Impact of the Resistance Responses to Stress Conditions Encountered in Food and Food Processing Environments on the Virulence and Growth Fitness of Non-Typhoidal Salmonellae
Abstract
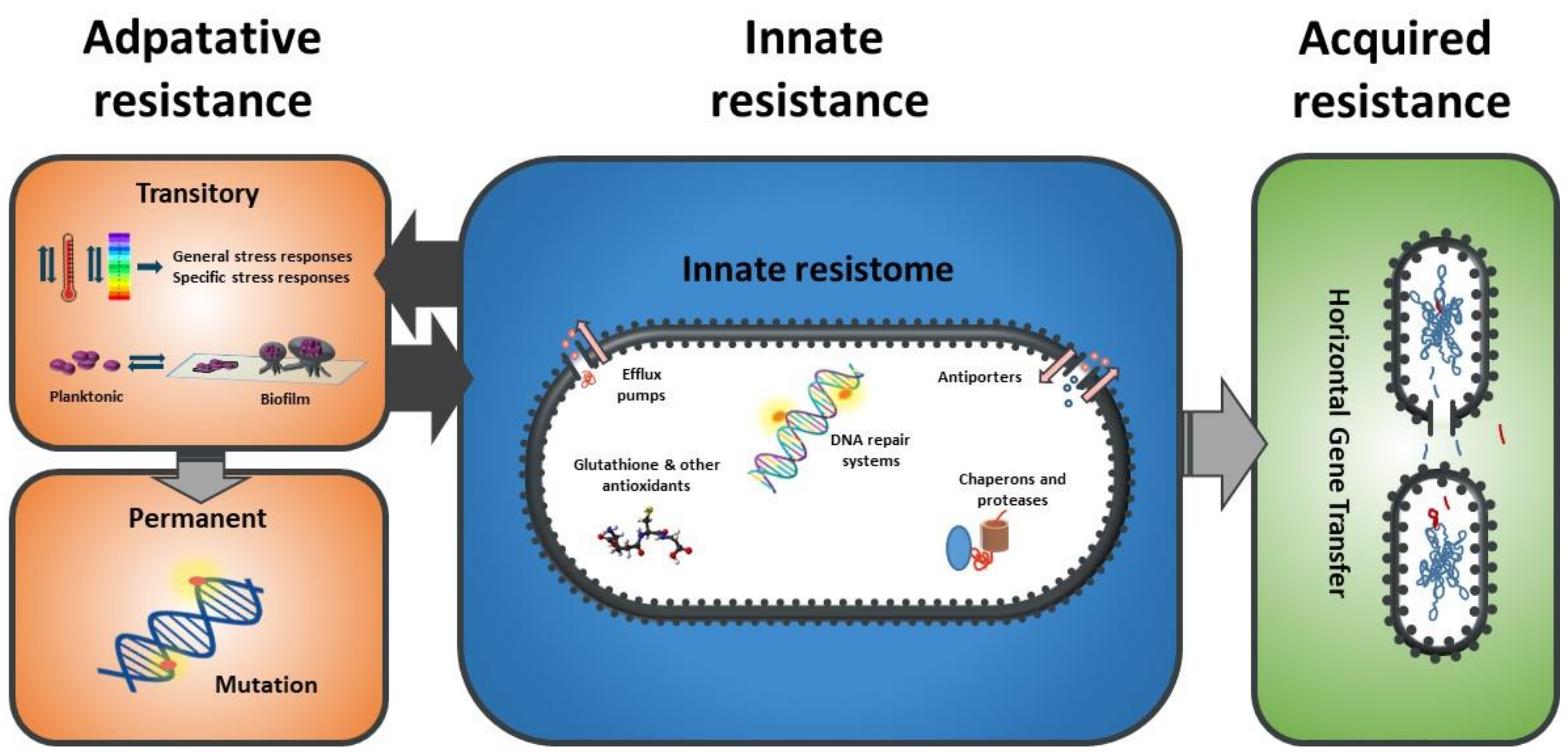
1. Introduction
2. Bacterial Stress Resistance, Virulence, and Growth Fitness
3. Salmonella Stress Resistance Mechanisms: Impact on Virulence and Growth Fitness
3.1. Non-Specific Stress Responses
3.1.1. The General Stress Response (GSR)
3.1.2. The Extracytoplasmic Stress Response (ESR)
3.2. Specific Stress Responses
3.2.1. Acid Stress
3.2.2. Osmotic Stress
3.2.3. Oxidative Stress
3.2.4. Starvation
3.2.5. Modified Atmospheres
3.2.6. Chemical Stressors: Detergents and Disinfectants
3.2.7. Chilling
3.2.8. Heat
3.2.9. Non-Thermal Technologies
3.3. Other Stress Responses
3.3.1. Viable but Non-Culturable (VBNC) and Persister Cells
3.3.2. Biofilms
4. Impact of Stress Resistance Responses on Other Aspects of Salmonella Physiology
5. Variability among Salmonella Strains and Serovars
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abee, T.; Wouters, J.A. Microbial Stress Response in Minimal Processing. Int. J. Food Microbiol. 1999, 50, 65–91. [Google Scholar] [CrossRef]
- Dodd, C.E.R.; Aldsworth, T.G. The Importance of RpoS in the Survival of Bacteria through Food Processing. Int. J. Food Microbiol. 2002, 74, 189–194. [Google Scholar] [CrossRef]
- Woolhouse, M.E.J.; Webster, J.P.; Domingo, E.; Charlesworth, B.; Levin, B.R. Biological and Biomedical Implications of the Co-Evolution of Pathogens and Their Hosts. Nat. Genet. 2002, 32, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef] [PubMed]
- Felipe-lópez, A.; Hensel, M. Bacterial responses to the host cell. In Bacterial Stress Responses, 2nd ed.; Storz, G., Hengge, R., Eds.; ASM Press: Washington, DC, USA, 2011; ISBN 978-1-55581-684-1. [Google Scholar]
- Zambrano, M.M.; Siegele, D.A.; Almirón, M.; Tormo, A.; Kolter, R. Microbial Competition: Escherichia Coli Mutants That Take over Stationary Phase Cultures. Science 1993, 259, 1757–1760. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic Resistance and Its Cost: Is It Possible to Reverse Resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- Marder, E.P.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Hurd, S.; Jervis, R.; Lathrop, S.; Muse, A.; Ryan, P.; Smith, K.; et al. Preliminary Incidence and Trends of Infections with Pathogens Transmitted Commonly through Food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2006–2017. Morb. Mortal. Wkly. Rep. 2018, 67, 324–328. [Google Scholar] [CrossRef]
- De Cesare, A. Salmonella in Foods: A Reemerging Problem. Adv. Food Nutr. Res. 2018, 86, 137–179. [Google Scholar] [CrossRef]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same Species, Different Diseases: How and Why Typhoidal and Non-Typhoidal Salmonella Enterica Serovars Differ. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Fatica, M.K.; Schneider, K.R. Salmonella and Produce: Survival in the Plant Environment and Implications in Food Safety. Virulence 2011, 2, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Podolak, R.; Enache, E.; Stone, W.; Black, D.G.; Elliott, P.H. Sources and Risk Factors for Contamination, Survival, Persistence, and Heat Resistance of Salmonella in Low-Moisture Foods. J. Food Prot. 2010, 73, 1919–1936. [Google Scholar] [CrossRef]
- Tessari, E.N.C.; Kanashiro, A.M.I.; Stoppa, G.F.Z.; Luciano, R.L.; Castro, A.G.M.D.; Cardoso, A.L.S.P. Important aspects of Salmonella in the poultry industry and in public health. In Salmonella—A Dangerous Foodborne Pathogen; Mahmoud, B.S.M., Ed.; InTech Europa: Rijeka, Croatia, 2012; pp. 181–206. ISBN 10.5772/30812. [Google Scholar]
- Santos, S.A.O.; Martins, C.; Pereira, C.; Silvestre, A.J.D.; Rocha, S.M. Current Challenges and Perspectives for the Use of Aqueous Plant Extracts in the Management of Bacterial Infections: The Case-Study of Salmonella Enterica Serovars. Int. J. Mol. Sci. 2019, 20, 940. [Google Scholar] [CrossRef]
- McMahon, M.A.S.; Xu, J.; Moore, J.E.; Blair, I.S.; McDowell, D.A. Environmental Stress and Antibiotic Resistance in Food-Related Pathogens. Appl. Environ. Microbiol. 2007, 73, 211–217. [Google Scholar] [CrossRef]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The Complex Relationship between Virulence and Antibiotic Resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef]
- Olivares, J.; Bernardini, A.; Garcia-Leon, G.; Corona, F.; Sanchez, M.B.; Martinez, J.L. The Intrinsic Resistome of Bacterial Pathogens. Front. Microbiol. 2013, 4, 103. [Google Scholar] [CrossRef]
- McDowell, D.A. Food processing stresses in the spread of antibiotic resistance. In Safety Assurance during Food Processing; Smulders, F.J.M., Collins, J.D., Eds.; Wageningen Academic Publishers: Wageningen, The Netherlands, 2004; ISBN 978-90-8686-522-2. [Google Scholar]
- Batt, C.A. Virulence. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 978-0-08-100596-5. [Google Scholar]
- Daigle, F. Typhi Genes Expressed during Infection or Involved in Pathogenesis. J. Infect. Dev. Ctries. 2008, 2, 431–437. [Google Scholar] [CrossRef]
- Sabbagh, S.C.; Forest, C.G.; Lepage, C.; Leclerc, J.-M.; Daigle, F. So Similar, yet so Different: Uncovering Distinctive Features in the Genomes of Salmonella Enterica Serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 2010, 305, 1–13. [Google Scholar] [CrossRef]
- Lee, M.D.; Curtiss, R.; Peay, T. The Effect of Bacterial Surface Structures on the Pathogenesis of Salmonella typhimurium Infection in Chickens. Avian Dis. 1996, 40, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Mambu, J.; Virlogeux-Payant, I.; Holbert, S.; Grépinet, O.; Velge, P.; Wiedemann, A. An Updated View on the Rck Invasin of Salmonella: Still Much to Discover. Front. Cell. Infect. Microbiol. 2017, 7. [Google Scholar] [CrossRef]
- Ilyas, B.; Tsai, C.N.; Coombes, B.K. Evolution of Salmonella-Host Cell Interactions through a Dynamic Bacterial Genome. Front. Cell. Infect. Microbiol. 2017, 7, 428. [Google Scholar] [CrossRef] [PubMed]
- Jajere, S.M. A Review of Salmonella Enterica with Particular Focus on the Pathogenicity and Virulence Factors, Host Specificity and Antimicrobial Resistance Including Multidrug Resistance. Vet. World 2019, 12, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.; Saxena, A.; Kumar, R.; KumarSaxena, M. Virulence System of Salmonella with Special Reference to Salmonella enterica. Salmonella-Re-Emerg. Pathog. 2018. [Google Scholar] [CrossRef]
- Ibarra, J.A.; Steele-Mortimer, O. Salmonella—The Ultimate Insider. Salmonella Virulence Factors That Modulate Intracellular Survival. Cell. Microbiol. 2009, 11, 1579–1586. [Google Scholar] [CrossRef]
- Erhardt, M.; Dersch, P. Regulatory Principles Governing Salmonella and Yersinia Virulence. Front. Microbiol. 2015, 6, 949. [Google Scholar] [CrossRef]
- Hengge, R. Stationary-Phase Gene Regulation in Escherichia coli. EcoSal Plus 2011, 4. [Google Scholar] [CrossRef]
- Österberg, S.; del Peso-Santos, T.; Shingler, V. Regulation of Alternative Sigma Factor Use. Annu. Rev. Microbiol. 2011, 65, 37–55. [Google Scholar] [CrossRef]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-Mediated General Stress Response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Lago, M.; Monteil, V.; Douche, T.; Guglielmini, J.; Criscuolo, A.; Maufrais, C.; Matondo, M.; Norel, F. Proteome Remodelling by the Stress Sigma Factor RpoS/σS in Salmonella: Identification of Small Proteins and Evidence for Post-Transcriptional Regulation. Sci. Rep. 2017, 7, 2127. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. Proteolysis of σS (RpoS) and the General Stress Response in Escherichia coli. Res. Microbiol. 2009, 160, 667–676. [Google Scholar] [CrossRef]
- Altuvia, S.; Almirón, M.; Huisman, G.; Kolter, R.; Storz, G. The Dps Promoter Is Activated by OxyR during Growth and by IHF and Sigma S in Stationary Phase. Mol. Microbiol. 1994, 13, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Lacour, S.; Landini, P. σS-Dependent Gene Expression at the Onset of Stationary Phase in Escherichia coli: Function of σS-Dependent Genes and Identification of Their Promoter Sequences. J. Bacteriol. 2004, 186, 7186–7195. [Google Scholar] [CrossRef] [PubMed]
- Halsey, T.A.; Vazquez-Torres, A.; Gravdahl, D.J.; Fang, F.C.; Libby, S.J. The Ferritin-like Dps Protein Is Required for Salmonella enterica Serovar Typhimurium Oxidative Stress Resistance and Virulence. Infect. Immun. 2004, 72, 1155–1158. [Google Scholar] [CrossRef]
- Chen, C.Y.; Eckmann, L.; Libby, S.J.; Fang, F.C.; Okamoto, S.; Kagnoff, M.F.; Fierer, J.; Guiney, D.G. Expression of Salmonella typhimurium rpoS and rpoS-Dependent Genes in the Intracellular Environment of Eukaryotic Cells. Infect. Immun. 1996, 64, 4739–4743. [Google Scholar] [CrossRef]
- Ibañez-Ruiz, M.; Robbe-Saule, V.; Hermant, D.; Labrude, S.; Norel, F. Identification of RpoS (Sigma(S))-Regulated Genes in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2000, 182, 5749–5756. [Google Scholar] [CrossRef] [PubMed]
- Visick, J.E.; Clarke, S. RpoS- and OxyR-Independent Induction of HPI Catalase at Stationary Phase in Escherichia coli and Identification of RpoS Mutations in Common Laboratory Strains. J. Bacteriol. 1997, 179, 4158–4163. [Google Scholar] [CrossRef]
