau, R. Functional properties of spinach (Spinacia oleracea L.) phytochemicals and bioactives. Food Funct. 2016, 7, 3337–3353. [Google Scholar] [CrossRef]
- Puerta-Gomez, A.; Kim, J.; Moreira, R.; Klutke, G.-A.; Castell-Perez, M. Quantitative assessment of the effectiveness of intervention steps to reduce the risk of contamination of ready-to-eat baby spinach with Salmonella. Food Control 2012, 31, 410–418. [Google Scholar] [CrossRef]
- Gelting, R.J.; Baloch, M.; Zarate-Bermudez, M.A.; Selman, C. Irrigation water issues potentially related to the 2006 multistate E. coli O157:H7 outbreak associated with spinach. Agric. Water Manag. 2011, 98, 1395–1402. [Google Scholar] [CrossRef]
- Espenhain, L.; Riess, M.; Müller, L.; Colombe, S.; Ethelberg, S.; Litrup, E.; Jernberg, C.; Kühlmann-Berenzon, S.; Lindblad, M.; Hove, N.K.; et al. Cross-border outbreak of Yersinia enterocolitica O3 associated with imported fresh spinach, Sweden and Denmark, March 2019. Eurosurveillance 2019, 24, 1900368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirumdas, R.; Kothakota, A.; Annapure, U.; Siliveru, K.; Blundell, R.; Gatt, R.; Valdramidis, V.P. Plasma activated water (PAW): Chemistry, physico-chemical properties, applications in food and agriculture. Trends Food Sci. Technol. 2018, 77, 21–31. [Google Scholar] [CrossRef]
- Soni, A.; Choi, J.; Brightwell, G. Plasma-Activated Water (PAW) as a Disinfection Technology for Bacterial Inactivation with a Focus on Fruit and Vegetables. Foods 2021, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Herianto, S.; Hou, C.; Lin, C.; Chen, H. Nonthermal plasma-activated water: A comprehensive review of this new tool for enhanced food safety and quality. Compr. Rev. Food Sci. Food Saf. 2021, 20, 583–626. [Google Scholar] [CrossRef] [PubMed]
- Vaka, M.R.; Sone, I.; Álvarez, R.G.; Walsh, J.L.; Prabhu, L.; Sivertsvik, M.; Fernández, E.N. Towards the Next-Generation Disinfectant: Composition, Storability and Preservation Potential of Plasma Activated Water on Baby Spinach Leaves. Foods 2019, 8, 692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacher, J.A. Senescence and Postharvest Physiology. Annu. Rev. Plant Physiol. 1973, 24, 197–224. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Making Sense of Senescence (Molecular Genetic Regulation and Manipulation of Leaf Senescence). Plant Physiol. 1997, 113, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auria, M.D.; Scienze, D.; Basilicata, U.; Lucano, A. Increase of the shelf-life of vegetables. Int. J. Res. Stud. Sci. Eng. Technol. 2021, 8, 9–11. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2021, 61, 1448–1469. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Huerta, O.D.; Uhlig, S.; Ivanova, L.; Dang, T.T.; Rode, T.M.; Fernández, E.N.; Fæste, C.K. Metabolomics workflow for quality control of differently-processed pre-cooked chicken fillets. Food Chem. 2022, 370, 131006. [Google Scholar] [CrossRef] [PubMed]
- Griess, P. Bemerkungen zu der Abhandlung der HH. Weselsky und Benedikt Ueber einige Azoverbindungen. Eur. J. Inorg. Chem. 1879, 12, 426–428. [Google Scholar] [CrossRef] [Green Version]
