Development of a Systematic qPCR Array for Screening GM Soybeans
Abstract
1. Introduction
2. Materials and Methods
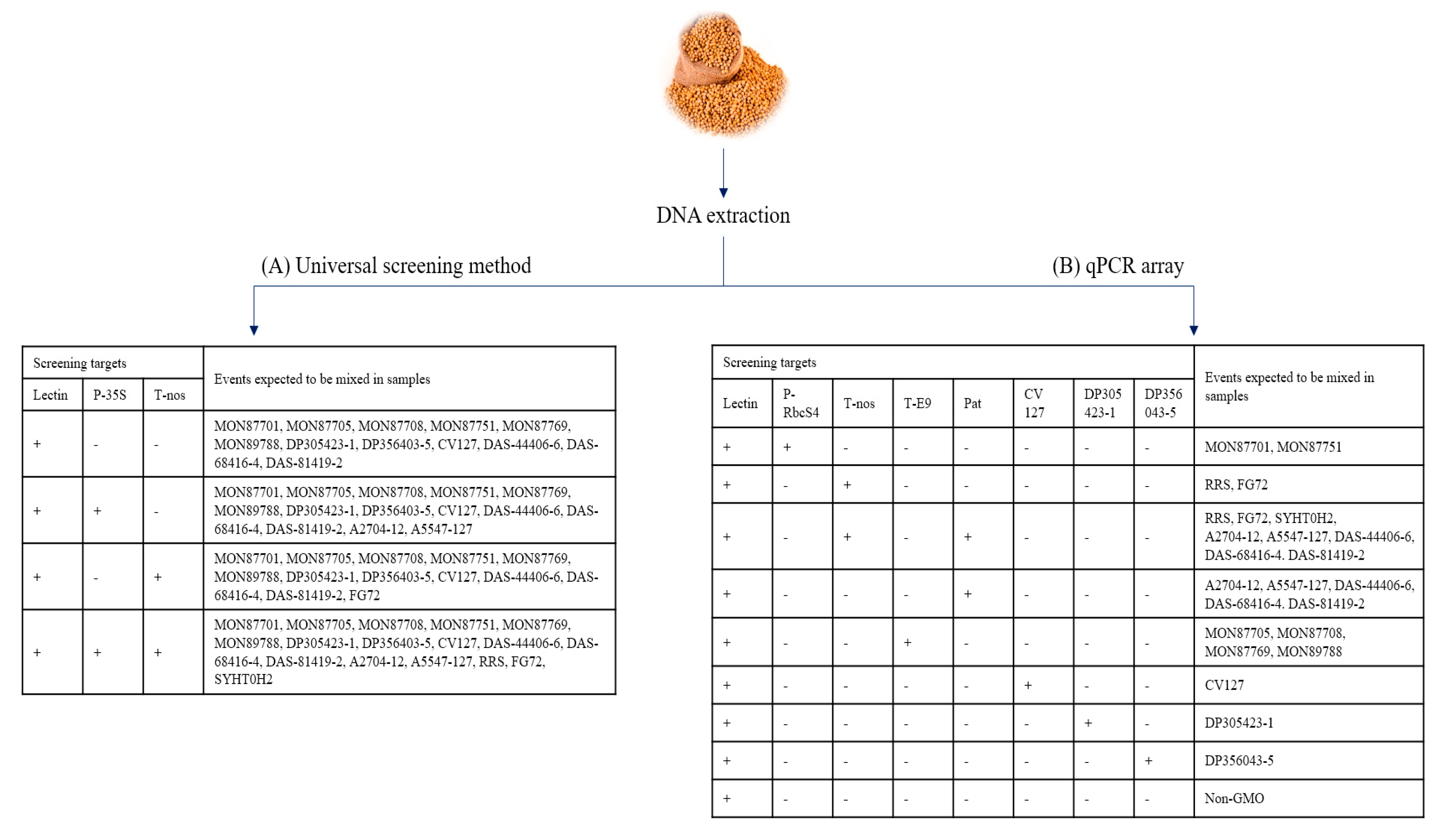
2.1. Preparation of Samples
2.2. DNA Extraction
2.3. qPCR Array
2.4. Target Specific Primers and Probes
2.5. PCR Condition
2.6. Estimation of the Limit of Detection (LOD)
3. Results
3.1. Development of the Soybean-Specific Screening qPCR Array
3.2. The Specificity and Sensitivity of Screening Assays
3.3. The Applicability Test Using Processed Foods
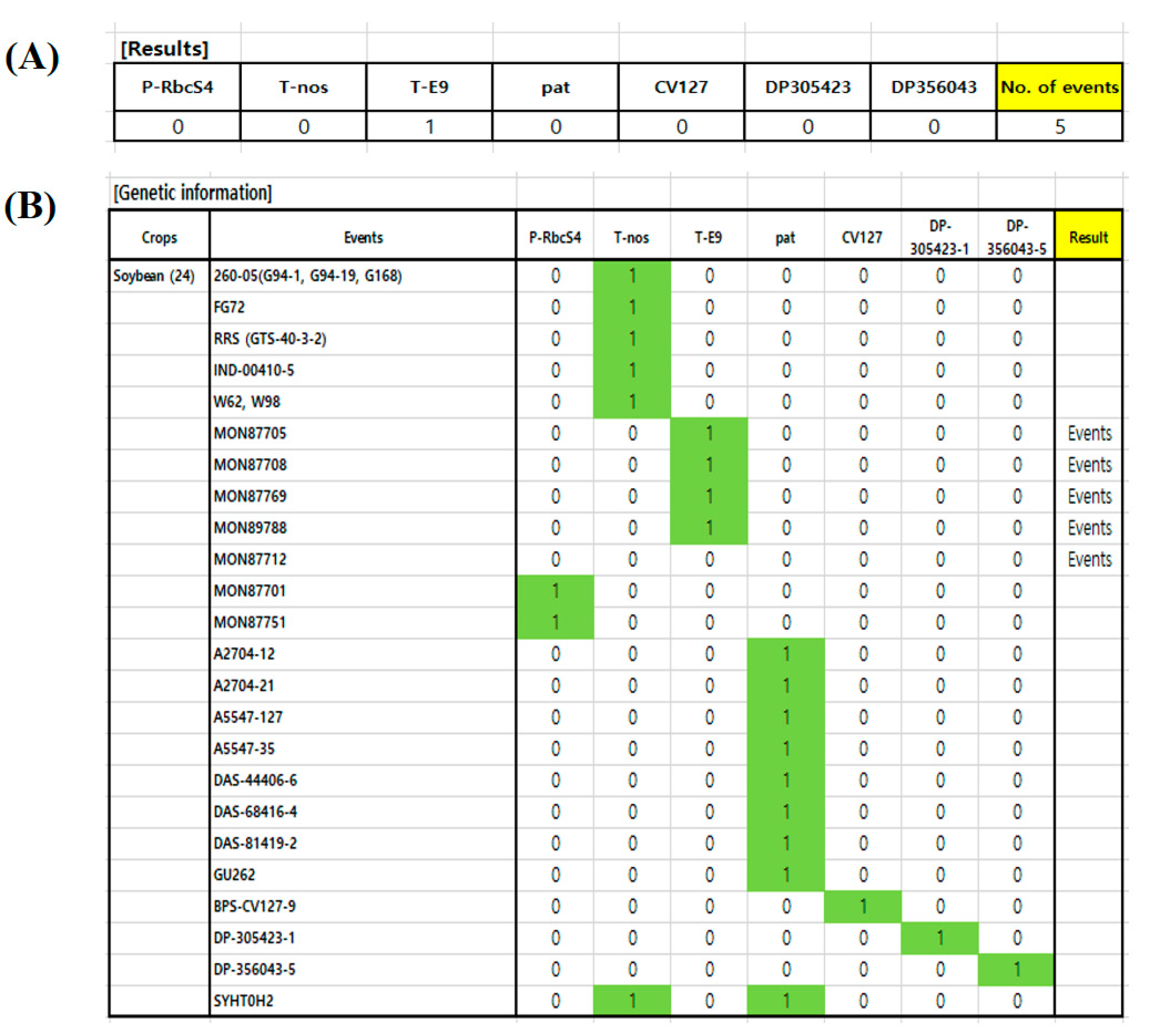
3.4. The Prediction System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Block, A.; Debode, F.; Grohmann, L.; Hulin, J.; Taverniers, I.; Kluga, L.; Barbau-Piednoir, E.; Boreders, S.; Huber, I.; Bulcke, M.; et al. The GMOseek matrix: A decision support tool for optimizing the detection of genetically modified plants. BMC Bioinform. 2013, 14, 256. [Google Scholar] [CrossRef] [PubMed]
- Datukishvili, N.; Kutateladze, T.; Gabriadze, I.; Bitskinashvili, K.; Vishnepolsky, B. New multiplex PCR methods for rapid screening of genetically modified organisms in foods. Front. Microbiol. 2015, 6, 757. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Zhang, D.; Kim, H.Y. Detection of sixteen genetically modified maize events in processed foods using four event-specific pentaplex PCR systems. Food Control 2014, 35, 345–353. [Google Scholar] [CrossRef]
- Park, S.B.; Kim, H.Y.; Kim, J.H. Multiplex PCR system to track authorized and unauthorized genetically modified soybean events in food and feed. Food Control 2015, 54, 47–52. [Google Scholar] [CrossRef]
- Randhawa, G.J.; Chhabra, R.; Singh, M. Multiplex PCR-based simultaneous amplification of selectable marker and reporter genes for the screening of genetically modified crops. J. Agric. Food Chem. 2009, 57, 5167–5172. [Google Scholar] [CrossRef] [PubMed]
- Park, S.B.; Kim, H.Y.; Kim, J.H. Development of a screening method for the monitoring 38 genetically modified maize events in food and feed in South Korea. Food Control 2017, 73, 1459–1465. [Google Scholar] [CrossRef]
- Singh, M.; Bhoge, R.; Randhawa, G. Crop-specific GMO matrix-multiplex PCR: A cost-efficient screening strategy for genetically modified maize and cotton events approved globally. Food Control 2016, 70, 271–280. [Google Scholar] [CrossRef]
