Influence of Bacterial Competitors on Salmonella enterica and Enterohemorrhagic Escherichia coli Growth in Microbiological Media and Attachment to Vegetable Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Vegetable Seeds
2.2. Competitive Growth between Salmonella or EHEC and Selected Plant Pathogen, Probiotic Strain, or Biocontrol Agents in Microbiological Media
2.3. Competitive Attachment between Salmonella or EHEC and Selected Plant Pathogen, Probiotic Strain, or Biocontrol Agents to Vegetable Seeds
2.4. Effect of Metabolites in Cell-Free Supernatants (CFS) of the Spend Cultures of Biocontrol Agents and Probiotic Strain on Salmonella and EHEC
2.5. Statistical Analysis
3. Results
3.1. Competitive Growth between Salmonella/EHEC and Bacterial Competitors
3.2. Competitive Attachment to Vegetable Seeds by Salmonella/EHEC as Affected by Bacterial Competitors
3.3. Inhibition of Salmonella and EHEC by the CFS of L. rhamnousus GG Spent Cultures
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Como-Sabetti, K.; Reagan, S.; Allaire, S.; Parrott, K.; Simonds, C.; Hrabowy, S.; Ritter, B.; Hall, W.; Altamirano, J.; Martin, R. Outbreaks of Escherichia coli O157:H7 infection associated with eating alfalfa sprouts—Michigan and Virginia, June–July 1997. Morb. Mortal. Wkly. Rep. 1997, 46, 741–744. [Google Scholar]
- Mahon, B.E.; Pönkä, A.; Hall, W.N.; Komatsu, K.; Dietrich, S.E.; Siitonen, A.; Cage, G.; Hayes, P.S.; Lambert-Fair, M.A.; Bean, N.H. An international outbreak of Salmonella infections caused by alfalfa sprouts grown from contaminated seeds. J. Infect. Dis. 1997, 175, 876–882. [Google Scholar] [CrossRef] [Green Version]
- Winthrop, K.; Palumbo, M.; Farrar, J.; Mohle-Boetani, J.; Abbott, S.; Beatty, M.; Inami, G.; Werner, S. Alfalfa sprouts and Salmonella Kottbus infection: A multistate outbreak following inadequate seed disinfection with heat and chlorine. J. Food Prot. 2003, 66, 13–17. [Google Scholar] [CrossRef]
- Taormina, P.J.; Beuchat, L.R.; Slutsker, L. Infections associated with eating seed sprouts: An international concern. Emerg. Inf. Dis. 1999, 5, 626. [Google Scholar] [CrossRef]
- Howard, M.B.; Hutcheson, S.W. Growth dynamics of Salmonella enterica strains on alfalfa sprouts and in waste seed irrigation water. Appl. Environ. Microbiol. 2003, 69, 548–553. [Google Scholar] [CrossRef] [Green Version]
- Deering, A.J.; Jack, D.R.; Pruitt, R.E.; Mauer, L.J. Movement of Salmonella serovar Typhimurium and E. coli O157:H7 to ripe tomato fruit following various routes of contamination. Microorganisms 2015, 3, 809–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, W.; Hammack, T.; Amaguana, R. Chapter 5, Salmonella. In Bacteriological Analytical Manual. US Food and Drug Administration. 2007. Available online: http://www.cfsan.fda.gov/ebam/bam-5.htm (accessed on 8 October 2020).
- Feng, P.; Weagant, S.; Jinneman, K. Bacteriological Analytical Manual, Chapter 4A, Diarrheagenic Escherichia coli. Available online: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070080.htm (accessed on 11 December 2019).
- Fett, W. Factors affecting the efficacy of chlorine against Escherichia coli O157:H7 and Salmonella on alfalfa seed. Food Microbiol. 2002, 19, 135–149. [Google Scholar] [CrossRef]
- Cooley, M.B.; Miller, W.G.; Mandrell, R.E. Colonization of Arabidopsis thaliana with Salmonella enterica and enterohemorrhagic Escherichia coli O157:H7 and competition by Enterobacter asburiae. Appl. Environ. Microbiol. 2003, 69, 4915–4926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, P.M.; Harrison, M.A. Resistance and adaptation to food antimicrobials, sanitizers, and other process controls. Food Technol. 2002, 56, 69–78. [Google Scholar]
- Kamilova, F.; Kravchenko, L.V.; Shaposhnikov, A.I.; Makarova, N.; Lugtenberg, B. Effects of the tomato pathogen Fusarium oxysporum f. sp. radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudate. Mol. Plant-Microbe Interact. 2006, 19, 1121–1126. [Google Scholar] [PubMed] [Green Version]
- Kamilova, F.; Validov, S.; Azarova, T.; Mulders, I.; Lugtenberg, B. Enrichment for enhanced competitive plant root tip colonizers selects for a new class of biocontrol bacteria. Environ. Microbiol. 2005, 7, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Fessehaie, A.; Walcott, R. Biological control to protect watermelon blossoms and seed from infection by Acidovorax avenae subsp. citrulli. Phytopathology 2005, 95, 413–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, C.H. Growth of Salmonella on sprouting alfalfa seeds as affected by the inoculum size, native microbial load and Pseudomonas fluorescens 2–79. Lett. Appl. Microbiol. 2008, 46, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Kostrzynska, M.; Dunfield, K.; Warriner, K. Control of Salmonella on sprouting mung bean and alfalfa seeds by using a biocontrol preparation based on antagonistic bacteria and lytic bacteriophages. J. Food Prot. 2010, 73, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [Green Version]