- Balaji, B.; O’Connor, K.; Lucas, J.R.; Anderson, J.M.; Csonka, L.N. Timing of Induction of Osmotically Controlled Genes in Salmonella enterica Serovar Typhimurium, Determined with Quantitative Real-Time Reverse Transcription-PCR. Appl. Environ. Microbiol. 2005, 71, 8273–8283. [Google Scholar] [CrossRef]
- Lévi-Meyrueis, C.; Monteil, V.; Sismeiro, O.; Dillies, M.-A.; Monot, M.; Jagla, B.; Coppée, J.-Y.; Dupuy, B.; Norel, F. Expanding the RpoS/σS-Network by RNA Sequencing and Identification of σS-Controlled Small RNAs in Salmonella. PLoS ONE 2014, 9, e96918. [Google Scholar] [CrossRef]
- Hengge-Aronis, R.; Klein, W.; Lange, R.; Rimmele, M.; Boos, W. Trehalose Synthesis Genes Are Controlled by the Putative Sigma Factor Encoded by RpoS and Are Involved in Stationary-Phase Thermotolerance in Escherichia coli. J. Bacteriol. 1991, 173, 7918–7924. [Google Scholar] [CrossRef] [PubMed]
- Bearson, B.L.; Wilson, L.; Foster, J.W. A Low PH-Inducible, PhoPQ-Dependent Acid Tolerance Response Protects Salmonella typhimurium against Inorganic Acid Stress. J. Bacteriol. 1998, 180, 2409–2417. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.J.; Ramachandran, V.K.; Shearer, N.; Thompson, A. Transcriptional and Post-Transcriptional Modulation of SPI1 and SPI2 Expression by PpGpp, RpoS and DksA in Salmonella enterica Sv Typhimurium. PLoS ONE 2015, 10, e0127523. [Google Scholar] [CrossRef]
- Velásquez, J.C.; Hidalgo, A.A.; Villagra, N.; Santiviago, C.A.; Mora, G.C.; Fuentes, J.A. SPI-9 of Salmonella enterica Serovar Typhi Is Constituted by an Operon Positively Regulated by RpoS and Contributes to Adherence to Epithelial Cells in Culture. Microbiology 2016, 162, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, C.A.; Curtiss, R. Role of Sigma Factor RpoS in Initial Stages of Salmonella typhimurium Infection. Infect. Immun. 1997, 65, 1814–1823. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.A.; Doyle, T.J.; Gulig, P.A. Exponential-Phase Expression of SpvA of the Salmonella typhimurium Virulence Plasmid: Induction in Intracellular Salts Medium and Intracellularly in Mice and Cultured Mammalian Cells. Microbiology 1997, 143, 3827–3839. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Matlock, B.C.; Heffernan, B.J.; Maloy, S.R. Genomic Analysis and Growth-Phase-Dependent Regulation of the SEF14 Fimbriae of Salmonella enterica Serovar EnteritidisThe GenBank Accession Number for the Sequence Reported in This Paper is AF239978. Microbiology 2001, 147, 2705–2715. [Google Scholar] [CrossRef][Green Version]
- Fang, F.C.; Libby, S.J.; Buchmeier, N.A.; Loewen, P.C.; Switala, J.; Harwood, J.; Guiney, D.G. The Alternative Sigma Factor KatF (RpoS) Regulates Salmonella Virulence. Proc. Natl. Acad. Sci. USA 1992, 89, 11978–11982. [Google Scholar] [CrossRef] [PubMed]
- Gulig, P.A.; Danbara, H.; Guiney, D.G.; Lax, A.J.; Norel, F.; Rhen, M. Molecular Analysis of Spv Virulence Genes of the Salmonella Virulence Plasmids. Mol. Microbiol. 1993, 7, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Guiney, D.G.; Fierer, J. The Role of the Spv Genes in Salmonella Pathogenesis. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef]
- Kowarz, L.; Coynault, C.; Robbe-Saule, V.; Norel, F. The Salmonella typhimurium KatF (RpoS) Gene: Cloning, Nucleotide Sequence, and Regulation of SpvR and SpvABCD Virulence Plasmid Genes. J. Bacteriol. 1994, 176, 6852–6860. [Google Scholar] [CrossRef] [PubMed]
- Wilmes-Riesenberg, M.R.; Foster, J.W.; Curtiss, R. An Altered RpoS Allele Contributes to the Avirulence of Salmonella typhimurium LT2. Infect. Immun. 1997, 65, 203–210. [Google Scholar] [CrossRef]
- Robbe-Saule, V.; Algorta, G.; Rouilhac, I.; Norel, F. Characterization of the RpoS Status of Clinical Isolates of Salmonella enterica. Appl. Environ. Microbiol. 2003, 69, 4352–4358. [Google Scholar] [CrossRef]
- Krogfelt, K.A.; Hjulgaard, M.; Sørensen, K.; Cohen, P.S.; Givskov, M. RpoS Gene Function Is a Disadvantage for Escherichia coli BJ4 during Competitive Colonization of the Mouse Large Intestine. Infect. Immun. 2000, 68, 2518–2524. [Google Scholar] [CrossRef][Green Version]
- Dong, T.; Schellhorn, H.E. Role of RpoS in Virulence of Pathogens. Infect. Immun. 2010, 78, 887–897. [Google Scholar] [CrossRef]
- Robbe-Saule, V.; Jaumouillé, V.; Prévost, M.-C.; Guadagnini, S.; Talhouarne, C.; Mathout, H.; Kolb, A.; Norel, F. Crl Activates Transcription Initiation of RpoS-Regulated Genes Involved in the Multicellular Behavior of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2006, 188, 3983–3994. [Google Scholar] [CrossRef]
- Amar, A.; Pezzoni, M.; Pizarro, R.A.; Costa, C.S. New Envelope Stress Factors Involved in σE Activation and Conditional Lethality of RpoE Mutations in Salmonella enterica. Microbiol. Read. Engl. 2018, 164, 1293–1307. [Google Scholar] [CrossRef]
- Rowley, G.; Spector, M.; Kormanec, J.; Roberts, M. Pushing the Envelope: Extracytoplasmic Stress Responses in Bacterial Pathogens. Nat. Rev. Microbiol. 2006, 4, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Bang, I.-S.; Velayudhan, J.; Karlinsey, J.; Papenfort, K.; Vogel, J.; Fang, F.C. Acid Stress Activation of the Sigma(E) Stress Response in Salmonella enterica Serovar Typhimurium. Mol. Microbiol. 2009, 71, 1228–1238. [Google Scholar] [CrossRef]
- Hews, C.L.; Cho, T.; Rowley, G.; Raivio, T.L. Maintaining Integrity under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 313. [Google Scholar] [CrossRef] [PubMed]
- Humphreys, S.; Stevenson, A.; Bacon, A.; Weinhardt, A.B.; Roberts, M. The Alternative Sigma Factor, SigmaE, Is Critically Important for the Virulence of Salmonella typhimurium. Infect. Immun. 1999, 67, 1560–1568. [Google Scholar] [CrossRef]
- Testerman, T.L.; Vazquez-Torres, A.; Xu, Y.; Jones-Carson, J.; Libby, S.J.; Fang, F.C. The Alternative Sigma Factor SigmaE Controls Antioxidant Defences Required for Salmonella Virulence and Stationary-Phase Survival. Mol. Microbiol. 2002, 43, 771–782. [Google Scholar] [CrossRef]
- Crouch, M.-L.; Becker, L.A.; Bang, I.-S.; Tanabe, H.; Ouellette, A.J.; Fang, F.C. The Alternative Sigma Factor Sigma Is Required for Resistance of Salmonella enterica Serovar Typhimurium to Anti-Microbial Peptides. Mol. Microbiol. 2005, 56, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Osborne, S.E.; Coombes, B.K. RpoE Fine Tunes Expression of a Subset of SsrB-Regulated Virulence Factors in Salmonella enterica Serovar Typhimurium. BMC Microbiol. 2009, 9, 45. [Google Scholar] [CrossRef]
- Li, J.; Overall, C.C.; Nakayasu, E.S.; Kidwai, A.S.; Jones, M.B.; Johnson, R.C.; Nguyen, N.T.; McDermott, J.E.; Ansong, C.; Heffron, F.; et al. Analysis of the Salmonella Regulatory Network Suggests Involvement of SsrB and H-NS in σE-Regulated SPI-2 Gene Expression. Front. Microbiol. 2015, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Sydenham, M.; Douce, G.; Bowe, F.; Ahmed, S.; Chatfield, S.; Dougan, G. Salmonella enterica Serovar Typhimurium SurA Mutants Are Attenuated and Effective Live Oral Vaccines. Infect. Immun. 2000, 68, 1109–1115. [Google Scholar] [CrossRef]
- Humphreys, S.; Rowley, G.; Stevenson, A.; Kenyon, W.J.; Spector, M.P.; Roberts, M. Role of Periplasmic Peptidylprolyl Isomerases in Salmonella enterica Serovar Typhimurium Virulence. Infect. Immun. 2003, 71, 5386–5388. [Google Scholar] [CrossRef]
- Rowley, G.; Skovierova, H.; Stevenson, A.; Rezuchova, B.; Homerova, D.; Lewis, C.; Sherry, A.; Kormanec, J.; Roberts, M. The Periplasmic Chaperone Skp Is Required for Successful Salmonella typhimurium Infection in a Murine Typhoid Model. Microbiol. Read. Engl. 2011, 157, 848–858. [Google Scholar] [CrossRef]
- Srikumar, S.; Kröger, C.; Hébrard, M.; Colgan, A.; Owen, S.V.; Sivasankaran, S.K.; Cameron, A.D.S.; Hokamp, K.; Hinton, J.C.D. RNA-Seq Brings New Insights to the Intra-Macrophage Transcriptome of Salmonella typhimurium. PLoS Pathog. 2015, 11, e1005262. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhang, H.; Zheng, Y.; Li, A.; Wang, M.; Zhou, H.; Zhu, X.; Schneider, Z.; Chen, L.; Kreiswirth, B.N.; et al. RpoE Is a Putative Antibiotic Resistance Regulator of Salmonella Enteric Serovar Typhi. Curr. Microbiol. 2016, 72, 457–464. [Google Scholar] [CrossRef]
- Zhang, H.; Jia, Y.; Xie, X.; Wang, M.; Zheng, Y.; Xu, S.; Zhang, W.; Wang, Q.; Huang, X.; Du, H. RpoE Promotes Invasion and Intracellular Survival by Regulating SPI-1 and SPI-2 in Salmonella enterica Serovar Typhi. Future Microbiol. 2016, 11, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, S.; Müller, V.S.; Hering, N.A.; Mollenkopf, H.; Becker, D.; Heroven, A.K.; Dersch, P.; Pohlmann, A.; Tedin, K.; Porwollik, S.; et al. Contribution of the Cpx Envelope Stress System to Metabolism and Virulence Regulation in Salmonella enterica Serovar Typhimurium. PLoS ONE 2019, 14, e0211584. [Google Scholar] [CrossRef]
- Humphreys, S.; Rowley, G.; Stevenson, A.; Anjum, M.F.; Woodward, M.J.; Gilbert, S.; Kormanec, J.; Roberts, M. Role of the Two-Component Regulator CpxAR in the Virulence of Salmonella enterica Serotype Typhimurium. Infect. Immun. 2004, 72, 4654–4661. [Google Scholar] [CrossRef] [PubMed]
- Shetty, D.; Abrahante, J.E.; Chekabab, S.M.; Wu, X.; Korber, D.R.; Vidovic, S. Role of CpxR in Biofilm Development: Expression of Key Fimbrial, O-Antigen and Virulence Operons of Salmonella Enteritidis. Int. J. Mol. Sci. 2019, 20, 5146. [Google Scholar] [CrossRef] [PubMed]
- Booth, I.R.; Stratford, M. Acidulants and low pH. In Food Preservatives; Russell, N.J., Gould, G.W., Eds.; Springer US: Boston, MA, USA, 2003; pp. 25–47. ISBN 978-1-4757-1006-9. [Google Scholar]