- Debode, F.; Janssen, E.; Berben, G. Development of 10 new screening PCR assays for GMO detection targeting promoters (pFMV, pNOS, pSSUAra, pTA29, pUbi, pRice actin) and terminators (t35S, tE9, tOCS, tg7). Eur. Food Res. Technol. 2013, 236, 659–669. [Google Scholar] [CrossRef]
- Randhawa, G.J.; Singh, M. Multiplex, construct-specific and real-time PCR-based analytical methods for Bt rice with cry1Ac gene. J. AOAC Int. 2012, 95, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Huber, I.; Block, A.; Sebah, D.; Debode, F.; Morisset, D.; Grohmann, L.; Berben, G.; Stebih, D.; Milavec, M.; Zel, J.; et al. Development and validation of duplex, triplex, and pentaplex real-time PCR screening assays for the detection of genetically modified organisms in food and feed. J. Agric. Food Chem. 2013, 61, 10293–10301. [Google Scholar] [CrossRef] [PubMed]
- Debode, F.; Huber, I.; Macarthur, R.; Rischitor, P.E.; Mazzara, M.; Herau, V.; Sebah, D.; Dobnik, D.; Broeders, S.; Roosens, N.H.; et al. Inter-laboratory studies for the validation of two singlepex (tE9 and pea lectin) and one duplex (pat/bar) real-time PCR methods for GMO detection. Food Control 2017, 73, 452–461. [Google Scholar] [CrossRef]
- Mano, J.; Shuko, H.; Yasuaki, N.; Futo, S.; Takabatake, R.; Kitta, K. Highly sensitive GMO detection using real-time PCR with a large amount of DNA template: Single-laboratory validation. J. Food Compos. Anal. 2018, 101, 2. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.; Gatto, F.; Querci, M.; Kreysa, J. Development of a ready-to-use, multi-target screening pre-spotted plate (sPSP) for GMO detection. JRC Tech. Rep. 2013. [Google Scholar]
- Querci, M.; Bulcke, M.; Eede, G.; Broll, H. New approaches in GMO detection. Anal. Bioanal. Chem. 2009, 396, 1991–2002. [Google Scholar] [CrossRef] [PubMed]
- Gatto, F.; Bssani, N.; Rosa, S.; Lievens, A.; Brustio, R.; Kreysa, J.; Querci, M. Semi-quantification of GM maize using ready-to-use RTi-PCR plates. Food Anal. Methods 2017, 10, 549–558. [Google Scholar] [CrossRef]
- Mano, J.; Shigemitsu, N.; Futo, S.; Akiyama, H.; Teshima, R.; Hino, A.; Furui, S.; Kitta, K. Real-time PCR array as a universal platform for the detection of genetically modified crops and its application in identifying unapproved genetically modified crops in Japan. J. Agric. Food Chem. 2009, 57, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Waiblinger, H.U.; Grohmann, L.; Mankertz, J.; Engelbert, D.; Pietsh, K. A practical approach to screen for authorized and unauthorized genetically modified plants. Anal. Bioanal. Chem. 2010, 396, 2065–2072. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, L.; Busch, U.; Pecoraro, S. GMOfinder—A GMO screening database. Food Anal. Methods 2012, 5, 1368–1376. [Google Scholar] [CrossRef]
- Wilkese, T.; Hall, L.; Burns, M. A brief review of current bioinformatics decision support system (DSS) tools for screening for GMOs in the EU using PCR-based approaches. J. Assoc. Publ. Anal. 2017, 45, 023–040. [Google Scholar]
- Angers-Loustau, A.; Petrillo, M.; Bonfini, L.; Gatto, F.; Rosa, S.; Patak, A.; Kreysa, J. JRC GMO-Matrix: A web application to support Genetically Modified Organisms detection strategies. BMC Bioinform. 2014, 15, 417. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, M.; Angers-Loustau, A.; Henriksson, P.; Bonfini, L.; Patak, A.; Kreysa, J. JRC GMO-Amplicons: A collection of nucleic acid sequences related to genetically modified organisms. Database 2015, 2015, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.F.; Gatto, F.; Angers-Loustau, A.; Petrillo, M.; Kreysa, J.; Querci, M. Development and applicability of a ready-to-use PCR system for GMO screening. Food Chem. 2016, 201, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Food and Drug Safety. Korean Food Code. MFDS Notification; No. 55; MFDS: Cheongju, Korea, 2020.
- Kim, M.J.; Yoo, I.; Lee, S.Y.; Kim, H.Y. Quantitative detection of pork in commercial meat products by TaqMan® real-time assay targeting the mitochondrial D-loop region. Food Chem. 2016, 201, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Kitpipit, T.; Sittichan, K.; Thanakiatkrai, P. Direct-multiplex PCR assay for meat species identification in food products. Food Chem. 2014, 163, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.F.; Hwang, D.F. A multiplex PCR assay for species identification of raw and cooked bonito. Food Control 2008, 19, 879–885. [Google Scholar] [CrossRef]
- Ministry of Agriculture and Forestry of Korea. Guidelines for Labeling of Genetically Modified Agricultural Products. MAF Notification; No. 31; MAF: Sejong, Korea, 2000.
- European Commission Regulation (EC) 1829/2003 and 1830/2003. Off. J. Eur. Communities 2003, L268, 1–28.
| Soybean Events | Genetic Elements | ||