- Cabrefiga, J.; Bonaterra, A.; Montesinos, E. Mechanisms of antagonism of Pseudomonas fluorescens EPS62e against Erwinia amylovora, the causal agent of fire blight. Int. Microbiol. 2007, 10, 123. [Google Scholar]
- Bredholt, S.; Nesbakken, T.; Holck, A. Industrial application of an antilisterial strain of Lactobacillus sakei as a protective culture and its effect on the sensory acceptability of cooked, sliced, vacuum-packaged meats. Int. J. Food Microbiol. 2001, 66, 191–196. [Google Scholar] [CrossRef]
- Nitschke, M.; Araújo, L.; Costa, S.; Pires, R.; Zeraik, A.; Fernandes, A.; Freire, D.; Contiero, J. Surfactin reduces the adhesion of food-borne pathogenic bacteria to solid surfaces. Lett. Appl. Microbiol. 2009, 49, 241–247. [Google Scholar] [CrossRef]
- De Keersmaecker, S.C.; Verhoeven, T.L.; Desair, J.; Marchal, K.; Vanderleyden, J.; Nagy, I. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol. Lett. 2006, 259, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Walcott, R.; Chen, J. Differential attachment of Salmonella enterica and enterohemorrhagic Escherichia coli to alfalfa, fenugreek, lettuce, and tomato seeds. Appl. Environ. Microbiol. 2017, 83, e03170-16. [Google Scholar] [CrossRef] [Green Version]
- Fayol-Messaoudi, D.; Berger, C.N.; Coconnier-Polter, M.-H.; Lievin-Le Moal, V.; Servin, A.L. pH-, lactic acid-, and non-lactic acid-dependent activities of probiotic lactobacilli against Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 2005, 71, 6008–6013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pithva, S.; Ambalam, P.; Dave, J.; Vyas, B. Antimicrobial Peptides of Probiotic Lactobacillus Strains. 2011. Available online: https://www.researchgate.net/publication/258338154_Antimicrobial_Peptides_of_Probiotic_Lactobacillus_strains (accessed on 8 October 2020).
- Alexandre, Y.; Le Berre, R.; Barbier, G.; Le Blay, G. Screening of Lactobacillus spp. for the prevention of Pseudomonas aeruginosa pulmonary infections. BMC Microbiol. 2014, 14, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tharmaraj, N.; Shah, N.P. Antimicrobial effects of probiotics against selected pathogenic and spoilage bacteria in cheese-based dips. Int. Food. Res. J. 2009, 16, 261–276. [Google Scholar]
- Deepika, G.; Karunakaran, E.; Hurley, C.R.; Biggs, C.A.; Charalampopoulos, D. Influence of fermentation conditions on the surface properties and adhesion of Lactobacillus rhamnosus GG. Microb. Cell Factories 2012, 11, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Østlie, H.M.; Treimo, J.; Narvhus, J.A. Effect of temperature on growth and metabolism of probiotic bacteria in milk. Int. Dairy J. 2005, 15, 989–997. [Google Scholar] [CrossRef]
- Arias, O.A.; Reyes, M.M.; Navarro, V.M.L.; Solis, C.Y.; Márquez, G.M.; Sanchez, S.G.; Snell, C.R.; Zuñiga, R.R. Antagonistic Effect of Probiotic Strains against Two Pathogens: Salmonella Typhimurium and E. coli O157:H7 Resistant to Antibiotics. Available online: https://www.redalyc.org/pdf/730/73029399005.pdf (accessed on 8 October 2020).
- Breidt, F., Jr.; Kay, K.; Cook, J.; Osborne, J.; Ingham, B.; Arritt, F. Determination of 5-log reduction times for Escherichia coli O157:H7, Salmonella enterica, or Listeria monocytogenes in acidified foods with pH 3.5 or 3.8. J. Food Prot. 2013, 76, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Berry, E.D.; Cutter, C.N. Effects of acid adaptation of Escherichia coli O157:H7 on efficacy of acetic acid spray washes to decontaminate beef carcass tissue. Appl. Environ. Microbiol. 2000, 66, 1493–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brackett, R.; Hao, Y.-Y.; Doyle, M. Ineffectiveness of hot acid sprays to decontaminate Escherichia coli O157:H7 on beef. J. Food Prot. 1994, 57, 198–203. [Google Scholar] [CrossRef]
- Vanderhoof, J.A.; Whitney, D.B.; Antonson, D.L.; Hanner, T.L.; Lupo, J.V.; Young, R.J. Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J. Pediatr. 1999, 135, 564–568. [Google Scholar] [CrossRef]
- Stockwell, V.; Johnson, K.; Sugar, D.; Loper, J. Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 2010, 100, 1330–1339. [Google Scholar] [CrossRef] [Green Version]
