Convolutional Neural Networks Using Enhanced Radiographs for Real-Time Detection of Sitophilus zeamais in Maize Grain
Abstract
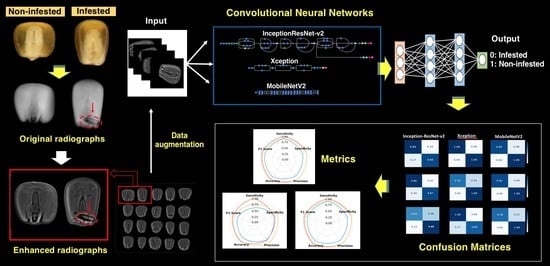
:1. Introduction
2. Materials and Methods
2.1. Insect Infestation
2.2. X-ray Imaging for Classes of Infested and Non-Infested Grains
2.3. Confirmation of Grains Infested by Eggs
2.4. Datasets
2.5. Data Augmentation
2.6. Transfer Learning and Architecture Approaches
2.7. Confusion Matrix and Metrics
3. Results
3.1. Classes of Infested and Non-Infested Grains
3.2. Training, Validation, and Test Sets
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dhliwayo, T.; Pixley, K.V. Divergent selection for resistance to maize weevil in six maize populations. Crop Breed. Genet. Cytol. 2003, 43, 2043–2049. [Google Scholar] [CrossRef]
- Devi, S.R.; Thomas, A.; Rebijith, K.B.; Ramamurthy, V.V. Biology, morphology and molecular characterization of Sitophilus oryzae and S. zeamais (Coleoptera: Curculionidae). J. Stored Prod. Res. 2017, 73, 135–141. [Google Scholar] [CrossRef]
- Stuhl, C.J. Does prior feeding behavior by previous generations of the maize weevil (Coleoptera: Curculionidae) determine future descendants feeding preference and ovipositional suitability? Fla. Entomol. 2019, 102, 366–372. [Google Scholar] [CrossRef] [Green Version]
- Danho, M.; Gaspar, C.; Haubruge, E. The impact of grain quantity on the biology of Sitophilus zeamais Motschulsky (Coleoptera: Curculionidae): Oviposition, distribution of eggs, adult emergence, body weight and sex ratio. J. Stored Prod. Res. 2002, 38, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Trematerra, P. Preferences of Sitophilus zeamais to different types of Italian commercial rice and cereal pasta. Bull. Insectology 2009, 62, 103–106. [Google Scholar]
- Neethirajan, S.; Karunakaran, C.; Jayas, D.S.; White, N.D.G. Detection techniques for stored-product insects in grain. Food Control 2007, 18, 157–162. [Google Scholar] [CrossRef]
- Banga, K.S.; Kotwaliwale, N.; Mohapatra, D.; Giri, S.K. Techniques for insect detection in stored food grains: An overview. Food Control 2018, 94, 167–176. [Google Scholar] [CrossRef]
- Karunakaran, C.; Jayas, D.S.; White, N.D.G. Soft X-ray inspection of wheat kernels infested by Sitophilus oryzae. Trans. ASAE 2003, 46, 739–745. [Google Scholar] [CrossRef]
- Pisano, E.D.; Cole, E.B.; Hemminger, B.M.; Yaffe, M.J.; Aylward, S.R.; Maidment, A.D.A.; Johnston, R.E.; Williams, M.B.; Niklason, L.T.; Conant, E.F.; et al. Image processing algorithms for digital mammography: A pictorial essay. Imaging Ther. Technol. 2000, 20, 1479–1491. [Google Scholar] [CrossRef]
- Warren, L.M.; Given-Wilson, R.M.; Wallis, M.G.; Cooke, J.; Halling-Brown, M.D.; Mackenzie, A.; Chakraborty, D.P.; Bosmans, H.; Dance, D.R.; Young, K.C. The effect of image processing on the detection of cancers in digital mammography. Med. Phys. Inform. 2014, 203, 387–393. [Google Scholar] [CrossRef]