- Álvarez-Ordóñez, A.; Begley, M.; Prieto, M.; Messens, W.; López, M.; Bernardo, A.; Hill, C. Salmonella Spp. Survival Strategies within the Host Gastrointestinal Tract. Microbiology 2011, 157, 3268–3281. [Google Scholar] [CrossRef]
- Hall, H.K.; Foster, J.W. The Role of Fur in the Acid Tolerance Response of Salmonella typhimurium Is Physiologically and Genetically Separable from Its Role in Iron Acquisition. J. Bacteriol. 1996, 178, 5683–5691. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.W. Microbial responses to acid stress. In Bacterial Stress Responses, 2nd ed.; Storz, G., Hengge, R., Eds.; ASM Press: Washington, DC, USA, 2011; pp. 99–115. ISBN 978-1-55581-684-1. [Google Scholar]
- Curtiss, R.; Wanda, S.-Y.; Gunn, B.M.; Zhang, X.; Tinge, S.A.; Ananthnarayan, V.; Mo, H.; Wang, S.; Kong, W. Salmonella enterica Serovar Typhimurium Strains with Regulated Delayed Attenuation in Vivo. Infect. Immun. 2009, 77, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Karasova, D.; Sebkova, A.; Vrbas, V.; Havlickova, H.; Sisak, F.; Rychlik, I. Comparative Analysis of Salmonella enterica Serovar Enteritidis Mutants with a Vaccine Potential. Vaccine 2009, 27, 5265–5270. [Google Scholar] [CrossRef] [PubMed]
- Ellermeier, J.R.; Slauch, J.M. Fur Regulates Expression of the Salmonella Pathogenicity Island 1 Type III Secretion System through HilD. J. Bacteriol. 2008, 190, 476–486. [Google Scholar] [CrossRef] [PubMed]
- Prost, L.R.; Daley, M.E.; Le Sage, V.; Bader, M.W.; Le Moual, H.; Klevit, R.E.; Miller, S.I. Activation of the Bacterial Sensor Kinase PhoQ by Acidic PH. Mol. Cell 2007, 26, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Alpuche Aranda, C.M.; Swanson, J.A.; Loomis, W.P.; Miller, S.I. Salmonella typhimurium Activates Virulence Gene Transcription within Acidified Macrophage Phagosomes. Proc. Natl. Acad. Sci. USA 1992, 89, 10079–10083. [Google Scholar] [CrossRef]
- Merighi, M.; Ellermeier, C.D.; Slauch, J.M.; Gunn, J.S. Resolvase-in Vivo Expression Technology Analysis of the Salmonella enterica Serovar Typhimurium PhoP and PmrA Regulons in BALB/c Mice. J. Bacteriol. 2005, 187, 7407–7416. [Google Scholar] [CrossRef]
- Martin-Orozco, N.; Touret, N.; Zaharik, M.L.; Park, E.; Kopelman, R.; Miller, S.; Finlay, B.B.; Gros, P.; Grinstein, S. Visualization of Vacuolar Acidification-Induced Transcription of Genes of Pathogens inside Macrophages. Mol. Biol. Cell 2006, 17, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.I.; Kukral, A.M.; Mekalanos, J.J. A Two-Component Regulatory System (PhoP PhoQ) Controls Salmonella typhimurium Virulence. Proc. Natl. Acad. Sci. USA 1989, 86, 5054–5058. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-Y.; Cho, S.-A.; Lee, I.-S.; Park, J.-H.; Seok, S.-H.; Baek, M.-W.; Kim, D.-J.; Lee, S.-H.; Hur, S.-J.; Ban, S.-J.; et al. Evaluation of PhoP and RpoS Mutants of Salmonella enterica Serovar Typhi as Attenuated Typhoid Vaccine Candidates: Virulence and Protective Immune Responses in Intranasally Immunized Mice. FEMS Immunol. Med. Microbiol. 2007, 51, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Bernal, G.; Tierrez, A.; Bartolomé, A.; Martínez-Pulgarín, S.; Salguero, F.J.; Orden, J.A.; de la Fuente, R. Salmonella enterica Serovar Choleraesuis Derivatives Harbouring Deletions in rpoS and phoP Regulatory Genes Are Attenuated in Pigs, and Survive and Multiply in Porcine Intestinal Macrophages and Fibroblasts, Respectively. Vet. Microbiol. 2008, 130, 298–311. [Google Scholar] [CrossRef]
- Bang, I.S.; Kim, B.H.; Foster, J.W.; Park, Y.K. OmpR Regulates the Stationary-Phase Acid Tolerance Response of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2000, 182, 2245–2252. [Google Scholar] [CrossRef]
- Bang, I.S.; Audia, J.P.; Park, Y.K.; Foster, J.W. Autoinduction of the OmpR Response Regulator by Acid Shock and Control of the Salmonella enterica Acid Tolerance Response. Mol. Microbiol. 2002, 44, 1235–1250. [Google Scholar] [CrossRef]
- Zhao, B.; Houry, W.A. Acid Stress Response in Enteropathogenic Gammaproteobacteria: An Aptitude for Survival. Biochem. Cell Biol. Biochim. Biol. Cell. 2010, 88, 301–314. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.K.; Detweiler, C.S.; Falkow, S. OmpR Regulates the Two-Component System SsrA-SsrB in Salmonella Pathogenicity Island 2. J. Bacteriol. 2000, 182, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.C.; Falkow, S. Delineation of Upstream Signaling Events in the Salmonella Pathogenicity Island 2 Transcriptional Activation Pathway. J. Bacteriol. 2004, 186, 4694–4704. [Google Scholar] [CrossRef] [PubMed]
- Fass, E.; Groisman, E.A. Control of Salmonella Pathogenicity Island-2 Gene Expression. Curr. Opin. Microbiol. 2009, 12, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Deiwick, J.; Nikolaus, T.; Erdogan, S.; Hensel, M. Environmental Regulation of Salmonella Pathogenicity Island 2 Gene Expression. Mol. Microbiol. 1999, 31, 1759–1773. [Google Scholar] [CrossRef]
- Tu, X.; Latifi, T.; Bougdour, A.; Gottesman, S.; Groisman, E.A. The PhoP/PhoQ Two-Component System Stabilizes the Alternative Sigma Factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. USA 2006, 103, 13503–13508. [Google Scholar] [CrossRef]
- Ryan, D.; Pati, N.B.; Ojha, U.K.; Padhi, C.; Ray, S.; Jaiswal, S.; Singh, G.P.; Mannala, G.K.; Schultze, T.; Chakraborty, T.; et al. Global Transcriptome and Mutagenic Analyses of the Acid Tolerance Response of Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2015, 81, 8054–8065. [Google Scholar] [CrossRef]
- Kim, S.; Ryu, K.; Biswas, D.; Ahn, J. Survival, Prophage Induction, and Invasive Properties of Lysogenic Salmonella typhimurium Exposed to Simulated Gastrointestinal Conditions. Arch. Microbiol. 2014, 196, 655–659. [Google Scholar] [CrossRef]
- Coombes, B.K.; Brown, N.F.; Valdez, Y.; Brumell, J.H.; Finlay, B.B. Expression and Secretion of Salmonella Pathogenicity Island-2 Virulence Genes in Response to Acidification Exhibit Differential Requirements of a Functional Type III Secretion Apparatus and SsaL. J. Biol. Chem. 2004, 279, 49804–49815. [Google Scholar] [CrossRef] [PubMed]
- Jia, K.; Wang, G.; Liang, L.; Wang, M.; Wang, H.; Xu, X. Preliminary Transcriptome Analysis of Mature Biofilm and Planktonic Cells of Salmonella Enteritidis Exposure to Acid Stress. Front. Microbiol. 2017, 8, 1861. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, D.; McCabe, E.M.; McCusker, M.P.; Martins, M.; Fanning, S.; Duffy, G. Acid Environments Affect Biofilm Formation and Gene Expression in Isolates of Salmonella enterica Typhimurium DT104. Int. J. Food Microbiol. 2015, 206, 7–16. [Google Scholar] [CrossRef]
- Kitamoto, S.; Nagao-Kitamoto, H.; Kuffa, P.; Kamada, N. Regulation of Virulence: The Rise and Fall of Gastrointestinal Pathogens. J. Gastroenterol. 2016, 51, 195–205. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Hocking, P.M.; Jørgensen, F.; Mattick, K.; Leach, S.; Humphrey, T.J. Effects of Repeated Cycles of Acid Challenge and Growth on the Phenotype and Virulence of Salmonella enterica. J. Appl. Microbiol. 2008, 105, 1640–1648. [Google Scholar] [CrossRef]
- Grogan, D.W.; Cronan, J.E. Cyclopropane Ring Formation in Membrane Lipids of Bacteria. Microbiol. Mol. Biol. Rev. 1997, 61, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Y.; Cronan, J.E. The Growth Phase-Dependent Synthesis of Cyclopropane Fatty Acids in Escherichia coli Is the Result of an RpoS(KatF)-Dependent Promoter plus Enzyme Instability. Mol. Microbiol. 1994, 11, 1009–1017. [Google Scholar] [CrossRef]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knøchel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela, S.; Tresse, O. The Response of Foodborne Pathogens to Osmotic and Desiccation Stresses in the Food Chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef]
- Rychlik, I.; Barrow, P.A. Salmonella Stress Management and Its Relevance to Behaviour during Intestinal Colonisation and Infection. FEMS Microbiol. Rev. 2005, 29, 1021–1040. [Google Scholar] [CrossRef]
- Finn, S.; Händler, K.; Condell, O.; Colgan, A.; Cooney, S.; McClure, P.; Amézquita, A.; Hinton, J.C.D.; Fanning, S. ProP Is Required for the Survival of Desiccated Salmonella enterica Serovar Typhimurium Cells on a Stainless Steel Surface. Appl. Environ. Microbiol. 2013, 79, 4376–4384. [Google Scholar] [CrossRef]
- Deng, X.; Li, Z.; Zhang, W. Transcriptome Sequencing of Salmonella enterica Serovar Enteritidis under Desiccation and Starvation Stress in Peanut Oil. Food Microbiol. 2012, 30, 311–315. [Google Scholar] [CrossRef]
- Gruzdev, N.; McClelland, M.; Porwollik, S.; Ofaim, S.; Pinto, R.; Saldinger-Sela, S. Global Transcriptional Analysis of Dehydrated Salmonella enterica Serovar Typhimurium. Appl. Environ. Microbiol. 2012, 78, 7866–7875. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xu, H.; Sun, X.; Ohkusu, K.; Kawamura, Y.; Ezaki, T. Genome-Wide Scan of the Gene Expression Kinetics of Salmonella enterica Serovar Typhi during Hyperosmotic Stress. Int. J. Mol. Sci. 2007, 8, 116–135. [Google Scholar] [CrossRef]