- Eisenberg, G. Colorimetric Determination of Hydrogen Peroxide. Ind. Eng. Chem. Anal. Ed. 1943, 15, 327–328. [Google Scholar] [CrossRef]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Fraisier-Vannier, O.; Chervin, J.; Cabanac, G.; Puech-Pages, V.; Fournier, S.; Durand, V.; Amiel, A.; André, O.; Benamar, O.A.; Dumas, B.; et al. MS-CleanR: A Feature-Filtering Workflow for Untargeted LC–MS Based Metabolomics. Anal. Chem. 2020, 92, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Nakabayashi, R.; Mori, T.; Yamada, Y.; Takahashi, M.; Rai, A.; Sugiyama, R.; Yamamoto, H.; Nakaya, T.; Yamazaki, M.; et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms. Nat. Methods 2019, 16, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Dührkop, K.; Nothias, L.-F.; Fleischauer, M.; Reher, R.; Ludwig, M.; Hoffmann, M.A.; Petras, D.; Gerwick, W.H.; Rousu, J.; Dorrestein, P.C.; et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 2021, 39, 462–471. [Google Scholar] [CrossRef]
- Feunang, Y.D.; Eisner, R.; Knox, C.; Chepelev, L.; Hastings, J.; Owen, G.; Fahy, E.; Steinbeck, C.; Subramanian, S.; Bolton, E.; et al. ClassyFire: Automated chemical classification with a comprehensive, computable taxonomy. J. Chemin. 2016, 8, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; Morais, D.A.D.L.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference re-source for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- UN. The Sustainable Development Goals Report 2021; United Nations: New York, NY, USA, 2021; pp. 1–68. [Google Scholar]
- Pereira, C.; Dias, M.I.; Petropoulos, S.A.; Plexida, S.; Chrysargyris, A.; Tzortzakis, N.; Calhelha, R.C.; Ivanov, M.; Stojković, D.; Soković, M.; et al. The Effects of Biostimulants, Biofertilizers and Water-Stress on Nutritional Value and Chemical Composition of Two Spinach Genotypes (Spinacia oleracea L.). Molecules 2019, 24, 4494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munekata, P.E.S.; Domínguez, R.; Pateiro, M.; Lorenzo, J.M. Influence of Plasma Treatment on the Polyphenols of Food Products—A Review. Foods 2020, 9, 929. [Google Scholar] [CrossRef]
- Yamauchi, N.; Watada, A.E. Regulated Chlorophyll Degradation in Spinach Leaves during Storage. J. Am. Soc. Hortic. Sci. 1991, 116, 58–62. [Google Scholar] [CrossRef] [Green Version]
- López-Ayerra, B.; Murcia, M.A.; Garcia-Carmona, F. Lipid peroxidation and chlorophyll levels in spinach during refrigerated storage and after industrial processing. Food Chem. 1998, 61, 113–118. [Google Scholar] [CrossRef]
- Conte, A.; Conversa, G.; Scrocco, C.; Brescia, I.; Laverse, J.; Elia, A.; Del Nobile, M. Influence of growing periods on the quality of baby spinach leaves at harvest and during storage as minimally processed produce. Postharvest Biol. Technol. 2008, 50, 190–196. [Google Scholar] [CrossRef]
- Min, K.; Chen, K.; Arora, R. Short versus prolonged freezing differentially impacts freeze-thaw injury in spinach leaves: Mechanistic insights through metabolite profiling. Physiol. Plant. 2020, 168, 777–789. [Google Scholar] [CrossRef]