- Samelis, J.; Sofos, J.N. Role of glucose in enhancing the temperature-dependent growth inhibition of Escherichia coli O157:H7 ATCC 43895 by a Pseudomonas sp. Appl. Environ. Microbiol. 2002, 68, 2600–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacon, C.W.; Hinton, D.M. Endophytic and biological control potential of Bacillus mojavensis and related species. Biol. Control 2002, 23, 274–284. [Google Scholar] [CrossRef] [Green Version]
- Snook, M.E.; Mitchell, T.; Hinton, D.M.; Bacon, C.W. Isolation and characterization of Leu7-surfactin from the endophytic bacterium Bacillus mojavensis RRC 101, a biocontrol agent for Fusarium verticillioides. J. Agric. Food Chem. 2009, 57, 4287–4292. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, E.Z. Antimicrobial activity of a biosurfactant produced by Bacillus licheniformis strain M104 grown on whey. Braz. Arch. Biol. Technol. 2013, 56, 259–268. [Google Scholar] [CrossRef] [Green Version]
- Mireles, J.R.; Toguchi, A.; Harshey, R.M. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: Surfactin inhibits biofilm formation. J. Bacteriol. 2001, 183, 5848–5854. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumar, P.P.; Muriana, P.M. Inhibition of Listeria monocytogenes on ready-to-eat meats using bacteriocin mixtures based on mode-of-action. Foods 2017, 6, 22. [Google Scholar] [CrossRef] [Green Version]
- Bonaterra, A.; Badosa, E.; Cabrefiga, J.; Francés, J.; Montesinos, E. Prospects and limitations of microbial pesticides for control of bacterial and fungal pomefruit tree diseases. Trees 2012, 26, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Nisbet, D. Defined competitive exclusion cultures in the prevention of enteropathogen colonisation in poultry and swine. Antonie Leeuwenhoek 2002, 81, 481–486. [Google Scholar] [CrossRef]
- Alakomi, H.-L.; Skyttä, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef] [Green Version]
- Charkowski, A.O.; Sarreal, C.Z.; Mandrell, R.E. Wrinkled alfalfa seeds harbor more aerobic bacteria and are more difficult to sanitize than smooth seeds. J. Food Prot. 2001, 64, 1292–1298. [Google Scholar] [CrossRef]
- Mujumdar, S.; Bashetti, S.; Pardeshi, S.; Thombre, R.S. Industrial applications of biosurfactants. In Industrial Biotechnology: Sustainable Production and Bioresource Utilization, 1st ed.; Apple Academic Press: Palm Bay, FL, USA, 2016; p. 484. [Google Scholar]
- Dekkers, L.C.; Bloemendaal, C.J.P.; de Weger, L.A.; Wijffelman, C.A.; Spaink, H.P.; Lugtenberg, B.J. A two-component system plays an important role in the root-colonizing ability of Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 1998, 11, 45–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aruscavage, D.; Miller, S.A.; Lewis Ivey, M.L.; Lee, K.; LeJeune, J.T. Survival and dissemination of Escherichia coli O157:H7 on physically and biologically damaged lettuce plants. J. Food Prot. 2008, 71, 2384–2388. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; Chao, D.; Mandrell, R.E. Escherichia coli O157:H7 survival and growth on lettuce is altered by the presence of epiphytic bacteria. J. Food Prot. 2006, 69, 2329–2335. [Google Scholar] [CrossRef] [PubMed]
- Esseili, M.A.; Chin, A.; Saif, L.; Miller, S.A.; Qu, F.; Lewis Ivey, M.L.; Wang, Q. Postharvest survival of porcine sapovirus, a human norovirus surrogate, on phytopathogen-infected leafy greens. J. Food Prot. 2015, 78, 1472–1480. [Google Scholar] [CrossRef]
- Potnis, N.; Soto-Arias, J.P.; Cowles, K.N.; van Bruggen, A.H.; Jones, J.B.; Barak, J.D. Xanthomonas perforans colonization influences Salmonella enterica in the tomato phyllosphere. Appl. Environ. Microbiol. 2014, 80, 3173–3180. [Google Scholar] [CrossRef] [Green Version]
- Poza-Carrion, C.; Suslow, T.; Lindow, S. Resident bacteria on leaves enhance survival of immigrant cells of Salmonella enterica. Phytopathology 2013, 103, 341–351. [Google Scholar] [CrossRef] [Green Version]
 ) and the mixed cultures with P. fluorescenes A506 (
) and the mixed cultures with P. fluorescenes A506 (  ), B. subtilis ATCC 6051 (
), B. subtilis ATCC 6051 (  ), B. mojavensis RRC 101 (
), B. mojavensis RRC 101 (  ), Cocktail 1 (
), Cocktail 1 (  ), Cocktail 2 (
), Cocktail 2 (  ), L. rhamnousus GG (
), L. rhamnousus GG (  ) and P. syringae pv. tomato DC3000 (
) and P. syringae pv. tomato DC3000 (  ) after 6, 12, 24, 48 and 72 h of co-incubation at 25 °C.
) after 6, 12, 24, 48 and 72 h of co-incubation at 25 °C.
 ) and the mixed cultures with P. fluorescenes A506 (
) and the mixed cultures with P. fluorescenes A506 (  ), B. subtilis ATCC 6051 (
), B. subtilis ATCC 6051 (  ), B. mojavensis RRC 101 (
), B. mojavensis RRC 101 (  ), Cocktail 1 (
), Cocktail 1 (  ), Cocktail 2 (
), Cocktail 2 (  ), L. rhamnousus GG (
), L. rhamnousus GG (  ) and P. syringae pv. tomato DC3000 (
) and P. syringae pv. tomato DC3000 (  ) after 6, 12, 24, 48 and 72 h of co-incubation at 25 °C.