- Li, H.; Bhaskara, A.; Megalis, C.; Tortorello, M.L. Transcriptomic Analysis of Salmonella Desiccation Resistance. Foodborne Pathog. Dis. 2012, 9, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Kröger, C.; Colgan, A.; Srikumar, S.; Händler, K.; Sivasankaran, S.K.; Hammarlöf, D.L.; Canals, R.; Grissom, J.E.; Conway, T.; Hokamp, K.; et al. An Infection-Relevant Transcriptomic Compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 2013, 14, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Lang, E.; Guyot, S.; Alvarez-Martin, P.; Perrier-Cornet, J.-M.; Gervais, P. Caco-2 Invasion by Cronobacter Sakazakii and Salmonella enterica Exposed to Drying and Heat Treatments in Dried State in Milk Powder. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Yoon, H.; Park, B.-Y.; Oh, M.-H.; Choi, K.-H.; Yoon, Y. Effect of NaCl on Heat Resistance, Antibiotic Susceptibility, and Caco-2 Cell Invasion of Salmonella. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Finn, S.; Condell, O.; McClure, P.; Amézquita, A.; Fanning, S. Mechanisms of Survival, Responses and Sources of Salmonella in Low-Moisture Environments. Front. Microbiol. 2013, 4, 331. [Google Scholar] [CrossRef] [PubMed]
- Marcén, M.; Cebrián, G.; Ruiz-Artiga, V.; Condón, S.; Mañas, P. Cellular Events Involved in E. coli Cells Inactivation by Several Agents for Food Preservation: A Comparative Study. Food Microbiol. 2019, 84, 103246. [Google Scholar] [CrossRef] [PubMed]
- Rhen, M. Salmonella and Reactive Oxygen Species: A Love-Hate Relationship. J. Innate Immun. 2019, 11, 216–226. [Google Scholar] [CrossRef]
- Frick, K.; Schulte, M.; Friedrich, T. Reactive Oxygen Species Production by Escherichia coli Respiratory Complex I. Biochemistry 2015, 54, 2799–2801. [Google Scholar] [CrossRef]
- Van der Heijden, J.; Reynolds, L.A.; Deng, W.; Mills, A.; Scholz, R.; Imami, K.; Foster, L.J.; Duong, F.; Finlay, B.B. Salmonella Rapidly Regulates Membrane Permeability to Survive Oxidative Stress. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Robbiani, R.; Walker, A.W.; Westendorf, A.M.; Barthel, M.; Kremer, M.; Chaffron, S.; Macpherson, A.J.; Buer, J.; Parkhill, J.; et al. Salmonella enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota. PLoS Biol. 2007, 5, e244. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Thiennimitr, P.; Winter, M.G.; Butler, B.P.; Huseby, D.L.; Crawford, R.W.; Russell, J.M.; Bevins, C.L.; Adams, L.G.; Tsolis, R.M.; et al. Gut Inflammation Provides a Respiratory Electron Acceptor for Salmonella. Nature 2010, 467, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Bäumler, A.J. A Breathtaking Feat. Gut Microbes 2011, 2, 58–60. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Craig, M.; Imlay, J.A.; Slauch, J.M. Differences in Enzymatic Properties Allow SodCI but Not SodCII to Contribute to Virulence in Salmonella enterica Serovar Typhimurium Strain 14028. J. Bacteriol. 2004, 186, 5230–5238. [Google Scholar] [CrossRef]
- Eriksson, S.; Lucchini, S.; Thompson, A.; Rhen, M.; Hinton, J.C.D. Unravelling the Biology of Macrophage Infection by Gene Expression Profiling of Intracellular Salmonella enterica. Mol. Microbiol. 2003, 47, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Golubeva, Y.A.; Slauch, J.M. Salmonella enterica Serovar Typhimurium Periplasmic Superoxide Dismutase SodCI Is a Member of the PhoPQ Regulon and Is Induced in Macrophages. J. Bacteriol. 2006, 188, 7853–7861. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Spiro, S. Sensing and responding to reactive oxygen and nitrogen species. In Bacterial Stress Responses, 2nd ed.; Storz, G., Hengge, R., Eds.; ASM Press: Washington, DC, USA, 2011; pp. 157–173. ISBN 978-1-68367-121-3. [Google Scholar]
- Pomposiello, P.J.; Demple, B. Identification of SoxS-Regulated Genes in Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2000, 182, 23–29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Taylor, P.D.; Inchley, C.J.; Gallagher, M.P. The Salmonella typhimurium AhpC Polypeptide Is Not Essential for Virulence in BALB/c Mice but Is Recognized as an Antigen during Infection. Infect. Immun. 1998, 66, 3208–3217. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.R.; Morgan, E.; Peters, S.E.; Pleasance, S.J.; Hudson, D.L.; Davies, H.M.; Wang, J.; van Diemen, P.M.; Buckley, A.M.; Bowen, A.J.; et al. Comprehensive Assignment of Roles for Salmonella typhimurium Genes in Intestinal Colonization of Food-Producing Animals. PLoS Genet. 2013, 9, e1003456. [Google Scholar] [CrossRef]
- Karlinsey, J.E.; Bang, I.-S.; Becker, L.A.; Frawley, E.R.; Porwollik, S.; Robbins, H.F.; Thomas, V.C.; Urbano, R.; McClelland, M.; Fang, F.C. The NsrR Regulon in Nitrosative Stress Resistance of Salmonella enterica Serovar Typhimurium. Mol. Microbiol. 2012, 85, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Phillippy, A.M.; Deng, K.; Rui, X.; Li, Z.; Tortorello, M.L.; Zhang, W. Transcriptomic Responses of Salmonella enterica Serovars Enteritidis and Typhimurium to Chlorine-Based Oxidative Stress. Appl. Environ. Microbiol. 2010, 76, 5013–5024. [Google Scholar] [CrossRef]
- Cadena, M.; Froenicke, L.; Britton, M.; Settles, M.L.; Durbin-Johnson, B.; Kumimoto, E.; Gallardo, R.A.; Ferreiro, A.; Chylkova, T.; Zhou, H.; et al. Transcriptome Analysis of Salmonella Heidelberg after Exposure to Cetylpyridinium Chloride, Acidified Calcium Hypochlorite, and Peroxyacetic Acid. J. Food Prot. 2019, 82, 109–119. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Randall, L.P.; Webber, M.; Piddock, L.J.V.; Humphrey, T.J.; Woodward, M.J.; Coldham, N.G. Phenotypic and Proteomic Characterization of Multiply Antibiotic-Resistant Variants of Salmonella enterica Serovar Typhimurium Selected Following Exposure to Disinfectants. Appl. Environ. Microbiol. 2008, 74, 1508–1516. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Webber, M.A.; Jorgensen, F.; Woodward, M.J.; Piddock, L.J.V.; Humphrey, T.J. Prolonged Treatment of Salmonella enterica Serovar Typhimurium with Commercial Disinfectants Selects for Multiple Antibiotic Resistance, Increased Efflux and Reduced Invasiveness. J. Antimicrob. Chemother. 2007, 60, 947–955. [Google Scholar] [CrossRef]
- O’Neal, C.R.; Gabriel, W.M.; Turk, A.K.; Libby, S.J.; Fang, F.C.; Spector, M.P. RpoS Is Necessary for Both the Positive and Negative Regulation of Starvation Survival Genes during Phosphate, Carbon, and Nitrogen Starvation in Salmonella typhimurium. J. Bacteriol. 1994, 176, 4610–4616. [Google Scholar] [CrossRef] [PubMed]
- Potts, A.H.; Guo, Y.; Ahmer, B.M.M.; Romeo, T. Role of CsrA in Stress Responses and Metabolism Important for Salmonella Virulence Revealed by Integrated Transcriptomics. PLoS ONE 2019, 14, e211430. [Google Scholar] [CrossRef] [PubMed]
- Spector, M.P.; Cubitt, C.L. Starvation-Inducible Loci of Salmonella typhimurium: Regulation and Roles in Starvation-Survival. Mol. Microbiol. 1992, 6, 1467–1476. [Google Scholar] [CrossRef]
- Henard, C.A.; Vázquez-Torres, A. DksA-Dependent Resistance of Salmonella enterica Serovar Typhimurium against the Antimicrobial Activity of Inducible Nitric Oxide Synthase. Infect. Immun. 2012, 80, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Henard, C.A.; Tapscott, T.; Crawford, M.A.; Husain, M.; Doulias, P.-T.; Porwollik, S.; Liu, L.; McClelland, M.; Ischiropoulos, H.; Vázquez-Torres, A. The 4-Cysteine Zinc-Finger Motif of the RNA Polymerase Regulator DksA Serves as a Thiol Switch for Sensing Oxidative and Nitrosative Stress. Mol. Microbiol. 2014, 91, 790–804. [Google Scholar] [CrossRef]
- Fitzsimmons, L.F.; Liu, L.; Kant, S.; Kim, J.-S.; Till, J.K.; Jones-Carson, J.; Porwollik, S.; McClelland, M.; Vazquez-Torres, A. SpoT Induces Intracellular Salmonella Virulence Programs in the Phagosome. mBio 2020, 11. [Google Scholar] [CrossRef]
- Cashell, M.; Rudd, K. The stringent response. In Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biolog; Neidhardt, F.C., Ingraham, J.L., Low, K.B., Magasanik, B., Schaechter, M., Umbarger, H.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1987; Volume 63, pp. 1410–1438. [Google Scholar]
- Kenyon, W.J.; Sayers, D.G.; Humphreys, S.; Roberts, M.; Spector, M.P. The Starvation-Stress Response of Salmonella enterica Serovar Typhimurium Requires Sigma(E)-, but Not CpxR-Regulated Extracytoplasmic Functions. Microbiology 2002, 148, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Cerdá, J.; Tedin, K. The Bacterial Signal Molecule, PpGpp, Regulates Salmonella Virulence Gene Expression. Mol. Microbiol. 2004, 52, 1827–1844. [Google Scholar] [CrossRef] [PubMed]
- Tapscott, T.; Kim, J.-S.; Crawford, M.A.; Fitzsimmons, L.; Liu, L.; Jones-Carson, J.; Vázquez-Torres, A. Guanosine Tetraphosphate Relieves the Negative Regulation of Salmonella Pathogenicity Island-2 Gene Transcription Exerted by the AT-Rich SsrA Discriminator Region. Sci. Rep. 2018, 8, 9465. [Google Scholar] [CrossRef] [PubMed]
- Altier, C.; Suyemoto, M.; Lawhon, S.D. Regulation of Salmonella enterica Serovar Typhimurium Invasion Genes by CsrA. Infect. Immun. 2000, 68, 6790–6797. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.C.; Yakhnin, H.; Camacho, M.I.; Georgellis, D.; Babitzke, P.; Puente, J.L.; Bustamante, V.H. Integration of a Complex Regulatory Cascade Involving the SirA/BarA and Csr Global Regulatory Systems That Controls Expression of the Salmonella SPI-1 and SPI-2 Virulence Regulons through HilD. Mol. Microbiol. 2011, 80, 1637–1656. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.M.; Bosecker, B.A.; Curtiss, R. Characterization and Protective Properties of Attenuated Mutants of Salmonella Choleraesuis. Infect. Immun. 1992, 60, 4881–4890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kelly, S.M.; Bollen, W.S.; Curtiss, R. Characterization and Immunogenicity of Salmonella typhimurium SL1344 and UK-1 Delta Crp and Delta Cdt Deletion Mutants. Infect. Immun. 1997, 65, 5381–5387. [Google Scholar] [CrossRef] [PubMed]