- Tajima, T.; Yamaguchi, A.; Matsushima, S.; Satoh, M.; Hayasaka, S.; Yoshimatsu, K.; Shioi, Y. Biochemical and molecular characterization of senescence-related cysteine protease-cystatin complex from spinach leaf. Physiol. Plant. 2010, 141, 97–116. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf—A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.-U.; Biehl, B. Activation of latent phenolase during spinach leaf senescence. Phytochemistry 1981, 20, 955–959. [Google Scholar] [CrossRef]
- Land, E.J.; Ramsden, C.A.; Riley, P.A.; Stratford, M.R. Evidence Consistent with the Requirement of Cresolase Activity for Suicide Inactivation of Tyrosinase. Tohoku J. Exp. Med. 2008, 216, 231–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzin, V.; Galili, G. New Insights into the Shikimate and Aromatic Amino Acids Biosynthesis Pathways in Plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Saltveit, M.E. Synthesis and metabolism of phenolic compounds. In Fruit and Vegetable Phytochemicals: Chemistry and Human Health, 2nd ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2017; pp. 89–100. ISBN 9781119158042. [Google Scholar]
- Su, Z.; Hu, M.; Gao, Z.; Li, M.; Yun, Z.; Pan, Y.; Zhang, Z.; Jiang, Y. Apple polyphenols delay senescence and maintain edible quality in litchi fruit during storage. Postharvest Biol. Technol. 2019, 157, 110976. [Google Scholar] [CrossRef]
- Hodges, D.M.; Forney, C. Postharvest Ascorbate Metabolism in Two Cultivars of Spinach Differing in Their Senescence Rates. J. Am. Soc. Hortic. Sci. 2003, 128, 930–935. [Google Scholar] [CrossRef] [Green Version]
- Lester, G.E.; Makus, D.J.; Hodges, D.M. Relationship between Fresh-Packaged Spinach Leaves Exposed to Continuous Light or Dark and Bioactive Contents: Effects of Cultivar, Leaf Size, and Storage Duration. J. Agric. Food Chem. 2010, 58, 2980–2987. [Google Scholar] [CrossRef]
- Tatjana, M.H.; Nesi, A.N.; Wagner, L.A.; Braun, H.-P. Amino Acid Catabolism in Plants. Mol. Plant 2015, 8, 1563–1579. [Google Scholar] [CrossRef] [Green Version]
- Soudry, E.; Ulitzur, S.; Gepstein, S. Accumulation and remobilization of amino acids during senescence of detached and attached leaves: In planta analysis of tryptophan levels by recombinant luminescent bacteria. J. Exp. Bot. 2004, 56, 695–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.; Park, S.; Natsagdorj, U.; Kim, Y.S.; Back, K. Methanol is an endogenous elicitor molecule for the synthesis of tryptophan and tryptophan-derived secondary metabolites upon senescence of detached rice leaves. Plant J. 2011, 66, 247–257. [Google Scholar] [CrossRef]
- Masclaux, C.; Quilleré, I.; Gallais, A.; Hirel, B. The challenge of remobilisation in plant nitrogen economy. A survey of physio-agronomic and molecular approaches. Ann. Appl. Biol. 2001, 138, 69–81. [Google Scholar] [CrossRef]
- Igarashi, D.; Tsuchida, H.; Miyao, M.; Ohsumi, C. Glutamate:Glyoxylate Aminotransferase Modulates Amino Acid Content during Photorespiration. Plant Physiol. 2006, 142, 901–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Zhang, C.; Suglo, P.; Sun, S.; Wang, M.; Su, T. l-Aspartate: An Essential Metabolite for Plant Growth and Stress Acclimation. Molecules 2021, 26, 1887. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.-E.; Kuppusamy, S.; Cho, K.M.; Kim, P.J.; Kwack, Y.-B.; Lee, Y.B. Influence of cold stress on contents of soluble sugars, vitamin C and free amino acids including gamma-aminobutyric acid (GABA) in spinach (Spinacia oleracea). Food Chem. 2017, 215, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.E.; Froese, C.D.; Madey, E.; Smith, M.; Hong, Y. Lipid metabolism during plant senescence. Prog. Lipid Res. 1998, 37, 119–141. [Google Scholar] [CrossRef]