) after 6, 12, 24, 48 and 72 h of co-incubation at 25 °C.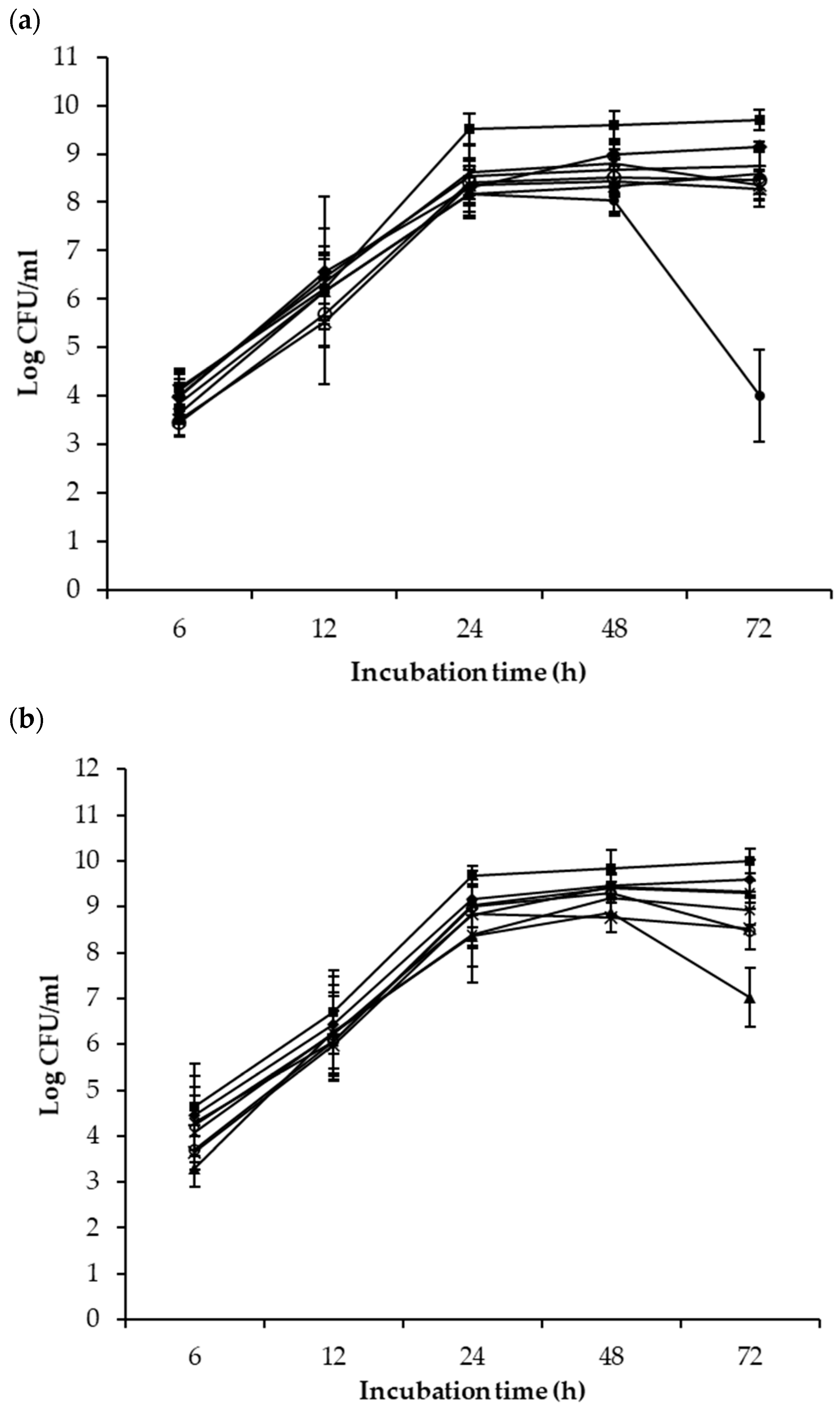
 ; E. coli
; E. coli  ) and 25 °C (S. enterica
) and 25 °C (S. enterica  ; E. coli
; E. coli  ) after 6, 12, 24, 48, and 72 h of incubation.
) after 6, 12, 24, 48, and 72 h of incubation.
 ; E. coli
; E. coli  ) and 25 °C (S. enterica
) and 25 °C (S. enterica  ; E. coli
; E. coli  ) after 6, 12, 24, 48, and 72 h of incubation.