|---|---|---|---|
| Promoter | Terminator | Gene | |
| RRS | 35S | Nos | Cp4 epsps |
| A2704-12 | 35S | 35S | pat, bla |
| A5547-127 | 35S | 35S | pat, bla |
| DAS-81419-2 | CsVMV, Ubi10_At | tr7, ORF23 | pat, cry1F, cry1Ac (synpro) |
| DAS-68416-4 | CsVMV, Ubi10_At | tr7, ORF23 | pat, aad-12 |
| DAS-44406-6 | CsVMV, Ubi10_At, h4a748 | tr7, ORF23, h4a748 | pat, aad-12, 2mepsps |
| MON87769 | 7Sα′, 7Sα | E9, tml | Δ6D_PJ, Fad3_NC |
| MON87705 | FMV/TSF1, 7Sα′ | E9, H6 | Cp4 epsps, fad2-1A, FATB1-A |
| MON89788 | FMV/TSF1 | E9 | Cp4 epsps |
| MON87701 | RbcS4 | 7Sα’ | cry1Ac |
| MON87751 | RbcS4, act2 | Mt, Pt1 | cry1A.105, cry2Ab2 |
| MON87708 | PCSV | E9 | dmo |
| DP305423-1 | SAMS, Kti3 | ALS_Gm, Kti3 | hra_Gm, fad2-1_Gm |
| DP356043-5 | SAMS, SCP1 | PinII, ALS_GM | gat4601, hra_Gm |
| SYHT0H2 | 35S, CMP, SMP | Nos | pat, avhppd |
| CV127 | AHASL | AHASL | csr1-2 |
| FG72 | h4a748 | nos, h4a748 | 2mepsps, hppdPfW336 |
| Targets | Primers/Probe | Sequences (5′→3′) | References |
|---|---|---|---|
| lectin | GM1-F | CCA GCT TCG CCG CTT CCT TC | [23] |
| GM1-R | GAA GGC AAG CCC ATC TGC AAG CC | ||
| GM1-P | FAM-CTT CAC CTT CTA TGC CCC TGA CAC-TAMRA | ||
| T-nos | NOS ter 2-5′ | GTC TTG CGA TG ATTA TCA TAT AAT TTC TG | [23] |
| NOS ter 2-3′ | CGC TAT ATT TTG TTT TCT ATC GCG T | ||
| NOS-Taq | FAM-AGA TGG GTT TTT ATG ATT AGA GTC CCG CAA-TAMRA | ||
| T-E9 | T-E9_MF | CGA CAA CGT TCG TCA AGT TC | This study |
| T-E9_MR | CCC AAT GCC ATA ATA CTC GAA C | ||
| T-E9_NP | FAM-AAT GCA TCA GTT TCA TTG CG-TAMRA | ||
| P-RbcS4 | P-RbcS4-F | AAG CAC CAC TCC ACC ATC AC | This study |
| P-RbcS4-R | AGG TGT TGA GAC CCT TAT CG | ||
| P-RbcS4-P | FAM-ACG TGG CAT TAT TCC AGC GG-TAMRA | ||
| pat | Pat-F | CGA TCC ATC TGT TAG GTT GC | [6] |
| Pat-R | CCT TGG AGG AGC TGG CAA CT | ||
| Pat-P | FAM-ATA CAA GCA TGG TGG ATG GC-TAMRA | This study | |
| CV127 | SE-127-f4 | AAC AGA AGT TTC CGT TGA GCT TTA AGA C | [23] |
| SE-127-r2 | CAT TCG TAG CTC GGA TCG TGT AC | ||
| SE-127-p3 | FAM-TTT GGG GAA GCT GTC CCA TGC CC-TAMRA | ||
| DP 305423-1 | DP305423-f1 | CGT GTT CTC TTT TTG GCT AGC | [23] |
| DP305423-r5 | GTG ACC AAT GAA TAC ATA ACA CAA ACT A | ||
| DP305423-1p | FAM-TGA CAC AAA TGA TTT TCA TAC AAA AGT CGA GA-TAMRA | ||
| DP 356043-5 | DP356-f1 | GTC GAA TAG GCT AGG TTT ACG AAA AA | [23] |
| DP356-r1 | TTT GAT ATT CTT GGA GTA GAC GAG AGT GT | ||