- Jia, Y.; Tao, F.; Li, W. Lipid Profiling Demonstrates That Suppressing Arabidopsis Phospholipase Dδ Retards ABA-Promoted Leaf Senescence by Attenuating Lipid Degradation. PLoS ONE 2013, 8, e65687. [Google Scholar] [CrossRef] [PubMed]
- Annunziata, M.G.; Ciarmiello, L.F.; Woodrow, P.; Dell’Aversana, E.; Carillo, P. Spatial and Temporal Profile of Glycine Betaine Accumulation in Plants Under Abiotic Stresses. Front. Plant Sci. 2019, 10, 230. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; He, M.; Zeng, J.; Chen, Z.; Hongxia, Q.; Duan, X.; Jiang, Y. Choline chloride alleviates the pericarp browning of harvested litchi fruit by inhibiting energy deficiency mediated programmed cell death. Postharvest Biol. Technol. 2020, 167, 111224. [Google Scholar] [CrossRef]
- Singh, J.; Jayaprakasha, G.K.; Patil, B.S. Extraction, Identification, and Potential Health Benefits of Spinach Flavonoids: A Review. In Advances in Plant Phenolics: From Chemistry to Human Health; American Chemical Society: Washington, DC, USA, 2018; pp. 107–136. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, J.; Li, X.; Zhang, Y. Salicylic Acid: Biosynthesis and Signaling. Annu. Rev. Plant Biol. 2021, 72, 761–791. [Google Scholar] [CrossRef]
- Sullivan, M.X. THE ORIGIN OF CREATININE IN SOILS.1. J. Am. Chem. Soc. 1911, 33, 2035–2042. [Google Scholar] [CrossRef]
- Bergquist, S.Å.M.; Gertsson, U.E.; Knuthsen, P.; Olsson, M.E. Flavonoids in Baby Spinach (Spinacia oleracea L.): Changes during Plant Growth and Storage. J. Agric. Food Chem. 2005, 53, 9459–9464. [Google Scholar] [CrossRef] [PubMed]
- Schlering, C.; Zinkernagel, J.; Dietrich, H.; Frisch, M.; Schweiggert, R. Alterations in the Chemical Composition of Spinach (Spinacia oleracea L.) as Provoked by Season and Moderately Limited Water Supply in Open Field Cultivation. Horticulturae 2020, 6, 25. [Google Scholar] [CrossRef] [Green Version]
- Pandjaitan, N.; Howard, L.R.; Morelock, A.T.; Gil, M.I. Antioxidant Capacity and Phenolic Content of Spinach As Affected by Genetics and Maturation. J. Agric. Food Chem. 2005, 53, 8618–8623. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.-A.; Von Roepenack-Lahaye, E.; Parr, A.; Daniels, M.J.; Dow, J.M. Induction of Hydroxycinnamoyl-Tyramine Conjugates in Pepper by Xanthomonas campestris, a Plant Defense Response Activated by hrp Gene-Dependent and hrp Gene-Independent Mechanisms. Mol. Plant-Microbe Interact. 2001, 14, 785–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaup, M.T.; Froese, C.D.; Thompson, J.E. A Role for Diacylglycerol Acyltransferase during Leaf Senescence. Plant Physiol. 2002, 129, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Hasperue, J.; Lemoine, M.L.; Chaves, A.; Martínez, G.A. Effect of time of day for harvest and postharvest treatments on the sugar metabolism of broccoli (Brassica oleracea var. italica). Agric. Food Sci. 2014, 23, 48–59. [Google Scholar] [CrossRef]