) after 6, 12, 24, 48, and 72 h of incubation.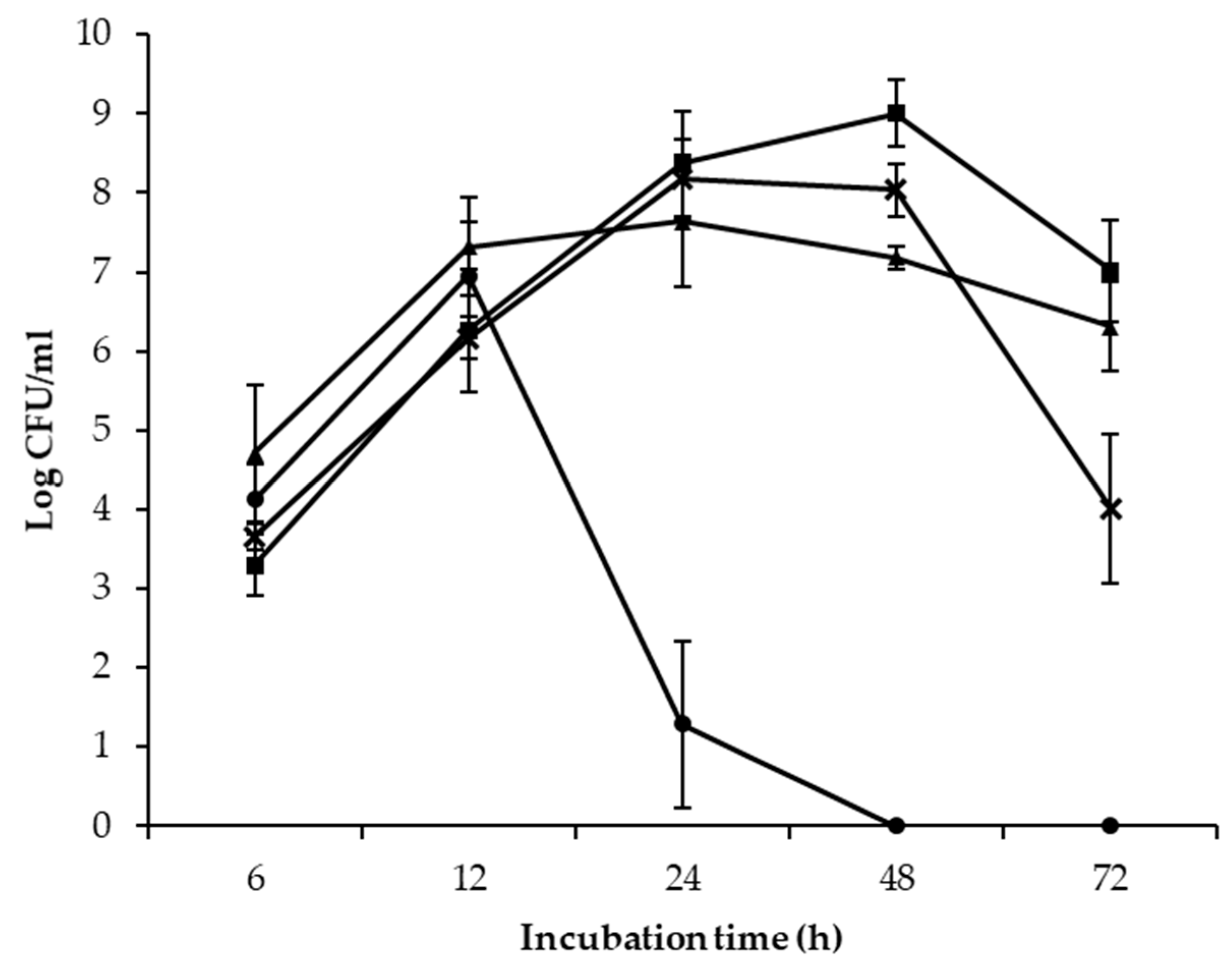
 ; EHEC
; EHEC  ), B. subtilis ATCC 6051 (S. enterica
), B. subtilis ATCC 6051 (S. enterica  ; EHEC
; EHEC  ), B. mojavensis RRC 101 (S. enterica
), B. mojavensis RRC 101 (S. enterica  ; EHEC
; EHEC  ) and L. rhamnousus GG (S. enterica
) and L. rhamnousus GG (S. enterica  ; EHEC
; EHEC  ) at the 2, 4, 8, 12, 24 and 48 h sampling points.
) at the 2, 4, 8, 12, 24 and 48 h sampling points.
 ; EHEC
; EHEC  ), B. subtilis ATCC 6051 (S. enterica
), B. subtilis ATCC 6051 (S. enterica  ; EHEC