- Rosu, V.; Chadfield, M.S.; Santona, A.; Christensen, J.P.; Thomsen, L.E.; Rubino, S.; Olsen, J.E. Effects of Crp Deletion in Salmonella enterica Serotype Gallinarum. Acta Vet. Scand. 2007, 49, 14. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Yin, J.; Kang, X.; Geng, S.; Hu, M.; Pan, Z.; Jiao, X. Safety and Protective Efficacy of a SpiC and Crp Deletion Mutant of Salmonella Gallinarum as a Live Attenuated Vaccine for Fowl Typhoid. Res. Vet. Sci. 2016, 107, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Moreno, M.; Wilmes-Riesenberg, M.; Curtiss, R.; Foster, J.W. Effects of DksA and ClpP Protease on Sigma S Production and Virulence in Salmonella typhimurium. Mol. Microbiol. 1999, 34, 112–123. [Google Scholar] [CrossRef]
- Khajanchi, B.K.; Xu, J.; Grim, C.J.; Ottesen, A.R.; Ramachandran, P.; Foley, S.L. Global Transcriptomic Analyses of Salmonella enterica in Iron-Depleted and Iron-Rich Growth Conditions. BMC Genom. 2019, 20, 490. [Google Scholar] [CrossRef] [PubMed]
- Dostal, A.; Gagnon, M.; Chassard, C.; Zimmermann, M.B.; O’Mahony, L.; Lacroix, C. Salmonella Adhesion, Invasion and Cellular Immune Responses Are Differentially Affected by Iron Concentrations in a Combined in Vitro Gut Fermentation-Cell Model. PLoS ONE 2014, 9, e93549. [Google Scholar] [CrossRef]
- Gogoi, M.; Shreenivas, M.M.; Chakravortty, D. Hoodwinking the Big-Eater to Prosper: The Salmonella-Macrophage Paradigm. J. Innate Immun. 2019, 11, 289–299. [Google Scholar] [CrossRef]
- Tan, Z.; Chekabab, S.M.; Yu, H.; Yin, X.; Diarra, M.S.; Yang, C.; Gong, J. Growth and Virulence of Salmonella typhimurium Mutants Deficient in Iron Uptake. ACS Omega 2019, 4, 13218–13230. [Google Scholar] [CrossRef]
- Chart, H.; Rowe, B. Iron Restriction and the Growth of Salmonella Enteritidis. Epidemiol. Infect. 1993, 110, 41–47. [Google Scholar] [CrossRef][Green Version]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial Iron Homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Bjarnason, J.; Southward, C.M.; Surette, M.G. Genomic Profiling of Iron-Responsive Genes in Salmonella enterica Serovar Typhimurium by High-Throughput Screening of a Random Promoter Library. J. Bacteriol. 2003, 185, 4973–4982. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vazquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef]
- Zaharik, M.L.; Vallance, B.A.; Puente, J.L.; Gros, P.; Finlay, B.B. Host–Pathogen Interactions: Host Resistance Factor Nramp1 up-Regulates the Expression of Salmonella Pathogenicity Island-2 Virulence Genes. Proc. Natl. Acad. Sci. USA 2002, 99, 15705–15710. [Google Scholar] [CrossRef] [PubMed]
- Erkmen, O. Modified-atmosphere storage of foods. In Progress in Food Preservation; Bhat, R., Alias, A.K., Paliyath, G., Eds.; John Wiley & Sons Ltd.: Oxford, UK, 2012; pp. 49–66. ISBN 978-1-119-96204-5. [Google Scholar]
- Encheva, V.; Shah, H.N.; Gharbia, S.E. Proteomic Analysis of the Adaptive Response of Salmonella enterica Serovar Typhimurium to Growth under Anaerobic Conditions. Microbiology 2009, 155, 2429–2441. [Google Scholar] [CrossRef]
- Ševčík, M.; Šebková, A.; Volf, J.; Rychlík, I. Transcription of ArcA and RpoS during Growth of Salmonella typhimurium under Aerobic and Microaerobic Conditions. Microbiology 2001, 147, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Fink, R.C.; Evans, M.R.; Porwollik, S.; Vazquez-Torres, A.; Jones-Carson, J.; Troxell, B.; Libby, S.J.; McClelland, M.; Hassan, H.M. FNR Is a Global Regulator of Virulence and Anaerobic Metabolism in Salmonella enterica Serovar Typhimurium (ATCC 14028s). J. Bacteriol. 2007, 189, 2262–2273. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Killoran, P.B.; Fang, F.C.; Riley, L.W. The Global Regulator ArcA Controls Resistance to Reactive Nitrogen and Oxygen Intermediates in Salmonella enterica Serovar Enteritidis. Infect. Immun. 2002, 70, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.R.; Fink, R.C.; Vazquez-Torres, A.; Porwollik, S.; Jones-Carson, J.; McClelland, M.; Hassan, H.M. Analysis of the ArcA Regulon in Anaerobically Grown Salmonella enterica Sv. Typhimurium. BMC Microbiol. 2011, 11, 58. [Google Scholar] [CrossRef]
- Lee, C.A.; Falkow, S. The Ability of Salmonella to Enter Mammalian Cells Is Affected by Bacterial Growth State. Proc. Natl. Acad. Sci. USA 1990, 87, 4304–4308. [Google Scholar] [CrossRef]
- Ernst, R.K.; Dombroski, D.M.; Merrick, J.M. Anaerobiosis, Type 1 Fimbriae, and Growth Phase Are Factors That Affect Invasion of HEp-2 Cells by Salmonella typhimurium. Infect. Immun. 1990, 58, 2014–2016. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Falkow, S. Identification and Characterization of a Salmonella typhimurium Oxygen-Regulated Gene Required for Bacterial Internalization. Infect. Immun. 1994, 62, 3745–3752. [Google Scholar] [CrossRef]
- Singh, R.D.; Khullar, M.; Ganguly, N.K. Role of Anaerobiosis in Virulence of Salmonella Typhimuirium. Mol. Cell. Biochem. 2000, 215, 39–46. [Google Scholar] [CrossRef]
- Gunn, J.S. Mechanisms of Bacterial Resistance and Response to Bile. Microbes Infect. 2000, 2, 907–913. [Google Scholar] [CrossRef]
- Begley, M.; Gahan, C.G.M.; Hill, C. The Interaction between Bacteria and Bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef]
- Merritt, M.E.; Donaldson, J.R. Effect of Bile Salts on the DNA and Membrane Integrity of Enteric Bacteria. J. Med. Microbiol. 2009, 58, 1533–1541. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, F.J.; Cloeckaert, A.; Grépinet, O.; Pinault, C.; Popoff, M.Y.; Waxin, H.; Pardon, P. Salmonella typhimurium AcrB-like Gene: Identification and Role in Resistance to Biliary Salts and Detergents and in Murine Infection. FEMS Microbiol. Lett. 1996, 135, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Van Velkinburgh, J.C.; Gunn, J.S. PhoP-PhoQ-Regulated Loci Are Required for Enhanced Bile Resistance in Salmonella Spp. Infect. Immun. 1999, 67, 1614–1622. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Morales, F.; Prieto, A.I.; Beuzón, C.R.; Holden, D.W.; Casadesús, J. Role for Salmonella enterica Enterobacterial Common Antigen in Bile Resistance and Virulence. J. Bacteriol. 2003, 185, 5328–5332. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.I.; Ramos-Morales, F.; Casadesús, J. Bile-Induced DNA Damage in Salmonella enterica. Genetics 2004, 168, 1787–1794. [Google Scholar] [CrossRef]
- Prouty, A.M.; Brodsky, I.E.; Manos, J.; Belas, R.; Falkow, S.; Gunn, J.S. Transcriptional Regulation of Salmonella enterica Serovar Typhimurium Genes by Bile. FEMS Immunol. Med. Microbiol. 2004, 41, 177–185. [Google Scholar] [CrossRef][Green Version]
- Prouty, A.M.; Gunn, J.S. Salmonella enterica Serovar Typhimurium Invasion Is Repressed in the Presence of Bile. Infect. Immun. 2000, 68, 6763–6769. [Google Scholar] [CrossRef]
- Urdaneta, V.; Hernández, S.B.; Casadesús, J. Mutational and Non Mutational Adaptation of Salmonella enterica to the Gall Bladder. Sci. Rep. 2019, 9, 5203. [Google Scholar] [CrossRef] [PubMed]
- Aryal, M.; Muriana, P.M. Efficacy of Commercial Sanitizers Used in Food Processing Facilities for Inactivation of Listeria monocytogenes, E. coli O157:H7, and Salmonella Biofilms. Foods 2019, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Kautz, M.J.M.; Dvorzhinskiy, A.; Frye, J.G.; Stevenson, N.; Herson, D.S. Pathogenicity of Dodecyltrimethylammonium Chloride-Resistant Salmonella enterica. Appl. Environ. Microbiol. 2013, 79, 2371–2376. [Google Scholar] [CrossRef][Green Version]
- Ricke, S.C.; Dawoud, T.M.; Kim, S.A.; Park, S.H.; Kwon, Y.M. Salmonella Cold Stress Response: Mechanisms and Occurrence in Foods. Adv. Appl. Microbiol. 2018, 104, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Bae, W.; Xia, B.; Inouye, M.; Severinov, K. Escherichia coli CspA-Family RNA Chaperones Are Transcription Antiterminators. Proc. Natl. Acad. Sci. USA 2000, 97, 7784–7789. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.-F.; Kuznedelov, K.; Mushegian, A.; Severinov, K.; Greenblatt, J. The α Subunit of E. coli RNA Polymerase Activates RNA Binding by NusA. Genes Dev. 2000, 14, 2664–2675. [Google Scholar] [CrossRef]
- Van Immerseel, F.; Eeckhaut, V.; Boyen, F.; Pasmans, F.; Haesebrouck, F.; Ducatelle, R. Mutations Influencing Expression of the Salmonella enterica Serovar Enteritidis Pathogenicity Island I Key Regulator HilA. Antonie Van Leeuwenhoek 2008, 94, 455. [Google Scholar] [CrossRef]
- Shah, J.; Desai, P.T.; Weimer, B.C. Genetic Mechanisms Underlying the Pathogenicity of Cold-Stressed Salmonella enterica Serovar Typhimurium in Cultured Intestinal Epithelial Cells. Appl. Environ. Microbiol. 2014, 80, 6943–6953. [Google Scholar] [CrossRef] [PubMed]
- Cebrián, G.; Condón, S.; Mañas, P. Physiology of the Inactivation of Vegetative Bacteria by Thermal Treatments: Mode of Action, Influence of Environmental Factors and Inactivation Kinetics. Foods 2017, 6, 107. [Google Scholar] [CrossRef]