- European Food Safety Authority Homepage. Available online: http://efsa.europa.eu (accessed on 21 September 2021).



| NO2 (mg/L) | NO3 (mg/L) | H2O2 (mg/L) | pH | ORP (mV) | |
|---|---|---|---|---|---|
| TAP | ND | ND | ND | 8.0 ± 0.1 | −45.3 ± 2.4 |
| PAW | 32.4 ± 5.6 | 462.3 ± 1.2 | 8.8 ± 0.4 | 2.3 ± 0.1 | 284.1 ± 11.5 |
| Type | Model | Time Points | LV | R2X | R2Y | Q2 | p-Value |
|---|---|---|---|---|---|---|---|
| PCA | all samples | all | 3 | 0.691 | 0.405 | ||
| OPLS-DA | C vs. TAP vs. PAW | D1 | 1 + 0 + 0 | 0.257 | 0.412 | −0.0244 | NS |
| OPLS-DA | C-S vs. TAP-S vs. PAW-S | D8 | 1 + 0 + 0 | 0.210 | 0.376 | −0.0176 | NS |
| OPLS-DA | C vs. C-S | D1 vs. D8 | 1 + 0 + 0 | 0.618 | 0.961 | 0.933 | 0.017 |
| OPLS-DA | TAP vs. TAP-S | D1 vs. D8 | 1 + 0 + 0 | 0.764 | 0.993 | 0.967 | 0.049 |
| OPLS-DA | PAW vs. PAW-S | D1 vs. D8 | 1 + 0 + 0 | 0.790 | 0.993 | 0.971 | 0.042 |
| LC Mode | Ionisation Mode | Average RT | Average m/z | Cluster Size a | Ion | Neutral Formula | Tentative Annotation (Spectral Match) | CANOPUS b Level | ClassyFire | Ann. Level c | D1 vs. D8 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C p(corr) d | C p( ) | TAP p(corr) d | TAP p( ) | PAW p(corr) d | PAW p( ) | |||||||||||
| Shared by All Processing Methods | ||||||||||||||||
| RPLC | pos | 1.97 | 166.0857 | 6 | [M + H]+ | C9H11NO2 | Phenylalanine | 7 | Phenylalanine and deriv. | 2 | 1.00 | 0.48 | 0.84 | 0.44 | 0.97 | 0.43 |
| HILIC | neg | 9.86 | 164.0714 | 3 | [M − H]− | C9H11NO2 | Phenylalanine | 7 | Phenylalanine and deriv. | 2 | 0.98 | 0.08 | 0.94 | 0.07 | 0.90 | 0.06 |
| RPLC | pos | 2.35 | 205.0968 | 16 | [M + H]+ | C11H12N2O2 | Tryptophan | 7 | α-amino acids | 2 | 0.98 | 0.24 | 0.92 | 0.27 | 0.97 | 0.27 |
| HILIC | neg | 12.27 | 175.0608 | 2 | [M − H]− | C7H12O5 | 2-Isopropylmalate | 6 | Meth. branched fatty acids | 2 | 0.99 | 0.21 | 0.84 | 0.20 | 0.92 | 0.21 |
| RPLC | neg | 2.66 | 175.0608 | 1 | [M − H]− | C7H12O5 | 2-Isopropylmalate | 6 | Meth. branched fatty acids | 2 | 0.99 | 0.12 | 0.66 | 0.11 | 0.81 | 0.13 |
| RPLC | pos | 1.20 | 182.0808 | 5 | [M + H]+ | C9H11NO3 | Tyrosine | 7 | Tyrosine and deriv. | 2 | 0.78 | 0.16 | 0.83 | 0.19 | 0.78 | 0.15 |
| HILIC | pos | 12.99 | 182.0808 | 3 | [M + H]+ | C9H11NO3 | Tyrosine | 7 | Tyrosine and deriv. | 2 | 0.95 | 0.13 | 0.95 | 0.12 | 0.74 | 0.09 |
| HILIC | neg | 15.64 | 117.0192 | 2 | [M − H]− | C4H6O4 | Succinate | Subclass | Dicarbox. acids and deriv. | 2 | 0.90 | 0.14 | 0.92 | 0.17 | 0.94 | 0.19 |
| HILIC | pos | 14.49 | 120.0652 | 2 | [M + H]+ | C4H9NO3 | L-(−)-Threonine | 7 | α-amino acids | 2 | 0.96 | 0.11 | 0.95 | 0.10 | 0.91 | 0.09 |
| HILIC | pos | 15.43 | 133.0605 | 1 | [M + H]+ | C4H8N2O3 | Asparagine | 7 | α-amino acids | 2 | 0.91 | 0.08 | 0.98 | 0.09 | 0.74 | 0.08 |
| HILIC | neg | 12.94 | 135.0305 | 2 | [M − H]− | C4H8O5 | Threonate | Subclass | β-hydroxy acids and deriv. | 2 | 0.94 | 0.08 | 0.96 | 0.10 | 0.94 | 0.10 |
| HILIC | pos | 3.88 | 679.2970 | 5 | [M + H]+ | C35H42N4O10 | Unknown | 6 | Oligopeptides | 3 | 0.89 | 0.08 | 0.77 | 0.08 | 0.74 | −0.05 |
| RPLC | pos | 1.14 | 245.0764 | 4 | [M + H]+ | C9H12N2O6 | Uridine | Class | Pyrimidine nucleosides | 2 | −0.83 | −0.05 | −0.89 | −0.07 | −0.89 | −0.08 |