; EHEC  ), B. mojavensis RRC 101 (S. enterica
), B. mojavensis RRC 101 (S. enterica  ; EHEC
; EHEC  ) and L. rhamnousus GG (S. enterica
) and L. rhamnousus GG (S. enterica  ; EHEC
; EHEC  ) at the 2, 4, 8, 12, 24 and 48 h sampling points.
) at the 2, 4, 8, 12, 24 and 48 h sampling points.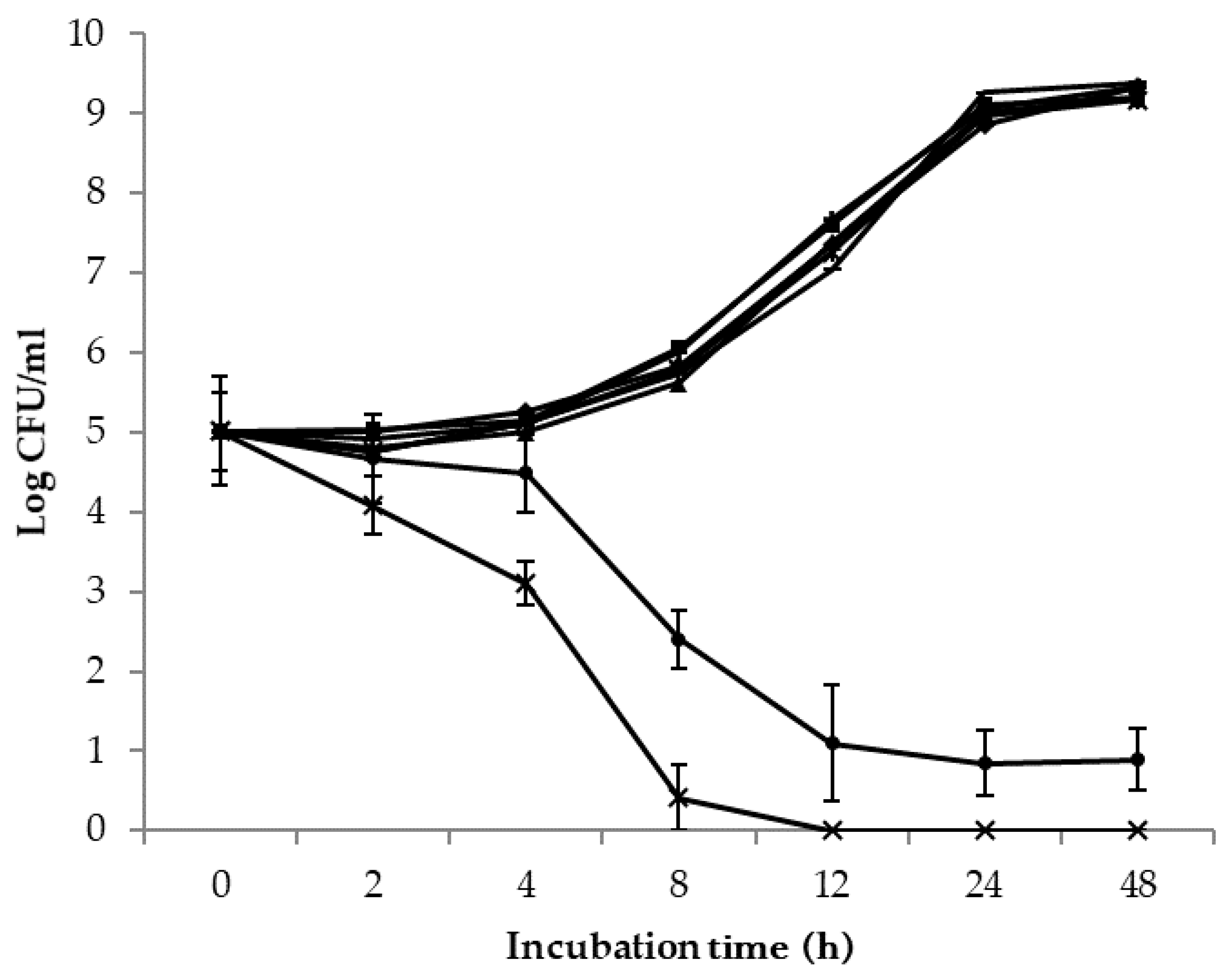
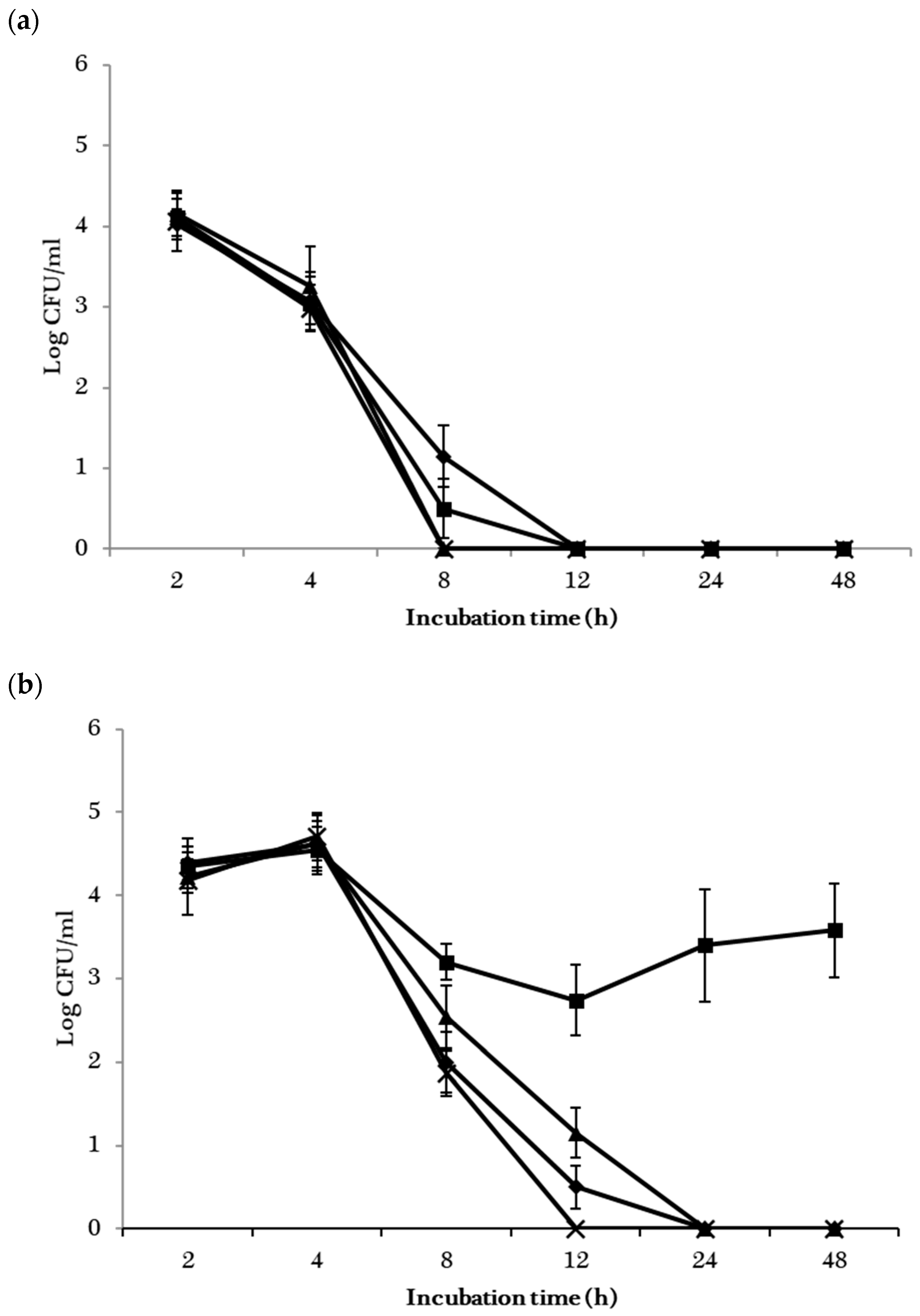
 ), S. Stanley and E. coli K4492 (
), S. Stanley and E. coli K4492 (  ), S. Cubana and E. coli H1730 (
), S. Cubana and E. coli H1730 (  ), as well as S. Baildon and E. coli BAA-2326 (
), as well as S. Baildon and E. coli BAA-2326 (  ).