| DP356-p | FAM-CTC TAG AGA TCC GTC AAC ATG GTG GAG CAC-TAMRA |
| Soybean Events | Ct Value ± SD * | |||||||
|---|---|---|---|---|---|---|---|---|
| lectin | P-RbcS4 | T-nos | T-E9 | pat | CV127 | DP305423-1 | DP356043-5 | |
| RRS | 25.42 ± 0.18 | - ** | 28.37 ± 0.06 | - | - | - | - | - |
| A2704-12 | 24.89 ± 0.02 | - | - | - | 20.85 ± 0.05 | - | - | - |
| A5547-127 | 25.78 ± 0.14 | - | - | - | 22.99 ± 0.01 | - | - | - |
| DAS-81419-2 | 26.43 ± 0.00 | - | - | - | 23.60 ± 0.00 | - | - | - |
| DAS-68416-4 | 26.63 ± 0.01 | - | - | - | 24.64 ± 0.03 | - | - | - |
| DAS-44406-6 | 25.31 ± 0.13 | - | - | - | 24.10 ± 0.00 | - | - | - |
| MON87769 | 25.59 ± 0.05 | - | - | 27.27 ± 0.10 | - | - | - | - |
| MON87705 | 25.61 ± 0.02 | - | - | 26.21 ± 0.08 | - | - | - | - |
| MON89788 | 26.07 ± 0.18 | - | - | 26.22 ± 0.07 | - | - | - | - |
| MON87701 | 25.25 ± 0.05 | 26.33 ± 0.05 | - | - | - | - | - | - |
| MON87751 | 25.66 ± 0.02 | 25.95 ± 0.01 | - | - | - | - | - | - |
| MON87708 | 25.32 ± 0.17 | - | - | 26.71 ± 0.09 | - | - | - | - |
| DP305423-1 | 24.94 ± 0.10 | - | - | - | - | - | 25.43 ± 0.08 | - |
| DP356043-5 | 25.67 ± 0.14 | - | - | - | - | - | - | 26.98 ± 0.01 |
| SYHT0H2 | 26.40 ± 0.01 | - | 24.91 ± 0.02 | - | 20.54 ± 0.01 | - | - | - |
| CV127 | 25.63 ± 0.05 | - | - | - | - | 25.95 ± 0.02 | - | - |
| FG72 | 26.64 ± 0.02 | - | 26.84 ± 0.01 | - | - | - | - | - |
| Target | Template | Positive Signal | ||
|---|---|---|---|---|
| Experimenter A | Experimenter B | Experimenter C | ||
| P-RbcS4 | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 9/9 | 9/9 | 9/9 | |
| T-nos | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 9/9 | 9/9 | 9/9 | |
| T-E9 | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 9/9 | 9/9 | 9/9 | |
| pat | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 9/9 | 9/9 | 9/9 | |
| CV127 | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 9/9 | 9/9 | 9/9 | |
| DP305423-1 | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 8/9 | 9/9 | 9/9 | |
| DP356043-5 | 0.1% | 9/9 | 9/9 | 9/9 |
| 0.05% | 9/9 | 9/9 | 9/9 | |
| 0.01% | 7/9 | 9/9 | 9/9 | |
| Product Type | Sample No. | Results | Events Expected to Be Mixed in Samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| lectin | P-RbcS4 | T-nos | T-E9 | Pat | CV127 | DP305 423-1 | DP356 043-5 | |||