| RPLC | pos | 2.80 | 374.1438 | 24 | [M + NH4]+ | C16H20O9 | 1-O-Feruloylglucose | 5 | Coumaric acids and deriv. | 2 | −0.98 | −0.12 | −0.92 | −0.10 | −0.69 | −0.05 |
| HILIC | pos | 14.73 | 148.0603 | 4 | [M + H]+ | C5H9NO4 | L-Glutamate | 7 | Glutamic acid and deriv. | 2 | −0.67 | −0.09 | −0.85 | −0.12 | −0.53 | −0.09 |
| RPLC | pos | 0.67 | 455.1137 | 1 | [M + Na]+ | C23H20N4O3S | Asperulosidic acid | 6 | Phenolic glycosides | 3 | −0.91 | −0.06 | −0.88 | −0.08 | −0.99 | −0.12 |
| RPLC | pos | 0.96 | 168.0652 | 1 | [M + H]+ | C8H9NO3 | Pyridoxal | 6 | Aryl-aldehydes | 2 | −0.92 | −0.08 | −0.90 | −0.09 | −0.96 | −0.10 |
| HILIC | pos | 7.39 | 168.0652 | 1 | [M + H]+ | C8H9NO3 | Pyridoxal | Subclass | Pyridine carboxaldehydes | 2 | −0.92 | −0.05 | −0.92 | −0.06 | −0.90 | −0.07 |
| HILIC | neg | 15.11 | 132.0300 | 1 | [M − H]− | C4H7NO4 | L-Aspartate | 8 | L-α-amino acids | 2 | −0.89 | −0.08 | −0.99 | −0.09 | −0.73 | −0.06 |
| RPLC | pos | 1.26 | 307.0830 | 1 | [M + H]+ | Unknown | Unknown | Subclass | 1-hydoxy-2-unsubstituted benzenoids | 4 | −0.89 | −0.08 | −0.97 | −0.09 | −0.87 | −0.09 |
| RPLC | neg | 0.71 | 133.0138 | 4 | [M − H]− | C4H6O5 | D-(+)-Malate | Subclass | β-hydroxy acids and deriv. | 2 | −0.90 | −0.14 | −0.55 | −0.07 | −0.64 | −0.06 |
| RPLC | pos | 0.55 | 146.1649 | 3 | [M + H]+ | C7H19N3 | Spermidine | 6 | Dialkylamines | 2 | −0.82 | −0.15 | −0.92 | −0.15 | −0.66 | −0.09 |
| HILIC | pos | 21.60 | 104.1067 | 2 | [M + H]+ | C5H14NO | Choline | 5 | Tetraalkylammonium salts | 2 | −0.62 | −0.23 | −0.73 | −0.18 | −0.97 | −0.24 |
| RPLC | pos | 1.56 | 268.1030 | 10 | [M + H]+ | C10H13N5O4 | Adenosine | Class | Purine nucleosides | 2 | −0.96 | −0.23 | −0.96 | −0.26 | −0.93 | −0.32 |
| Shared by Two Processing Methods | ||||||||||||||||
| TAP and PAW | ||||||||||||||||
| RPLC | pos | 1.13 | 101.0232 | 4 | [M + H]+ | C4H4O3 | Succinic anhydride | Subclass | Dicarbox. acids and deriv. | 2 | 0.86 | 0.04 | 0.85 | 0.05 | 0.94 | 0.06 |
| HILIC | pos | 10.96 | 132.1016 | 4 | [M + H]+ | C6H13NO2 | DL-Norleucine | 6 | β-amino acids and deriv. | 3 | 0.31 | 0.04 | 0.59 | 0.10 | 0.54 | 0.06 |
| RPLC | pos | 1.89 | 199.1913 | 1 | [M + H]+ | C10H22N4 | Unknown | Subclass | Guanidines | 4 | −0.11 | −0.01 | −0.97 | −0.20 | −0.78 | −0.19 |
| HILIC | pos | 11.98 | 130.0861 | 1 | [M + H]+ | C6H11NO2 | Unknown | 6 | Amino acids | 3 | 0.39 | 0.03 | −0.58 | −0.05 | −0.62 | −0.11 |
| HILIC | pos | 25.21 | 123.0550 | 1 | [M + H]+ | C6H6N2O | Nicotinamide | 6 | Nicotinamides | 2 | −0.61 | −0.03 | −0.81 | −0.05 | −0.79 | −0.06 |
| RPLC | pos | 5.19 | 200.2009 | 1 | [M + H]+ | C12H25NO | Unknown | Subclass | Fatty amides | 3 | −0.92 | −0.04 | −0.95 | −0.05 | −0.98 | −0.05 |
| C and TAP | ||||||||||||||||
| RPLC | pos | 3.70 | 679.2974 | 3 | [M + H]+ | C35H42N4O10 | Unknown | 6 | α-amino acids and deriv. | 3 | 0.73 | 0.06 | 0.62 | 0.07 | 0.40 | 0.02 |
| RPLC | pos | 4.53 | 615.2780 | 5 | [M + H]+ | - | Unknown | Subclass | Billirubins | 4 | 0.87 | 0.05 | 0.70 | 0.05 | 0.89 | 0.04 |
| HILIC | pos | 10.38 | 132.1016 | 3 | [M + H]+ | C6H13NO2 | DL-Norleucine | 7 | α-amino acids | 3 | 0.59 | 0.09 | 0.34 | 0.08 | −0.21 | −0.04 |