- Sirsat, S.A.; Burkholder, K.M.; Muthaiyan, A.; Dowd, S.E.; Bhunia, A.K.; Ricke, S.C. Effect of Sublethal Heat Stress on Salmonella typhimurium Virulence. J. Appl. Microbiol. 2011, 110, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Guisbert, E.; Yura, T.; Rhodius, V.A.; Gross, C.A. Convergence of Molecular, Modeling, and Systems Approaches for an Understanding of the Escherichia coli Heat Shock Response. Microbiol. Mol. Biol. Rev. 2008, 72, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, G.; Blankschien, M.; Herman, C.; Gross, C.A.; Rhodius, V.A. Regulon and Promoter Analysis of the E. coli Heat-Shock Factor, σ32, Reveals a Multifaceted Cellular Response to Heat Stress. Genes Dev. 2006, 20, 1776–1789. [Google Scholar] [CrossRef] [PubMed]
- Lund, P.A. Microbial Molecular Chaperones. Adv. Microb. Physiol. 2001, 44, 93–140. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, M.; Burgess, R.R. The Global Transcriptional Response of Escherichia coli to Induced σ32 Protein Involves σ32 Regulon Activation Followed by Inactivation and Degradation of σ32 in Vivo. J. Biol. Chem. 2005, 280, 17758–17768. [Google Scholar] [CrossRef]
- Wade, J.T.; Roa, D.C.; Grainger, D.C.; Hurd, D.; Busby, S.J.W.; Struhl, K.; Nudler, E. Extensive Functional Overlap between σ Factors in Escherichia coli. Nat. Struct. Mol. Biol. 2006, 13, 806–814. [Google Scholar] [CrossRef]
- Alba, B.M.; Gross, C.A. Regulation of the Escherichia coli σE-Dependent Envelope Stress Response. Mol. Microbiol. 2004, 52, 613–619. [Google Scholar] [CrossRef]
- Raivio, T.L. MicroReview: Envelope Stress Responses and Gram-Negative Bacterial Pathogenesis. Mol. Microbiol. 2005, 56, 1119–1128. [Google Scholar] [CrossRef]
- Ades, S.E. Regulation by Destruction: Design of the σE Envelope Stress Response. Curr. Opin. Microbiol. 2008, 11, 535–540. [Google Scholar] [CrossRef]
- Ades, S.E. Control of the Alternative Sigma Factor σE in Escherichia coli. Curr. Opin. Microbiol. 2004, 7, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Duguay, A.R.; Silhavy, T.J. Quality Control in the Bacterial Periplasm. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2004, 1694, 121–134. [Google Scholar] [CrossRef]
- Hayden, J.D.; Ades, S.E. The Extracytoplasmic Stress Factor, σE, Is Required to Maintain Cell Envelope Integrity in Escherichia coli. PLoS ONE 2008, 3, e1573. [Google Scholar] [CrossRef] [PubMed]
- Erickson, J.W.; Gross, C.A. Identification of the Sigma E Subunit of Escherichia coli RNA Polymerase: A Second Alternate Sigma Factor Involved in High-Temperature Gene Expression. Genes Dev. 1989, 3, 1462–1471. [Google Scholar] [CrossRef]
- Wang, Q.P.; Kaguni, J.M. A Novel Sigma Factor Is Involved in Expression of the RpoH Gene of Escherichia coli. J. Bacteriol. 1989, 171, 4248–4253. [Google Scholar] [CrossRef] [PubMed]
- Hiratsu, K.; Amemura, M.; Nashimoto, H.; Shinagawa, H.; Makino, K. The RpoE Gene of Escherichia coli, Which Encodes Sigma E, Is Essential for Bacterial Growth at High Temperature. J. Bacteriol. 1995, 177, 2918–2922. [Google Scholar] [CrossRef]
- Peñas, A.D.L.; Connolly, L.; Gross, C.A. SigmaE Is an Essential Sigma Factor in Escherichia coli. J. Bacteriol. 1997, 179, 6862–6864. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.; Raina, S. The Extracytoplasmic Function Sigma Factors: Role and Regulation. Mol. Microbiol. 1998, 28, 1059–1066. [Google Scholar] [CrossRef]
- Bury-Moné, S.; Nomane, Y.; Reymond, N.; Barbet, R.; Jacquet, E.; Imbeaud, S.; Jacq, A.; Bouloc, P. Global Analysis of Extracytoplasmic Stress Signaling in Escherichia coli. PLoS Genet. 2009, 5, e1000651. [Google Scholar] [CrossRef]
- Yang, Y.; Khoo, W.J.; Zheng, Q.; Chung, H.-J.; Yuk, H.-G. Growth Temperature Alters Salmonella Enteritidis Heat/Acid Resistance, Membrane Lipid Composition and Stress/Virulence Related Gene Expression. Int. J. Food Microbiol. 2014, 172, 102–109. [Google Scholar] [CrossRef]
- Dawoud, T.M.; Davis, M.L.; Park, S.H.; Kim, S.A.; Kwon, Y.M.; Jarvis, N.; O’Bryan, C.A.; Shi, Z.; Crandall, P.G.; Ricke, S.C. The Potential Link between Thermal Resistance and Virulence in Salmonella: A Review. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef]
- Katz, C.; Ron, E.Z. Dual Role of FtsH in Regulating Lipopolysaccharide Biosynthesis in Escherichia coli. J. Bacteriol. 2008, 190, 7117–7122. [Google Scholar] [CrossRef] [PubMed]
- Alix, E.; Blanc-Potard, A.-B. Peptide-Assisted Degradation of the Salmonella MgtC Virulence Factor. EMBO J. 2008, 27, 546–557. [Google Scholar] [CrossRef]
- Horne, S.M.; Kottom, T.J.; Nolan, L.K.; Young, K.D. Decreased Intracellular Survival of an FkpA Mutant of Salmonella typhimurium Copenhagen. Infect. Immun. 1997, 65, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Ünal, C.M.; Steinert, M. Microbial Peptidyl-Prolyl Cis/Trans Isomerases (PPIases): Virulence Factors and Potential Alternative Drug Targets. Microbiol. Mol. Biol. Rev. 2014, 78, 544–571. [Google Scholar] [CrossRef]
- Behrens-Kneip, S. The Role of SurA Factor in Outer Membrane Protein Transport and Virulence. Int. J. Med. Microbiol. 2010, 300, 421–428. [Google Scholar] [CrossRef] [PubMed]
- White-Ziegler, C.A.; Davis, T.R. Genome-Wide Identification of H-NS-Controlled, Temperature-Regulated Genes in Escherichia coli K-12. J. Bacteriol. 2009, 191, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C.D. H-NS Mediates the Silencing of Laterally Acquired Genes in Bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef] [PubMed]
- Olekhnovich, I.N.; Kadner, R.J. Role of Nucleoid-Associated Proteins Hha and H-NS in Expression of Salmonella enterica Activators HilD, HilC, and RtsA Required for Cell Invasion. J. Bacteriol. 2007, 189, 6882–6890. [Google Scholar] [CrossRef] [PubMed]
- Baños, R.C.; Vivero, A.; Aznar, S.; García, J.; Pons, M.; Madrid, C.; Juárez, A. Differential Regulation of Horizontally Acquired and Core Genome Genes by the Bacterial Modulator H-NS. PLoS Genet. 2009, 5, e1000513. [Google Scholar] [CrossRef] [PubMed]
- Martínez, L.C.; Banda, M.M.; Fernández-Mora, M.; Santana, F.J.; Bustamante, V.H. HilD Induces Expression of Salmonella Pathogenicity Island 2 Genes by Displacing the Global Negative Regulator H-NS from SsrAB. J. Bacteriol. 2014, 196, 3746–3755. [Google Scholar] [CrossRef] [PubMed]
- Duong, N.; Osborne, S.; Bustamante, V.H.; Tomljenovic, A.M.; Puente, J.L.; Coombes, B.K. Thermosensing Coordinates a Cis-Regulatory Module for Transcriptional Activation of the Intracellular Virulence System in Salmonella enterica Serovar Typhimurium. J. Biol. Chem. 2007, 282, 34077–34084. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.S.; Cowen, L.E. Thermal Control of Microbial Development and Virulence: Molecular Mechanisms of Microbial Temperature Sensing. mBio 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Steinmann, R.; Dersch, P. Thermosensing to Adjust Bacterial Virulence in a Fluctuating Environment. Future Microbiol. 2012, 8, 85–105. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.D.S.; Stoebel, D.M.; Dorman, C.J. DNA Supercoiling Is Differentially Regulated by Environmental Factors and FIS in Escherichia coli and Salmonella enterica. Mol. Microbiol. 2011, 80, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.D.S.; Dorman, C.J. A Fundamental Regulatory Mechanism Operating through OmpR and DNA Topology Controls Expression of Salmonella Pathogenicity Islands SPI-1 and SPI-2. PLoS Genet. 2012, 8, e1002615. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J.; Deighan, P. Regulation of Gene Expression by Histone-like Proteins in Bacteria. Curr. Opin. Genet. Dev. 2003, 13, 179–184. [Google Scholar] [CrossRef]
- Browning, D.F.; Grainger, D.C.; Busby, S.J. Effects of Nucleoid-Associated Proteins on Bacterial Chromosome Structure and Gene Expression. Curr. Opin. Microbiol. 2010, 13, 773–780. [Google Scholar] [CrossRef]
- Corry, J.E.L.; Roberts, T.A. A Note on the Development of Resistance to Heat and Gamma Radiation in Salmonella. J. Appl. Bacteriol. 1970, 33, 733–737. [Google Scholar] [CrossRef]
- Rudolph, B.; Gebendorfer, K.M.; Buchner, J.; Winter, J. Evolution of Escherichia coli for Growth at High Temperatures. J. Biol. Chem. 2010, 285, 19029–19034. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Muñoz, B.; Prats-Escriche, M.; Montagud-Martínez, R.; López-Cerdán, A.; Toft, C.; Aguilar-Rodríguez, J.; Wagner, A.; Fares, M.A. Fitness Trade-Offs Determine the Role of the Molecular Chaperonin GroEL in Buffering Mutations. Mol. Biol. Evol. 2015, 32, 2681–2693. [Google Scholar] [CrossRef] [PubMed]
- Ezemaduka, A.N.; Yu, J.; Shi, X.; Zhang, K.; Yin, C.-C.; Fu, X.; Chang, Z. A Small Heat Shock Protein Enables Escherichia coli to Grow at a Lethal Temperature of 50 °C Conceivably by Maintaining Cell Envelope Integrity. J. Bacteriol. 2014, 196, 2004–2011. [Google Scholar] [CrossRef]
- Troxell, B. Salmonella enterica Serovar Typhimurium Utilizes the ClpPX and Lon Proteases for Optimal Fitness in the Ceca of Chickens. Poult. Sci. 2016, 95, 1617–1623. [Google Scholar] [CrossRef] [PubMed]
- Maâlej, L.; Chatti, A.; Khefacha, S.; Salma, K.; Gottardi, D.; Vannini, L.; Guerzoni, M.E.; Hassen, A. UV-C Pre-Adaptation of Salmonella: Effect on Cell Morphology and Membrane Fatty Acids Composition. World J. Microbiol. Biotechnol. 2014, 30, 925–930. [Google Scholar] [CrossRef] [PubMed]
- Timmons, C. Elucidation of the Molecular Mechanisms of Foodborne Human Pathogen Inactivation by Cold Atmospheric Plasma through RNA-Seq Analysis. Ph.D. Thesis, Oklahoma State University, Stillwater, OK, USA, 2016. [Google Scholar]