).
 ), S. Stanley and E. coli K4492 (
), S. Stanley and E. coli K4492 (  ), S. Cubana and E. coli H1730 (
), S. Cubana and E. coli H1730 (  ), as well as S. Baildon and E. coli BAA-2326 (
), as well as S. Baildon and E. coli BAA-2326 (  ).
).
| Nalidixic Acid Resistant Derivate | Serovar/Strain | Source |
|---|---|---|
| Salmonella enterica | Stanley | Sprout-related outbreak, Finland, Canada, and the US, 1997 |
| Baildon | Tomato- and lettuce-related outbreaks, the US, 1999 | |
| Montevideo | Tomato-related outbreak, the US, 1993 | |
| Cubana | Sprout-related outbreak, the US, 2012 | |
| Enterohemorrhagic Escherichia coli | F4546 | Sprout-related outbreak, the US, 1997 |
| BAA-2326 | Fenugreek sprout-related outbreak, Germany, 2011 | |
| K4492 | Spinach-related outbreak, the US, 2006 | |
| H1730 | Lettuce-related outbreak, the US, 2003 | |
| Pseudomonas fluorescenes | A506 | Commercial biocontrol agent for Erwinia amylovora (fire blight) on fruits |
| Bacillus mojavensis | RRC 101 | Commercial biocontrol agent for Fusarium verticillioides in maize and other crops; surfactin-producing |
| Bacillus subtilis | ATCC 6051 | surfactin-producing strain |
| Lactobacillus rhamnosus | GG | Commercial probiotic |
| Pseudomonas syringae pv. tomato | DC 3000 | Frequent seed-borne pathogen on tomato plant |
| Main Effect | Mean Population of Salmonella or EHEC (log CFU/mL) as Influenced by | Main Effect | Mean Population (log CFU/mL) of Salmonella and EHEC as Influenced by | |
|---|---|---|---|---|
| S. enterica1 (n = 540) | E. coli2 (n = 540) | |||
| Competitive bacteria presence | S. enterica strains used | |||
| Control (n = 60) | 7.8 ± 0.26 A | 8.1 ± 0.26 A | S. Montevideo (n = 135) | 6.8 ± 0.19 A |
| P. syringae pv. tomato DC 3000 (n = 60) | 7.4 ± 0.24 B | 7.8 ± 0.26 B | S. Baildon (n = 135) | 6.6 ± 0.19 A |
| B. subtilis ATCC 6051 (n = 60) | 7.3 ± 0.22 BC | 7.6 ± 0.25 C | S. Stanley (n = 135) | 6.1 ± 0.20 B |
| B. mojavensis RRC 101 (n = 60) | 7.2 ± 0.21 C | 7.5 ± 0.24 C | S. Cubana (n = 135) | 6.1 ± 0.19 B |
| P. fluorescenes A506 (n = 60) | 7.0 ± 0.22 D | 7.4 ± 0.24 D | E. coli strains used | |
| Cocktail 1 3 (n = 60) | 6.9 ± 0.24 D | 7.3 ± 0.24 D | E. coli H1730 (n = 135) | 7.9 ± 0.12 A |
| Cocktail 2 4 (n = 60) | 6.9 ± 0.24 D | 7.2 ± 0.24 E | E. coli K4492 (n =135) | 7.7 ± 0.13 A |
| L. rhamnosus GG (25 °C; n = 60) | 6.0 ± 0.23 E | 6.8 ± 0.22 F | E. coli F4546 (n = 135) | 7.1 ± 0.17 B |
| L. rhamnosus GG (37 °C; n = 60) | 2.5 ± 0.31 F | 6.6 ± 0.15 G | E. coli BAA 2326 (n = 135) | 6.4 ± 0.17 C |
| Sampling points (h) used | ||||
| 24 (n = 108) | 7.5 ± 0.15 A | 8.7 ± 0.07 B | ||
| 48 (n = 108) | 7.5 ± 0.23 A | 9.0 ± 0.06 A | ||
| 72 (n = 108) | 6.9 ± 0.26 B | 8.4 ± 0.10 C | ||
| 12 (n = 108) | 6.2 ± 0.12 C | 6.3 ± 0.14 D | ||
| 6 (n = 108) | 3.8 ± 0.24 D | 4.1 ± 0.11 E | ||
| Salmonella Strains | Mean Population of Salmonella 1 (log CFU/mL) (n = 480) | |||||||
|---|---|---|---|---|---|---|---|---|
| P. fluorescens A506 | Cocktail 2 | Cocktail 1 | L. rhamnosus GG | Pst DC3000 | B. mojavensis RRC101 | Control | B. subtilis ATCC6051 | |