| RPLC | pos | 5.46 | 368.4239 | 1 | [M + H]+ | Unknown | - | - | - | 4 | −0.92 | −0.06 | −0.70 | −0.05 | 0.04 | 0.00 |
| HILIC | neg | 16.60 | 133.0139 | 6 | [M − H]− | C4H6O5 | D-(+)-Malate | Subclass | β-hydroxy acids and deriv. | 3 | −0.56 | −0.11 | −0.72 | −0.13 | −0.36 | −0.06 |
| RPLC | pos | 5.14 | 279.1588 | 19 | [M + H]+ | C16H22O4 | Unknown | 5 | Benzoic acids | 4 | −0.84 | −0.20 | −0.70 | −0.17 | −0.34 | −0.08 |
| RPLC | pos | 0.70 | 360.1485 | 1 | [M + NH4]+ | C12H22O11 | - | 5 | Disaccharides | 3 | −0.70 | −0.05 | −0.82 | −0.05 | −0.81 | −0.04 |
| C and PAW | ||||||||||||||||
| HILIC | pos | 4.86 | 242.1283 | 1 | [M + H]+ | C14H15N3O | Unknown | Subclass | Cinnamic acid amides | 3 | 0.57 | 0.05 | 0.42 | 0.02 | 0.75 | 0.08 |
| HILIC | pos | 18.01 | 156.0765 | 1 | [M + H]+ | C6H9N3O2 | Histidine | 7 | Histidine and deriv. | 3 | 0.87 | 0.06 | 0.68 | 0.04 | 0.86 | 0.07 |
| HILIC | neg | 18.59 | 191.0193 | 2 | [M − H]− | C6H8O7 | Citrate | Subclass | Tricarboxylic acids and deriv. | 3 | −0.90 | −0.14 | −0.35 | −0.04 | −0.50 | −0.08 |
| HILIC | neg | 18.10 | 173.0091 | 1 | [M − H]− | C6H6O6 | Aconitate | Subclass | Tricarboxylic acids and deriv. | 3 | −0.65 | −0.05 | −0.62 | −0.04 | −0.63 | −0.05 |
| Unique for One Processing Method | ||||||||||||||||
| C | ||||||||||||||||
| RPLC | pos | 1.48 | 132.1016 | 4 | [M + H]+ | C6H13NO2 | DL-Norleucine | 7 | α-amino acids | 3 | −0.82 | −0.29 | −0.19 | −0.09 | −0.61 | −0.21 |
| TAP | ||||||||||||||||
| HILIC | pos | 9.26 | 114.0660 | 1 | [M + H]+ | C4H7N3O | Creatinine | 6 | α-amino acids and deriv. | 2 | 0.45 | 0.00 | −0.51 | −0.05 | −0.40 | −0.01 |
| RPLC | pos | 2.52 | 155.1541 | 1 | [M + H]+ | Unknown | - | - | - | 4 | −0.16 | 0.00 | −0.69 | −0.18 | −0.30 | 0.00 |
| RPLC | pos | 3.78 | 171.1489 | 1 | [M + H]+ | C9H18N2O | - | Class | Organic carbonic acids and deriv. | 3 | −0.80 | −0.01 | −0.61 | −0.14 | −0.67 | −0.01 |
| HILIC | pos | 10.76 | 118.0859 | 1 | [M + H]+ | C5H11NO2 | Glycine Betaine | - | - | 4 | −0.43 | −0.08 | −0.67 | −0.17 | 0.05 | 0.02 |
| PAW | ||||||||||||||||
| RPLC | pos | 3.12 | 242.1284 | 1 | [M + H]+ | C14H15N3O | - | Subclass | Cinnamic acid amides | 3 | 0.40 | 0.05 | 0.02 | 0.00 | 0.67 | 0.11 |
| HILIC | neg | 14.20 | 165.0192 | 1 | [M − H − H2O]− | C8H8O5 | - | Subclass | α-hydroxy acids and deriv. | 3 | 0.87 | 0.04 | 0.01 | 0.00 | 0.98 | 0.06 |
| RPLC | neg | 5.33 | 447.2512 | 1 | [M − H]− | Unknown | - | - | - | 4 | −0.48 | −0.01 | −0.71 | −0.03 | −0.89 | −0.06 |
| HILIC | neg | 3.46 | 277.2166 | 3 | [M − H]− | C18H30O2 | γ-Linolenic acid | Subclass | Glycerophosphocholine | 3 | 0.04 | 0.00 | 0.30 | 0.04 | −0.88 | −0.10 |
| HILIC | pos | 11.60 | 244.0924 | 1 | [M + H]+ | C9H13N3O5 | Cytidine | Class | Pyrimidine nucleosides | 2 | −0.73 | −0.03 | −0.93 | −0.04 | −0.85 | −0.05 |
| HILIC | pos | 8.98 | 136.0616 | 1 | [M + H]+ | C5H5N5 | Adenine | 5 | 6-aminopurines | 2 | −0.70 | −0.03 | −0.66 | −0.04 | −0.58 | −0.05 |
| RPLC | pos | 5.62 | 496.3395 | 2 | [M + H]+ | C24H50NO7P | Lysophosphatidylcholine (16:0) | 5 | Lysophosphatidylcholines | 2 | −0.55 | −0.04 | −0.02 | 0.00 | −0.77 | −0.07 |