- Davies, R.; Sinskey, A.J. Radiation-Resistant Mutants of Salmonella typhimurium LT2: Development and Characterization. J. Bacteriol. 1973, 113, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, J.J.; Nickerson, J.T.R.; Goldblith, S.A.; Shannon, C.A.; Bishop, W.W. Development of Radiation Resistance in Salmonella Cultures. Appl. Microbiol. 1969, 18, 24–30. [Google Scholar] [CrossRef]
- Licciardello, J.J.; Nickerson, J.T.R.; Goldblith, S.A.; Bishop, W.W.; Shannon, C.A. Effect of Repeated Irradiation on Various Characteristics of Salmonella. Appl. Microbiol. 1969, 18, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Previte, J.J.; Chang, Y.; Scrutchfield, W.; El-Bisi, H.M. Effects of Radiation Pasteurization on Salmonella. II. Influence of Repeated Radiation-Growth Cycles on Virulence and Resistance to Radiation and Antibiotics. Can. J. Microbiol. 1971, 17, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Gayán, E.; Mañas, P.; Álvarez, I.; Condón, S. Mechanism of the Synergistic Inactivation of Escherichia coli by UV-C Light at Mild Temperatures. Appl. Environ. Microbiol. 2013, 79, 4465–4473. [Google Scholar] [CrossRef]
- Child, M.; Strike, P.; Pickup, R.; Edwards, C. Salmonella typhimurium Displays Cyclical Patterns of Sensitivity to UV-C Killing during Prolonged Incubation in the Stationary Phase of Growth. FEMS Microbiol. Lett. 2002, 213, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Sagarzazu, N.; Cebrián, G.; Pagán, R.; Condón, S.; Mañas, P. Emergence of Pulsed Electric Fields Resistance in Salmonella enterica Serovar Typhimurium SL1344. Int. J. Food Microbiol. 2013, 166, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Puig, M.; Lázaro, E.; Armero, C.; Alvares, D.; Martínez, A.; Rodrigo, D.S. Typhimurium Virulence Changes Caused by Exposure to Different Non-Thermal Preservation Treatments Using C. Elegans. Int. J. Food Microbiol. 2017, 262, 49–54. [Google Scholar] [CrossRef]
- Sanz-Puig, M.; Velázquez-Moreira, A.; Torres, C.; Guerrero-Beltrán, J.Á.; Cunha, L.M.; Martinez, A.; Rodrigo, D. Resistance Changes in Salmonella enterica Serovar Typhimurium Treated by High Hydrostatic Pressure and Pulsed Electric Fields and Assessment of Virulence Changes by Using Caenorhabditis Elegans as a Test Organism. Innov. Food Sci. Emerg. Technol. 2019, 51, 51–56. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The Importance of the Viable but Non-Culturable State in Human Bacterial Pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Caro, A.; Got, P.; Lesne, J.; Binard, S.; Baleux, B. Viability and Virulence of Experimentally Stressed Nonculturable Salmonella typhimurium. Appl. Environ. Microbiol. 1999, 65, 3229–3232. [Google Scholar] [CrossRef]
- Baleux, B.; Caro, A.; Lesne, J.; Got, P.; Binard, S.; Delpeuch, B. Survie et maintien de la virulence de Salmonella typhimurium VNC exposée simultanément à trois facteurs stressants expérimentaux. Oceanol. Acta 1998, 21, 939–950. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Williams, T.; Oliver, J.D. Relationship between the Viable but Nonculturable State and Antibiotic Persister Cells. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef]
- Rahman, I.; Shahamat, M.; Kirchman, P.A.; Russek-Cohen, E.; Colwell, R.R. Methionine Uptake and Cytopathogenicity of Viable but Nonculturable Shigella Dysenteriae Type 1. Appl. Environ. Microbiol. 1994, 60, 3573–3578. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I.; Shahamat, M.; Chowdhury, M.A.; Colwell, R.R. Potential Virulence of Viable but Nonculturable Shigella Dysenteriae Type 1. Appl. Environ. Microbiol. 1996, 62, 115–120. [Google Scholar] [CrossRef]
- Pommepuy, M.; Butin, M.; Derrien, A.; Gourmelon, M.; Colwell, R.R.; Cormier, M. Retention of Enteropathogenicity by Viable but Nonculturable Escherichia coli Exposed to Seawater and Sunlight. Appl. Environ. Microbiol. 1996, 62, 4621–4626. [Google Scholar] [CrossRef]
- Colwell, R.R.; Brayton, P.; Herrington, D.; Tall, B.; Huq, A.; Levine, M.M. Viable but Non-Culturable Vibrio Cholerae O1 Revert to a Cultivable State in the Human Intestine. World J. Microbiol. Biotechnol. 1996, 12, 28–31. [Google Scholar] [CrossRef] [PubMed]
- Steinert, M.; Emödy, L.; Amann, R.; Hacker, J. Resuscitation of Viable but Nonculturable Legionella Pneumophila Philadelphia JR32 by Acanthamoeba Castellanii. Appl. Environ. Microbiol. 1997, 63, 2047–2053. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent Bacterial Infections and Persister Cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by Macrophages Induces Formation of Nonreplicating Persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef] [PubMed]
- Stapels, D.A.C.; Hill, P.W.S.; Westermann, A.J.; Fisher, R.A.; Thurston, T.L.; Saliba, A.-E.; Blommestein, I.; Vogel, J.; Helaine, S. Salmonella Persisters Undermine Host Immune Defenses during Antibiotic Treatment. Science 2018, 362, 1156–1160. [Google Scholar] [CrossRef]
- Kint, C.I.; Verstraeten, N.; Fauvart, M.; Michiels, J. New-Found Fundamentals of Bacterial Persistence. Trends Microbiol. 2012, 20, 577–585. [Google Scholar] [CrossRef]
- Wu, S.; Yu, P.-L.; Flint, S. Persister Cell Formation of Listeria Monocytogenes in Response to Natural Antimicrobial Agent Nisin. Food Control 2017, 77, 243–250. [Google Scholar] [CrossRef]
- Wang, C.; Cheng, T.; Li, X.; Jin, L. Metronidazole-Treated Porphyromonas Gingivalis Persisters Invade Human Gingival Epithelial Cells and Perturb Innate Responses. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Marin, C.; Hernandiz, A.; Lainez, M. Biofilm Development Capacity of Salmonella Strains Isolated in Poultry Risk Factors and Their Resistance against Disinfectants. Poult. Sci. 2009, 88, 424–431. [Google Scholar] [CrossRef]
- Scher, K.; Romling, U.; Yaron, S. Effect of Heat, Acidification, and Chlorination on Salmonella enterica Serovar Typhimurium Cells in a Biofilm Formed at the Air-Liquid Interface. Appl. Environ. Microbiol. 2005, 71, 1163–1168. [Google Scholar] [CrossRef] [PubMed]
- Joseph, B.; Otta, S.K.; Karunasagar, I.; Karunasagar, I. Biofilm Formation by Salmonella Spp. on Food Contact Surfaces and Their Sensitivity to Sanitizers. Int. J. Food Microbiol. 2001, 64, 367–372. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella Biofilms: An Overview on Occurrence, Structure, Regulation and Eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- MacKenzie, K.D.; Palmer, M.B.; Köster, W.L.; White, A.P. Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef]
- Simm, R.; Ahmad, I.; Rhen, M.; Le Guyon, S.; Römling, U. Regulation of Biofilm Formation in Salmonella enterica Serovar Typhimurium. Future Microbiol. 2014, 9, 1261–1282. [Google Scholar] [CrossRef]
- White, A.P.; Gibson, D.L.; Grassl, G.A.; Kay, W.W.; Finlay, B.B.; Vallance, B.A.; Surette, M.G. Aggregation via the Red, Dry, and Rough Morphotype Is Not a Virulence Adaptation in Salmonella enterica Serovar Typhimurium. Infect. Immun. 2008, 76, 1048–1058. [Google Scholar] [CrossRef]
- MacKenzie, K.D.; Wang, Y.; Shivak, D.J.; Wong, C.S.; Hoffman, L.J.L.; Lam, S.; Kröger, C.; Cameron, A.D.S.; Townsend, H.G.G.; Köster, W.; et al. Bistable Expression of CsgD in Salmonella enterica Serovar Typhimurium Connects Virulence to Persistence. Infect. Immun. 2015, 83, 2312–2326. [Google Scholar] [CrossRef] [PubMed]
- Adcox, H.E.; Vasicek, E.M.; Dwivedi, V.; Hoang, K.V.; Turner, J.; Gunn, J.S. Salmonella Extracellular Matrix Components Influence Biofilm Formation and Gallbladder Colonization. Infect. Immun. 2016, 84, 3243–3251. [Google Scholar] [CrossRef]
- MacKenzie, K.D.; Wang, Y.; Musicha, P.; Hansen, E.G.; Palmer, M.B.; Herman, D.J.; Feasey, N.A.; White, A.P. Parallel Evolution Leading to Impaired Biofilm Formation in Invasive Salmonella Strains. PLoS Genet. 2019, 15, e1008233. [Google Scholar] [CrossRef] [PubMed]
- Weening, E.H.; Barker, J.D.; Laarakker, M.C.; Humphries, A.D.; Tsolis, R.M.; Bäumler, A.J. The Salmonella enterica Serotype Typhimurium Lpf, Bcf, Stb, Stc, Std, and Sth Fimbrial Operons Are Required for Intestinal Persistence in Mice. Infect. Immun. 2005, 73, 3358–3366. [Google Scholar] [CrossRef]
- Bester, E.; Wolfaardt, G.; Joubert, L.; Garny, K.; Saftic, S. Planktonic-Cell Yield of a Pseudomonad Biofilm. Appl. Environ. Microbiol. 2005, 71, 7792–7798. [Google Scholar] [CrossRef]
- Ellwood, D.C.; Keevil, C.W.; Marsh, P.D.; Brown, C.M.; Wardell, J.N.; Le Roux, N.; Quayle, J.R.; Bull, A.T. Surface-Associated Growth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982, 297, 517–532. [Google Scholar] [CrossRef] [PubMed]
- Barton, A.J.; Sagers, R.D.; Pitt, W.G. Measurement of Bacterial Growth Rates on Polymers. J. Biomed. Mater. Res. 1996, 32, 271–278. [Google Scholar] [CrossRef]
- Knudsen, G.M.; Nielsen, M.-B.; Grassby, T.; Danino-Appleton, V.; Thomsen, L.E.; Colquhoun, I.J.; Brocklehurst, T.F.; Olsen, J.E.; Hinton, J.C.D. A Third Mode of Surface-Associated Growth: Immobilization of Salmonella enterica Serovar Typhimurium Modulates the RpoS-Directed Transcriptional Programme. Environ. Microbiol. 2012, 14, 1855–1875. [Google Scholar] [CrossRef]
- Aviles, B.; Klotz, C.; Eifert, J.; Williams, R.; Ponder, M. Biofilms Promote Survival and Virulence of Salmonella enterica Sv. Tennessee during Prolonged Dry Storage and after Passage through an In Vitro Digestion System. Int. J. Food Microbiol. 2013, 162, 252–259. [Google Scholar] [CrossRef]
