Application of NMR and Chemometrics for the Profiling and Classification of Ale and Lager American Craft Beer
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Beer Samples
2.2. Sample Preparation
2.3. Enzymatic Hydrolysis
2.4. NMR Analysis
2.5. Multivariate Data Analysis
3. Results
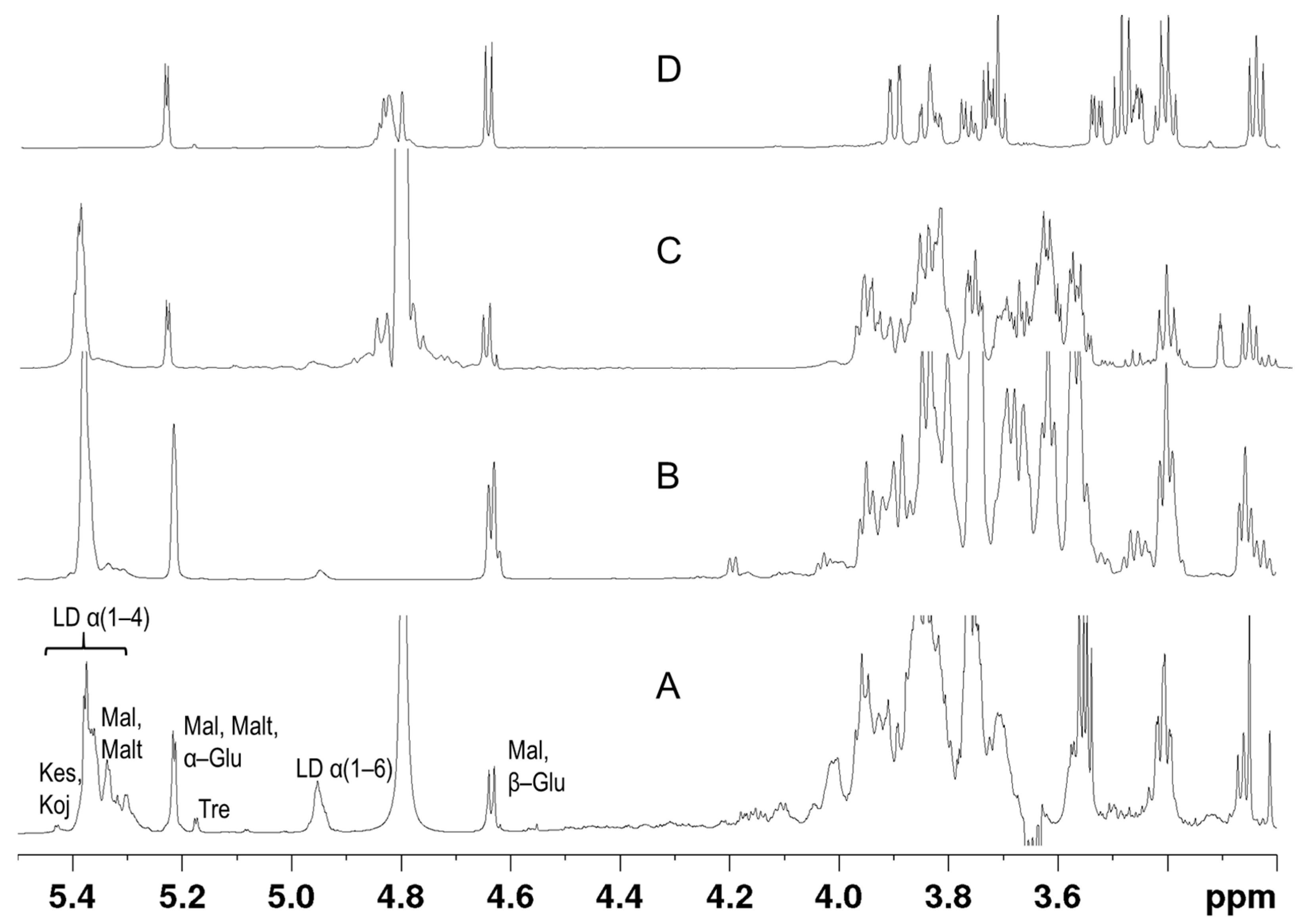
3.1. NMR Analysis
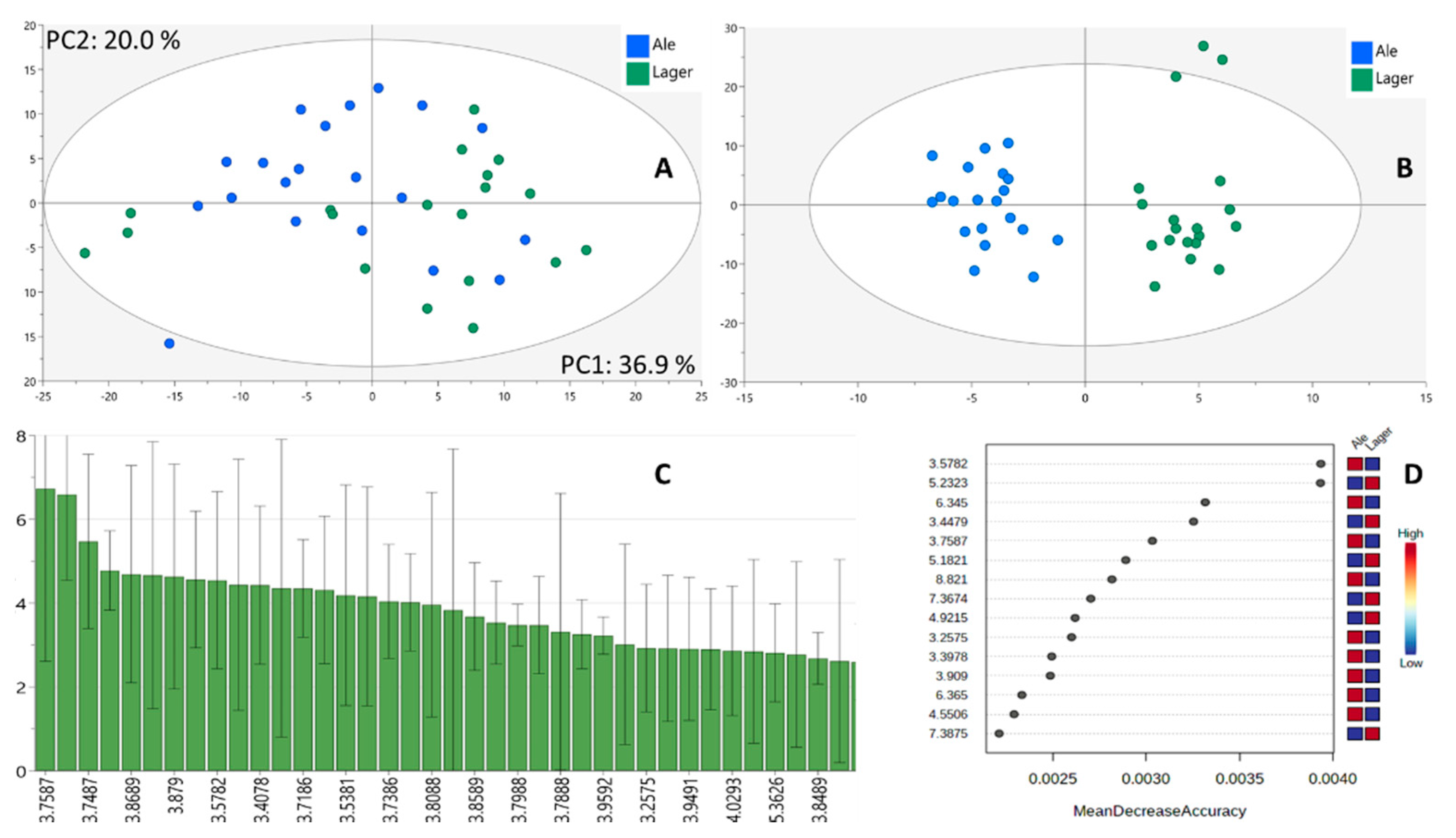
3.2. Chemometrics
3.2.1. Initial Untargeted Analysis
3.2.2. Model Improvement and Biomarker Selection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pires, E.; Brányik, T. The Brewing Yeast. In Biochemistry of Beer Fermentation; Pires, E., Brányik, T., Eds.; Springer Briefs in Biochemistry and Molecular Biology; Springer International Publishing: Cham, Switzerland, 2015; pp. 11–49. [Google Scholar] [CrossRef]
- Li, H.G.; Liu, F. Changes in Organic Acids during Beer Fermentation. J. Am. Soc. Brew. Chem. 2015, 73, 275–279. [Google Scholar] [CrossRef]
- Sammartino, M. Enzymes in Brewing. TQ 2015. [Google Scholar] [CrossRef]
- Brewers Association. Brewers Association Releases Annual Growth Report for 2019; Brewers Association: Boulder, CO, USA, 2020. [Google Scholar]
- Anderson, H.E.; Santos, I.C.; Hildenbrand, Z.L.; Schug, K.A. A Review of the Analytical Methods Used for Beer Ingredient and Finished Product Analysis and Quality Control. Anal. Chim. Acta 2019, 1085, 1–20. [Google Scholar] [CrossRef]
- Granato, D.; Putnik, P.; Kovačević, D.B.; Santos, J.S.; Calado, V.; Rocha, R.S.; Cruz, A.G.D.; Jarvis, B.; Rodionova, O.Y.; Pomerantsev, A. Trends in Chemometrics: Food Authentication, Microbiology, and Effects of Processing. Compr. Rev. Food Sci. Food