| Soybean Powder | 1 | + | - | - | - | - | - | - | - | - |
| 2 | + | - | - | + | - | - | - | - | MON89788, MON87705, MON87708, MON87769 | |
| 3 | + | - | - | - | - | - | - | - | - | |
| Dried Cereal | 4 | + | - | - | - | - | - | - | - | |
| 5 | + | - | - | - | - | - | - | - | ||
| 6 | + | - | - | - | - | - | - | - | ||
| 7 | + | - | - | + | - | - | - | - | MON89788, MON87705, MON87708, MON87769 | |
| 8 | + | - | - | - | - | - | - | - | ||
| 9 | + | - | - | - | - | - | - | - | ||
| 10 | + | - | - | - | - | - | - | - | ||
| 11 | + | - | - | + | - | - | - | - | MON89788, MON87705, MON87708, MON87769 | |
| Soy Milk | 12 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 |
| Tofu | 13 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 |
| 14 | + | - | - | - | - | - | - | - | - | |
| 15 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 | |
| 16 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 | |
| 17 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 | |
| 18 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 | |
| 19 | + | - | + | + | - | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769 | |
| 20 | + | - | + | + | + | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769, A2704-12, A5547-127, DAS-44406-6, DAS-68416-4, DAS-81419-2, SYHT0H2 | |
| 21 | + | - | + | + | - | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769 | |
| 22 | + | - | + | + | - | - | - | - | RRS, FG72, MON89788, MON87705, MON87708, MON87769 | |
| Snack | 23 | + | - | - | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-B.; Kim, J.-Y.; Lee, D.-G.; Kim, J.-H.; Shin, M.-K.; Kim, H.-Y. Development of a Systematic qPCR Array for Screening GM Soybeans. Foods 2021, 10, 610. https://doi.org/10.3390/foods10030610
Park S-B, Kim J-Y, Lee D-G, Kim J-H, Shin M-K, Kim H-Y. Development of a Systematic qPCR Array for Screening GM Soybeans. Foods. 2021; 10(3):610. https://doi.org/10.3390/foods10030610
Chicago/Turabian StylePark, Saet-Byul, Ji-Yeong Kim, Do-Geun Lee, Jae-Hwan Kim, Min-Ki Shin, and Hae-Yeong Kim. 2021. "Development of a Systematic qPCR Array for Screening GM Soybeans" Foods 10, no. 3: 610. https://doi.org/10.3390/foods10030610
APA StylePark, S.-B., Kim, J.-Y., Lee, D.-G., Kim, J.-H., Shin, M.-K., & Kim, H.-Y. (2021). Development of a Systematic qPCR Array for Screening GM Soybeans. Foods, 10(3), 610. https://doi.org/10.3390/foods10030610