- Deditius, J.A.; Felgner, S.; Spöring, I.; Kühne, C.; Frahm, M.; Rohde, M.; Weiß, S.; Erhardt, M. Characterization of Novel Factors Involved in Swimming and Swarming Motility in Salmonella enterica Serovar Typhimurium. PLoS ONE 2015, 10, e0135351. [Google Scholar] [CrossRef] [PubMed]
- Spöring, I.; Felgner, S.; Preuße, M.; Eckweiler, D.; Rohde, M.; Häussler, S.; Weiss, S.; Erhardt, M. Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella enterica Serovar Typhimurium. mBio 2018, 9. [Google Scholar] [CrossRef]
- Maserati, A.; Fink, R.C.; Lourenco, A.; Julius, M.L.; Diez-Gonzalez, F. General Response of Salmonella enterica Serovar Typhimurium to Desiccation: A New Role for the Virulence Factors SopD and SseD in Survival. PLoS ONE 2017, 12, e0187692. [Google Scholar] [CrossRef]
- Walker, S.L.; Sojka, M.; Dibb-Fuller, M.; Woodward, M.J. Effect of PH, Temperature and Surface Contact on the Elaboration of Fimbriae and Flagella by Salmonella Serotype Enteritidis. J. Med. Microbiol. 1999, 48, 253–261. [Google Scholar] [CrossRef] [PubMed]
- DeLisa, M.P.; Valdes, J.J.; Bentley, W.E. Mapping Stress-Induced Changes in Autoinducer AI-2 Production in Chemostat-Cultivated Escherichia coli K-12. J. Bacteriol. 2001, 183, 2918–2928. [Google Scholar] [CrossRef]
- Surette, M.G.; Bassler, B.L. Regulation of Autoinducer Production in Salmonella typhimurium. Mol. Microbiol. 1999, 31, 585–595. [Google Scholar] [CrossRef]
- Flynn, P.B.; Busetti, A.; Wielogorska, E.; Chevallier, O.P.; Elliott, C.T.; Laverty, G.; Gorman, S.P.; Graham, W.G.; Gilmore, B.F. Non-Thermal Plasma Exposure Rapidly Attenuates Bacterial AHL-Dependent Quorum Sensing and Virulence. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Ziuzina, D.; Boehm, D.; Patil, S.; Cullen, P.J.; Bourke, P. Cold Plasma Inactivation of Bacterial Biofilms and Reduction of Quorum Sensing Regulated Virulence Factors. PLoS ONE 2015, 10, 0138209. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Molina, A.; de Toro, M.; Ruiz, L.; López, M.; Prieto, M.; Alvarez-Ordóñez, A. Selection for Antimicrobial Resistance in Foodborne Pathogens through Exposure to UV Light and Nonthermal Atmospheric Plasma Decontamination Techniques. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.A.; Eade, C.R.; Wiedmann, M. Embracing Diversity: Differences in Virulence Mechanisms, Disease Severity, and Host Adaptations Contribute to the Success of Nontyphoidal Salmonella as a Foodborne Pathogen. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Guillén, S.; Marcén, M.; Mañas, P.; Cebrián, G. Differences in Resistance to Different Environmental Stresses and Non-Thermal Food Preservation Technologies among Salmonella enterica Subsp. Enterica Strains. Food Res. Int. 2020, 132, 109042. [Google Scholar] [CrossRef] [PubMed]
- Guillén, S.; Marcén, M.; Álvarez, I.; Mañas, P.; Cebrián, G. Stress Resistance of Emerging Poultry-Associated Salmonella Serovars. Int. J. Food Microbiol. 2020, 335, 108884. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.E.; Mazzotta, A.S. Review of Studies on the Thermal Resistance of Salmonellae. J. Food Prot. 2000, 63, 779–795. [Google Scholar] [CrossRef]
- Lianou, A.; Koutsoumanis, K.P. Evaluation of the Strain Variability of Salmonella enterica Acid and Heat Resistance. Food Microbiol. 2013, 34, 259–267. [Google Scholar] [CrossRef]
- Shah, D.H.; Casavant, C.; Hawley, Q.; Addwebi, T.; Call, D.R.; Guard, J. Salmonella Enteritidis Strains from Poultry Exhibit Differential Responses to Acid Stress, Oxidative Stress, and Survival in the Egg Albumen. Foodborne Pathog. Dis. 2012, 9, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Quintavalla, S.; Larini, S.; Mutti, P.; Barbuti, S. Evaluation of the Thermal Resistance of Different Salmonella Serotypes in Pork Meat Containing Curing Additives. Int. J. Food Microbiol. 2001, 67, 107–114. [Google Scholar] [CrossRef]
- Sherry, A.E.; Patterson, M.F.; Madden, R.H. Comparison of 40 Salmonella enterica Serovars Injured by Thermal, High-Pressure and Irradiation Stress. J. Appl. Microbiol. 2004, 96, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Saldaña, G.; Puértolas, E.; López, N.; García, D.; Álvarez, I.; Raso, J. Comparing the PEF Resistance and Occurrence of Sublethal Injury on Different Strains of Escherichia coli, Salmonella Typhimurium, Listeria monocytogenes and Staphylococcus aureus in Media of PH 4 and 7. Innov. Food Sci. Emerg. Technol. 2009, 10, 160–165. [Google Scholar] [CrossRef]
- Gill, A.; Tamber, S.; Yang, X. Relative Response of Populations of Escherichia coli and Salmonella enterica to Exposure to Thermal, Alkaline and Acidic Treatments. Int. J. Food Microbiol. 2019, 293, 94–101. [Google Scholar] [CrossRef]
- Kim, M.-J.; Yuk, H.-G. Antibacterial Mechanism of 405-Nanometer Light-Emitting Diode against Salmonella at Refrigeration Temperature. Appl. Environ. Microbiol. 2017, 83, e02582-16. [Google Scholar] [CrossRef]
- Abdullah, W.Z.W.; Mackey, B.M.; Karatzas, K.A.G. High Phenotypic Variability among Representative Strains of Common Salmonella enterica Serovars with Possible Implications for Food Safety. J. Food Prot. 2018, 81, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Andino, A.; Hanning, I. Salmonella enterica: Survival, Colonization, and Virulence Differences among Serovars. Sci. World J. 2015, 2015, 520179. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.H. RNA Sequencing Reveals Differences between the Global Transcriptomes of Salmonella enterica Serovar Enteritidis Strains with High and Low Pathogenicities. Appl. Environ. Microbiol. 2014, 80, 896–906. [Google Scholar] [CrossRef] [PubMed]

| Selection Agent | Strain | Effect in Virulence | Effect in Growth Fitness | Other Characteristics | References |
|---|---|---|---|---|---|
| Acid Stress | |||||
| pH 2.5 | S. Enteritidis 66045 | Lower colonization of spleens and livers | Reduced growth rate and yields | - | [105] |
| pH 2.5 | S. Typhimurium 30 | Lower virulence | Reduced growth rate and yields | Increased heat resistance | [105] |
| Osmotic stress | |||||
| NaCl | S. Typhimurium NCCP10812 | No changes in invasion | Not determined | Decreased atb resistance | [117] |
| NaCl | S. Enteritidis NCCP12243 | Increased invasion | Not determined | Antibiotic susceptibility | [117] |
| Oxidative stress, detergents and disinfectants | |||||
| Blend of oxidizing compounds | S. Typhimurium SL1344 | Decreased invasion | Reduced growth rate and yields | Decreased atb resistance Reduced motility | [136,137] |
| QA + FA + GA | S. Typhimurium SL1344 | Decreased invasion | Reduced growth rate and yields | Decreased atb resistance Reduced motility | [136,137] |
| Phenolic tar acids-based disinfectant | S. Typhimurium SL1344 | Decreased invasion | Reduced growth rate and yields | Decreased atb resistance Reduced motility | [136,137] |
| DTAC | S. Enteritidis ATCC 4931 | Decreased invasion | Not determined | Fewer fimbriae | [185] |
| Heat stress | |||||
| 55 °C | S. Typhimurium phage type l | Decreased virulence | Not determined | Increased roughness | [229] |
| Non-Thermal Technologies | |||||
| γ- radiation | S. Typhimurium phage type 2c | No change | Not determined | Increased roughness | [229] |
| γ- radiation | S. Typhimurium LT2 | No change | Grows poorly in minimal media | Increased cell size | [236] |
| Ionizing radiation | S. Typhimurium ATCC 7823 | Not determined | No change | - | [237,238] |
| Ionizing radiation | S. Newport ATCC 6962 | Not determined | Reduced growth rate at 10–20 °C. | - | [237,238] |
| Ionizing radiation | S. Thompson ATCC 8391 | Not determined | No change | - | [237,238] |
| Ionizing radiation | S. Heidelberg ATCC 8326 | Not determined | No change | - | [237,238] |
| Ionizing radiation | S. Typhimurium strain RIA | Decreased virulence | Not determined | - | [239] |
| UV-C | S. Typhimurium (various strains) | Not determined | Not determined | Increased atb resistance | [284] |
| PEF | S. Typhimurium SL1344 | Not determined | Not determined | - | [242] |
| PEF | S. Typhimurium CECT 443 | Less virulent in C. elegans | Not determined | - | [243,244] |
| HHP | S. Typhimurium CECT 443 | Less virulent in C. elegans | Not determined | - | [243,244] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guillén, S.; Nadal, L.; Álvarez, I.; Mañas, P.; Cebrián, G. Impact of the Resistance Responses to Stress Conditions Encountered in Food and Food Processing Environments on the Virulence and Growth Fitness of Non-Typhoidal Salmonellae. Foods 2021, 10, 617. https://doi.org/10.3390/foods10030617
Guillén S, Nadal L, Álvarez I, Mañas P, Cebrián G. Impact of the Resistance Responses to Stress Conditions Encountered in Food and Food Processing Environments on the Virulence and Growth Fitness of Non-Typhoidal Salmonellae. Foods. 2021; 10(3):617. https://doi.org/10.3390/foods10030617
Chicago/Turabian StyleGuillén, Silvia, Laura Nadal, Ignacio Álvarez, Pilar Mañas, and Guillermo Cebrián. 2021. "Impact of the Resistance Responses to Stress Conditions Encountered in Food and Food Processing Environments on the Virulence and Growth Fitness of Non-Typhoidal Salmonellae" Foods 10, no. 3: 617. https://doi.org/10.3390/foods10030617
APA StyleGuillén, S., Nadal, L., Álvarez, I., Mañas, P., & Cebrián, G. (2021). Impact of the Resistance Responses to Stress Conditions Encountered in Food and Food Processing Environments on the Virulence and Growth Fitness of Non-Typhoidal Salmonellae. Foods, 10(3), 617. https://doi.org/10.3390/foods10030617