- Emmert-Streib, F.; Yang, Z.; Feng, H.; Tripathi, S.; Dehmer, M. An introductory review of deep learning for prediction models with big data. Front. Artif. Intell. 2020, 1–23. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Yairi, T. A review on the application of deep learning in system health management. Mech. Syst. Signal Process. 2018, 107, 241–265. [Google Scholar] [CrossRef]
- Lawson, C.E.; Marti, J.M.; Radivojevic, T.; Jonnalagadda, S.V.R.; Gentz, R.; Hillson, N.J.; Peisert, S.; Kim, J.; Simmons, B.A.; Petzold, C.J.; et al. Machine learning for metabolic engineering: A review. Metab. Eng. 2021, 63, 34–60. [Google Scholar] [CrossRef]
- Sengupta, S.; Basak, S.; Saikia, P.; Sayak, P.; Tsalavoutis, V.; Atiah, F.; Ravi, V.; Peters, A. A review of deep learning with special emphasis on architectures, applications and recent trends. Knowl. Based Syst. 2019, 194, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Ponti, M.A.; Ribeiro, L.S.F.; Nazare, T.S.; Bui, T.; Collomosse, J. Everything you wanted to know about deep learning for computer vision but were afraid to ask. In Proceedings of the 30th SIBGRAPI Conference on Graphics, Patterns and Images Tutorials, Niterói, Brazil, 17–18 October 2017; pp. 17–41. [Google Scholar]
- Bhatia, Y.; Bajpayee, A.; Raghuvanshi, D.; Mittal, H. Image captioning using Google’s Inception-ResNet-v2 and Recurrent Neural Network. In Proceedings of the Twelfth International Conference on Contemporary Computing, NOIDA, India, 8–10 August 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Wan, X.; Ren, F.; Yong, D. Using Inception-Resnet v2 for face-based age recognition in scenic spots. In Proceedings of the IEEE 6th International Conference on Cloud Computing and Intelligence Systems, Singapore, 19–21 December 2019; pp. 159–163. [Google Scholar] [CrossRef]
- Lateef, F.; Ruichek, Y. Survey on semantic segmentation using deep learning techniques. Neurocomputing 2019, 338, 321–348. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Ouyang, W.; Wang, X.; Fieguth, P.; Chen, J.; Liu, X.; Pietikainen, M. Deep learning for generic object detection: A survey. Int. J. Comput. Vis. 2020, 128, 261–318. [Google Scholar] [CrossRef] [Green Version]
- Al-masni, M.A.; Kim, D.H.; Kim, T.S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Meth. Prog. Bio. 2020, 190, 1–12. [Google Scholar] [CrossRef]
- Medeiros, A.D.; Bernardes, R.C.; Silva, L.J.; Freitas, B.A.L.; Dias, D.C.F.S.; Barboza da Silva, C. Deep learning-based approach using X-ray images for classifying Crambe abyssinica seed quality. Ind. Crop. Prod. 2021, 164, 1–9. [Google Scholar] [CrossRef]
- Byng, J.W.; Critten, J.P.; Yaffe, M.J. Thickness-equalization processing for mammographic images. Radiology 1997, 203, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, J.; Zhang, X. A multiscale image enhancement method for calcification detection in screening mammograms. In Proceedings of the IEEE International Conference on Image Processing, Cairo, Egypt, 7–11 November 2009; pp. 677–680. [Google Scholar]
- Frankenfeld, J.C. Staining Methods for Detecting Weevil Infestation in Grain; USDA Bulletin of Entomology R Q. E-T Series; US Department of Agriculture, Bureau of Entomology and Plant Quarantine: Dublin, OH, USA, 1948; pp. 1–4.