Saf. 2018, 17, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Sobolev, A.P.; Circi, S.; Capitani, D.; Ingallina, C.; Mannina, L. Molecular Fingerprinting of Food Authenticity. Curr. Opin. Food Sci. 2017, 16, 59–66. [Google Scholar] [CrossRef]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, A.; Mannina, L.; Salvo, A. Multiple Assignment Recovered Analysis (MARA) NMR for a Direct Food Labeling: The Case Study of Olive Oils. Food Anal. Methods 2019, 12, 1238–1245. [Google Scholar] [CrossRef]
- da Silva, L.A.; Flumignan, D.L.; Pezza, H.R.; Pezza, L. 1H NMR Spectroscopy Combined with Multivariate Data Analysis for Differentiation of Brazilian Lager Beer According to Brewery. Eur. Food Res. Technol. 2019, 245, 2365–2372. [Google Scholar] [CrossRef]
- da Silva, L.A.; Flumignan, D.L.; Tininis, A.G.; Pezza, H.R.; Pezza, L. Discrimination of Brazilian Lager Beer by 1H NMR Spectroscopy Combined with Chemometrics. Food Chem. 2019, 272, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Mannina, L.; Marini, F.; Antiochia, R.; Cesa, S.; Magrì, A.; Capitani, D.; Sobolev, A.P. Tracing the Origin of Beer Samples by NMR and Chemometrics: Trappist Beers as a Case Study. Electrophoresis 2016, 37, 2710–2719. [Google Scholar] [CrossRef] [PubMed]
- Palmioli, A.; Alberici, D.; Ciaramelli, C.; Airoldi, C. Metabolomic Profiling of Beers: Combining 1H NMR Spectroscopy and Chemometric Approaches to Discriminate Craft and Industrial Products. Food Chem. 2020, 327, 127025. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Duarte, I.; Barros, A.; Rodrigues, J.; Spraul, M.; Gil, A. Composition of Beer by 1H NMR Spectroscopy: Effects of Brewing Site and Date of Production. J. Agric. Food Chem. 2006, 54, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Affine Transformation of Negative Values for NMR Metabolomics Using the mrbin R Package. J. Proteome Res. 2021, 20, 1397–1404. [CrossRef]
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A Comprehensive Server for Metabolomic Data Analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.R.; Soprano, S.E.; Wickham, L.M.; Fitzgerald, N.; Edwards, J.C. Nuclear Magnetic Resonance and Headspace Solid-Phase Microextraction Gas Chromatography as Complementary Methods for the Analysis of Beer Samples. Beverages 2017, 3, 21. [Google Scholar] [CrossRef]
- Pozo-Insfran, D.D.; Urias-Lugo, D.; Hernandez-Brenes, C.; Saldivar, S.O.S. Effect of Amyloglucosidase on Wort Composition and Fermentable Carbohydrate Depletion in Sorghum Lager Beers. J. Inst. Brew. 2004, 110, 124–132. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. CV-ANOVA for Significance Testing of PLS and OPLS® Models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Pérez-Ràfols, C.; Viñas, D.; Hernández-Cassou, S.; Saurina, J. Experimental Design for the Determination of Polyphenols by Liquid Chromatography: Application to the Chemometric Characterization and Classification of Beers. Anal. Methods 2015, 7, 3283–3290. [Google Scholar] [CrossRef]
- Duarte, I.; Barros, A.; Belton, P.S.; Righelato, R.; Spraul, M.; Humpfer, E.; Gil, A.M. High-Resolution Nuclear Magnetic Resonance Spectroscopy and Multivariate Analysis for the Characterization of Beer. J. Agric. Food Chem. 2002, 50, 2475–2481. [Google Scholar] [CrossRef]