| HILIC | neg | 4.68 | 519.0775 | 2 | [M − H]− | C23H20O14 | 5,3’,4’,-Trihydroxy-3-methoxy-6:7-methylenedioxyflavone 4’-βD-glucuronide | 7 | Flavonoid-7-O-glucuronides | 3 | 0.42 | 0.03 | −0.15 | −0.01 | −0.63 | −0.07 |
| Inverse | ||||||||||||||||
| RPLC | pos | 1.79 | 168.1014 | 3 | [M + H]+ | C9H13NO2 | Methoxytyramine | Subclass | Phenethyl amines | 2 | −0.86 | −0.08 | −0.86 | −0.07 | 0.71 | 0.07 |
| HILIC | pos | 15.03 | 147.0763 | 3 | [M + H]+ | C5H10N2O3 | Glutamine | 7 | α-amino acids | 2 | −0.55 | −0.09 | 0.19 | 0.05 | 0.37 | 0.06 |
| HILIC | pos | 12.22 | 118.0859 | 1 | [M + H]+ | C5H11NO2 | Valine | 7 | α-amino acids | 2 | 0.50 | 0.05 | 0.19 | 0.03 | −0.78 | −0.07 |
| RPLC | pos | 0.63 | 175.1184 | 1 | [M + H]+ | C6H14N4O2 | Arginine | 8 | L-α-amino acids | 2 | 0.78 | 0.11 | 0.32 | 0.04 | −0.50 | −0.07 |
| RPLC | pos | 5.71 | 282.2782 | 1 | [M + H]+ | C18H35NO | - | Subclass | Fatty amides | 2 | −0.42 | −0.03 | 0.45 | 0.07 | −0.61 | −0.05 |
| RPLC | neg | 3.12 | 279.0504 | 12 | [M − H]− | C13H12O7 | 2-(p-coumaroyl)malate | 6 | Coumaric acid esters | 3 | −0.85 | −0.07 | −0.74 | −0.05 | 0.58 | 0.04 |
| RPLC | pos | 1.38 | 138.0913 | 2 | [M + H]+ | C8H11NO | Tyramine | 5 | Aralkylamines | 2 | −0.95 | −0.07 | −0.69 | −0.05 | 0.66 | 0.03 |
| C | TAP | PAW |
|---|---|---|
| Alanine, aspartate and glutamate metabolism | Alanine, aspartate and glutamate metabolism | |
| Aminoacyl-tRNA biosynthesis | Aminoacyl-tRNA biosynthesis | |
| Ascorbate and aldarate metabolism | ||
| Butanoate metabolism | ||
| C5-branched dibasic acid metabolism | ||
| Cyanoamino acid metabolism | ||
| Cysteine and methionine metabolism | Cysteine and methionine metabolism | |
| Galactose metabolism | ||
| Phenylalanine, tyrosine and tryptophan biosynthesis | ||
| Propanoate metabolism | ||
| Starch and sucrose metabolism | ||
| Valine, leucine and isoleucine degradation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangel-Huerta, O.D.; Ivanova, L.; Uhlig, S.; Sivertsvik, M.; Sone, I.; Fernández, E.N.; Fæste, C.K. Impact of Plasma-Activated Water Treatment on Quality and Shelf-Life of Fresh Spinach Leaves Evaluated by Comprehensive Metabolomic Analysis. Foods 2021, 10, 3067. https://doi.org/10.3390/foods10123067
Rangel-Huerta OD, Ivanova L, Uhlig S, Sivertsvik M, Sone I, Fernández EN, Fæste CK. Impact of Plasma-Activated Water Treatment on Quality and Shelf-Life of Fresh Spinach Leaves Evaluated by Comprehensive Metabolomic Analysis. Foods. 2021; 10(12):3067. https://doi.org/10.3390/foods10123067
Chicago/Turabian StyleRangel-Huerta, Oscar Daniel, Lada Ivanova, Silvio Uhlig, Morten Sivertsvik, Izumi Sone, Estefanía Noriega Fernández, and Christiane Kruse Fæste. 2021. "Impact of Plasma-Activated Water Treatment on Quality and Shelf-Life of Fresh Spinach Leaves Evaluated by Comprehensive Metabolomic Analysis" Foods 10, no. 12: 3067. https://doi.org/10.3390/foods10123067
APA StyleRangel-Huerta, O. D., Ivanova, L., Uhlig, S., Sivertsvik, M., Sone, I., Fernández, E. N., & Fæste, C. K. (2021). Impact of Plasma-Activated Water Treatment on Quality and Shelf-Life of Fresh Spinach Leaves Evaluated by Comprehensive Metabolomic Analysis. Foods, 10(12), 3067. https://doi.org/10.3390/foods10123067