- Yu, Y.; Lin, H.; Meng, J.; Wei, X.; Guo, H.; Zhao, Z. Deep transfer learning for modality classification of medical images. Information 2017, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, J.R.C.P.; Romero, R.A.F. Transfer learning based model for classification of cocoa pods. In Proceedings of the International Joint Conference on Neural Networks, Rio de Janeiro, Brazil, 8–13 July 2018. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-ResNet and the impact of residual connections on learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; pp. 4278–4284. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar] [CrossRef] [Green Version]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. MobileNetV2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18 June 2018; pp. 4510–4520. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference for Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond accuracy, F-Score and ROC: A family of discriminant measures for performance evaluation. In AI 2006: Advances in Artificial Intelligence; Sattar, A., Kang, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4304, pp. 1015–1021. [Google Scholar]
- Tharwat, A. Classification assessment methods. Appl. Comput. Inform. 2021, 17, 168–192. [Google Scholar] [CrossRef]
- Deuner, C.; Rosa, K.C.; Meneghello, G.E.; Borges, C.T.; Almeida, A.D.S.; Bohn, A. Physiological performance during storage of corn seed treated with insecticides and fungicide. J. Seed Sci. 2014, 36, 204–212. [Google Scholar] [CrossRef]
- Karunakaran, C.; Jayas, D.S.; White, N.D.G. X-ray image analysis to detect infestations caused by insects in grain. Cereal Chem. 2003, 80, 553–557. [Google Scholar] [CrossRef]
- Bianchini, V.J.M.; Mascarin, G.M.; Silva, L.C.A.S.; Arthur, V.; Carstensen, J.M.; Boelt, B.; Barboza da Silva, C. Multispectral and X-ray images for characterization of Jatropha curcas L. seed quality. Plant Methods 2021, 17, 1–13. [Google Scholar] [CrossRef]
- Kotwaliwale, N.; Singh, K.; Kalne, A.; Jha, S.N.; Seth, N.; Kar, A. X-ray imaging methods for internal quality evaluation of agricultural produce. J. Food Sci. Technol. 2014, 51, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, M.L.M.; Leite, E.R.; Carvalho, G.A.; França-Silva, F.; Andrade, D.B.; Marques, E.R. The compared efficiency of the traditional method, radiography without contrast and radiography with contrast in the determination of infestation by weevil (Sitophilus zeamais) in maize seeds. Insects 2019, 10, 156. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Essen, B.C.V.; Awwal, A.A.S.; Asari, V.K. The history began from AlexNet: A comprehensive survey on deep learning approaches. arXiv 2018, arXiv:1803.01164. [Google Scholar]
- Phiphiphatphaisit, S.; Surinta, O. Food image classification with improved MobileNet architecture and data augmentation. In Proceedings of the 3rd International Conference on Information Science and System, Cambridge University, Cambridge, UK, 19–22 March 2020; pp. 51–56. [Google Scholar] [CrossRef]
- Nwosu, L.C. Maize and the maize weevil: Advances and innovations in postharvest control of the pest. Food Qual. Saf. 2018, 3, 145–152. [Google Scholar] [CrossRef] [Green Version]







| Inception-ResNet v2 | Xception | MobileNetV2 | |
|---|---|---|---|
| Size | 215 MB | 88 MB | 14 MB |
| Top-1 accuracy | 0.803 | 0.790 | 0.713 |
| Top-5 accuracy | 0.953 | 0.945 | 0.901 |
| Depth | 572 | 126 | 88 |
| Number of trainable parameters | 1537 | 2049 | 1281 |
| Number of non-trainable parameters | 54,336,736 | 20,861,480 | 2,257,984 |
| Total parameters | 54,338,273 | 20,863,529 | 2,259,265 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barboza da Silva, C.; Silva, A.A.N.; Barroso, G.; Yamamoto, P.T.; Arthur, V.; Toledo, C.F.M.; Mastrangelo, T.d.A. Convolutional Neural Networks Using Enhanced Radiographs for Real-Time Detection of Sitophilus zeamais in Maize Grain. Foods 2021, 10, 879. https://doi.org/10.3390/foods10040879
Barboza da Silva C, Silva AAN, Barroso G, Yamamoto PT, Arthur V, Toledo CFM, Mastrangelo TdA. Convolutional Neural Networks Using Enhanced Radiographs for Real-Time Detection of Sitophilus zeamais in Maize Grain. Foods. 2021; 10(4):879. https://doi.org/10.3390/foods10040879
Chicago/Turabian StyleBarboza da Silva, Clíssia, Alysson Alexander Naves Silva, Geovanny Barroso, Pedro Takao Yamamoto, Valter Arthur, Claudio Fabiano Motta Toledo, and Thiago de Araújo Mastrangelo. 2021. "Convolutional Neural Networks Using Enhanced Radiographs for Real-Time Detection of Sitophilus zeamais in Maize Grain" Foods 10, no. 4: 879. https://doi.org/10.3390/foods10040879
APA StyleBarboza da Silva, C., Silva, A. A. N., Barroso, G., Yamamoto, P. T., Arthur, V., Toledo, C. F. M., & Mastrangelo, T. d. A. (2021). Convolutional Neural Networks Using Enhanced Radiographs for Real-Time Detection of Sitophilus zeamais in Maize Grain. Foods, 10(4), 879. https://doi.org/10.3390/foods10040879