- Duarte, I.F.; Barros, A.; Almeida, C.; Spraul, M.; Gil, A.M. Multivariate Analysis of NMR and FTIR Data as a Potential Tool for the Quality Control of Beer. J. Agric. Food Chem. 2004, 52, 1031–1038. [Google Scholar] [CrossRef]
- Savorani, F.; Tomasi, G.; Engelsen, S.B. Icoshift: A Versatile Tool for the Rapid Alignment of 1D NMR Spectra. J. Magn. Reson. 2010, 202, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]

| Normalization Method | R2/Q2 Values | CV-ANOVA (p Value) |
|---|---|---|
| Total intensity | 0.87/0.51 | 0.013 |
| Total intensity/Log transformation | 0.94/0.45 | 0.011 |
| PQN | 0.90/0.64 | 0.002 |
| PQN/Log transformation | 0.99/0.63 | 0.052 |
| No normalization | 0.87/0.33 | 0.108 |
| Log transformation | 0.99/0.72 | 0.006 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasas, M.; Tang, F.; Hatzakis, E. Application of NMR and Chemometrics for the Profiling and Classification of Ale and Lager American Craft Beer. Foods 2021, 10, 807. https://doi.org/10.3390/foods10040807
Vasas M, Tang F, Hatzakis E. Application of NMR and Chemometrics for the Profiling and Classification of Ale and Lager American Craft Beer. Foods. 2021; 10(4):807. https://doi.org/10.3390/foods10040807
Chicago/Turabian StyleVasas, Morgan, Fenfen Tang, and Emmanuel Hatzakis. 2021. "Application of NMR and Chemometrics for the Profiling and Classification of Ale and Lager American Craft Beer" Foods 10, no. 4: 807. https://doi.org/10.3390/foods10040807
APA StyleVasas, M., Tang, F., & Hatzakis, E. (2021). Application of NMR and Chemometrics for the Profiling and Classification of Ale and Lager American Craft Beer. Foods, 10(4), 807. https://doi.org/10.3390/foods10040807