| Montevideo (n = 15) | 7.6 ± 0.39 aAB | 7.4 ± 0.44 aAB | 7.3 ± 0.44aAB | 6.5 ± 0.39 aB | 7.8 ± 0.45 aA | 7.6 ± 0.40 aAB | 8.0 ± 0.55 aA | 7.6 ± 0.41 aAB |
| Baildon (n = 15) | 7.2 ± 0.45 aA | 7.2 ± 0.46 aA | 7.4 ± 0.46 aA | 5.9 ± 0.53 aB | 7.8 ± 0.41 aA | 7.4 ± 0.39 aA | 8.0 ± 0.49 aA | 7.6 ± 0.38 aA |
| Stanley (n = 15) | 6.8 ± 0.47 aAB | 6.3 ± 0.46 aAB | 6.5 ± 0.48aAB | 5.7 ± 0.47 aB | 7.2 ± 0.46 aA | 7.0 ± 0.45 aAB | 7.5 ± 0.56 aA | 7.0 ± 0.49 aAB |
| Cubana (n = 15) | 6.5 ± 0.46 aAB | 6.5 ± 0.54 aAB | 6.5 ± 0.45 aAB | 5.9 ± 0.49 aB | 6.8 ± 0.54 aAB | 6.8 ± 0.45 aAB | 7.9 ± 0.50 aA | 6.8 ± 0.47 aAB |
| EHEC Strains | Mean Population of EHEC 1 (log CFU/mL) (n = 480) | |||||||
|---|---|---|---|---|---|---|---|---|
| P. fluorescens A506 | Cocktail 2 | Cocktail 1 | L. rhamnosus GG | Pst DC3000 | B. mojavensis RRC101 | Control | B. subtilis ATCC6051 | |
| H1730 (n = 15) | 8.2 ± 0.30 aAB | 7.5 ± 0.35 abAB | 7.7 ± 0.35 abAB | 7.4 ± 0.38 aB | 8.6 ± 0.33 aAB | 8.2 ± 0.33 aAB | 8.6 ± 0.42 abA | 8.3 ± 0.32 aAB |
| K4492 (n = 15) | 8.1 ± 0.40 aAB | 7.6 ± 0.43 aAB | 7.7 ± 0.42 aAB | 7.1 ± 0.46 aB | 8.4 ± 0.38 aA | 8.3 ± 0.38 aA | 8.7 ± 0.41 aA | 8.3 ± 0.39 aA |
| F4546 (n = 15) | 6.9 ± 0.56 abA | 7.3 ± 0.58 abA | 7.3 ± 0.56 abA | 6.7 ± 0.47 aA | 7.7 ± 0.52 abA | 7.3 ± 0.53 abA | 7.9 ± 0.52 abA | 7.2 ± 0.54 abA |
| BAA2326 (n = 15) | 6.3 ± 0.48 bA | 6.2 ± 0.53 bA | 6.6 ± 0.49 bA | 6.0 ± 0.43 aA | 6.6 ± 0.60 bA | 6.7 ± 0.57 bA | 7.3 ± 0.65 bA | 6.6 ± 0.59 bA |
| Percentage (%) of Attached Cells | ||
|---|---|---|
| Salmonella1 (n = 84) | EHEC (n = 84) | |
| As influenced by competitive strains | ||
| Control (n = 12) | 10.5 ± 0.38 A | 3.9 ± 0.14 A |
| L. rhamnosus GG (n = 12) | 9.4 ± 0.33 B | 3.4 ± 0.11 B |
| Cocktail 2 2 (n = 12) | 9.0 ± 0.29 C | 3.1 ± 0.18 C |
| B. mojavensis RRC 101 (n = 12) | 7.9 ± 0.21 D | 2.9 ± 0.16 C |
| B. subtilis ATCC 6051 (n = 12) | 8.1 ± 0.24 D | 2.8 ± 0.23 C |
| P. syringae pv. tomato DC 3000 (n = 12) | 8.2 ± 0.27 D | 3.5 ± 0.20 B |
| P. fluorescenes A506 (n = 12) | 7.0 ± 0.20 E | 2.4 ± 0.12 D |
| On different seed types | ||
| Fenugreek (n = 21) | 12.5 ± 0.19 A | 6.7 ± 0.11 A |
| Alfalfa (n = 21) | 11.8 ± 0.08 B | 2.0 ± 0.08 B |
| Lettuce (n = 21) | 8.9 ± 0.08 C | 1.7 ± 0.04 C |
| Tomato (n = 21) | ND 3 | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Walcott, R.; Mis Solval, K.; Chen, J. Influence of Bacterial Competitors on Salmonella enterica and Enterohemorrhagic Escherichia coli Growth in Microbiological Media and Attachment to Vegetable Seeds. Foods 2021, 10, 285. https://doi.org/10.3390/foods10020285
Liu D, Walcott R, Mis Solval K, Chen J. Influence of Bacterial Competitors on Salmonella enterica and Enterohemorrhagic Escherichia coli Growth in Microbiological Media and Attachment to Vegetable Seeds. Foods. 2021; 10(2):285. https://doi.org/10.3390/foods10020285
Chicago/Turabian StyleLiu, Da, Ronald Walcott, Kevin Mis Solval, and Jinru Chen. 2021. "Influence of Bacterial Competitors on Salmonella enterica and Enterohemorrhagic Escherichia coli Growth in Microbiological Media and Attachment to Vegetable Seeds" Foods 10, no. 2: 285. https://doi.org/10.3390/foods10020285





