Metagenetic Analysis for Microbial Characterization of Focaccia Doughs Obtained by Using Two Different Starters: Traditional Baker’s Yeast and a Selected Leuconostoc citreum Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Starter Strain and Culture Conditions
2.2. Liquid Sourdough and Dough Fermentation
2.3. Microbiological Analyses of Doughs
2.4. Characterization and Identification of LAB and Yeasts
2.5. Culture-Independent Community Identifications
2.6. Library Preparation and Sequencing
2.7. Bioinformatics
2.8. Statistics
2.9. Data Availability
3. Results
3.1. Culture-Dependent Microbiological Analyses
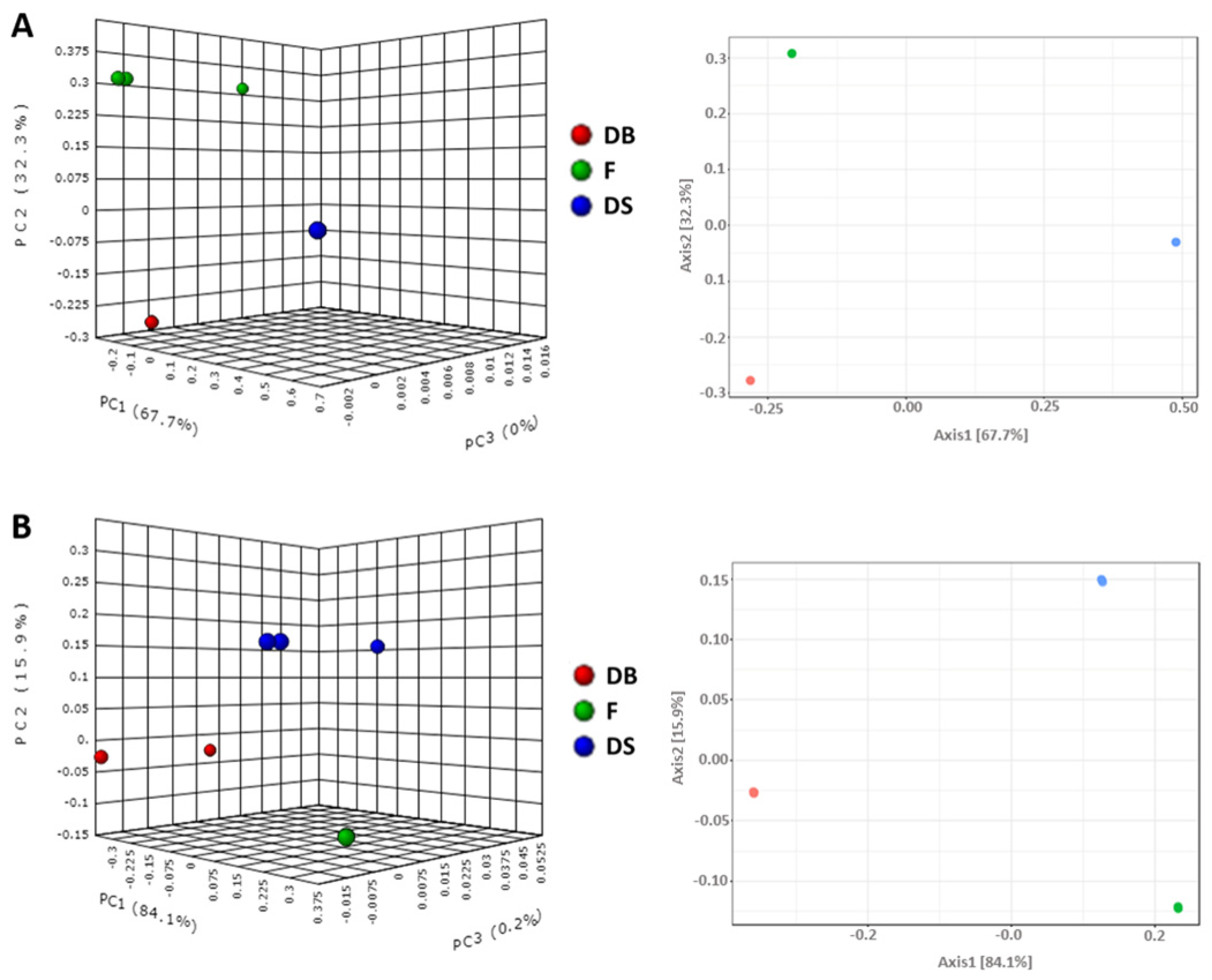
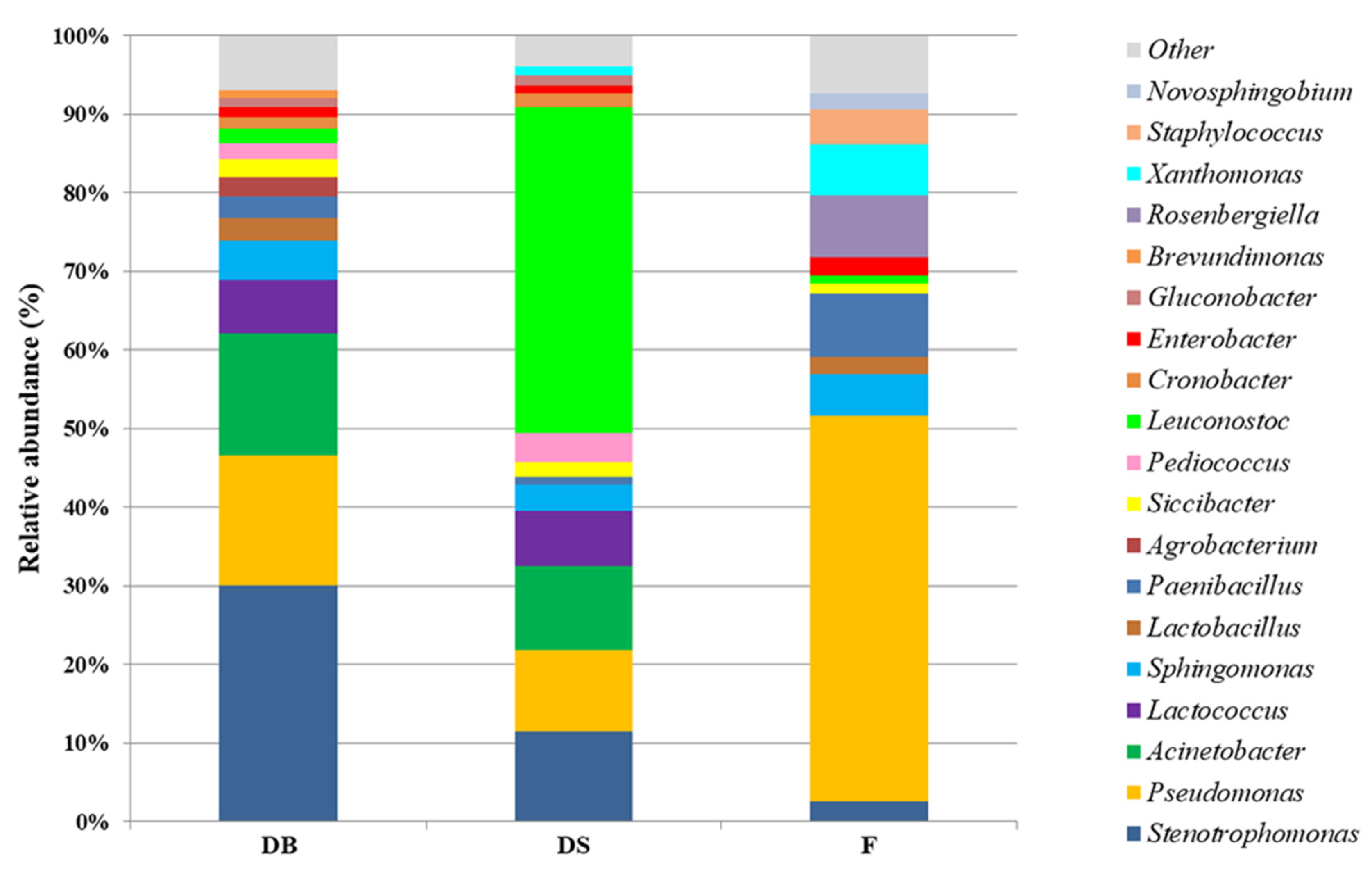
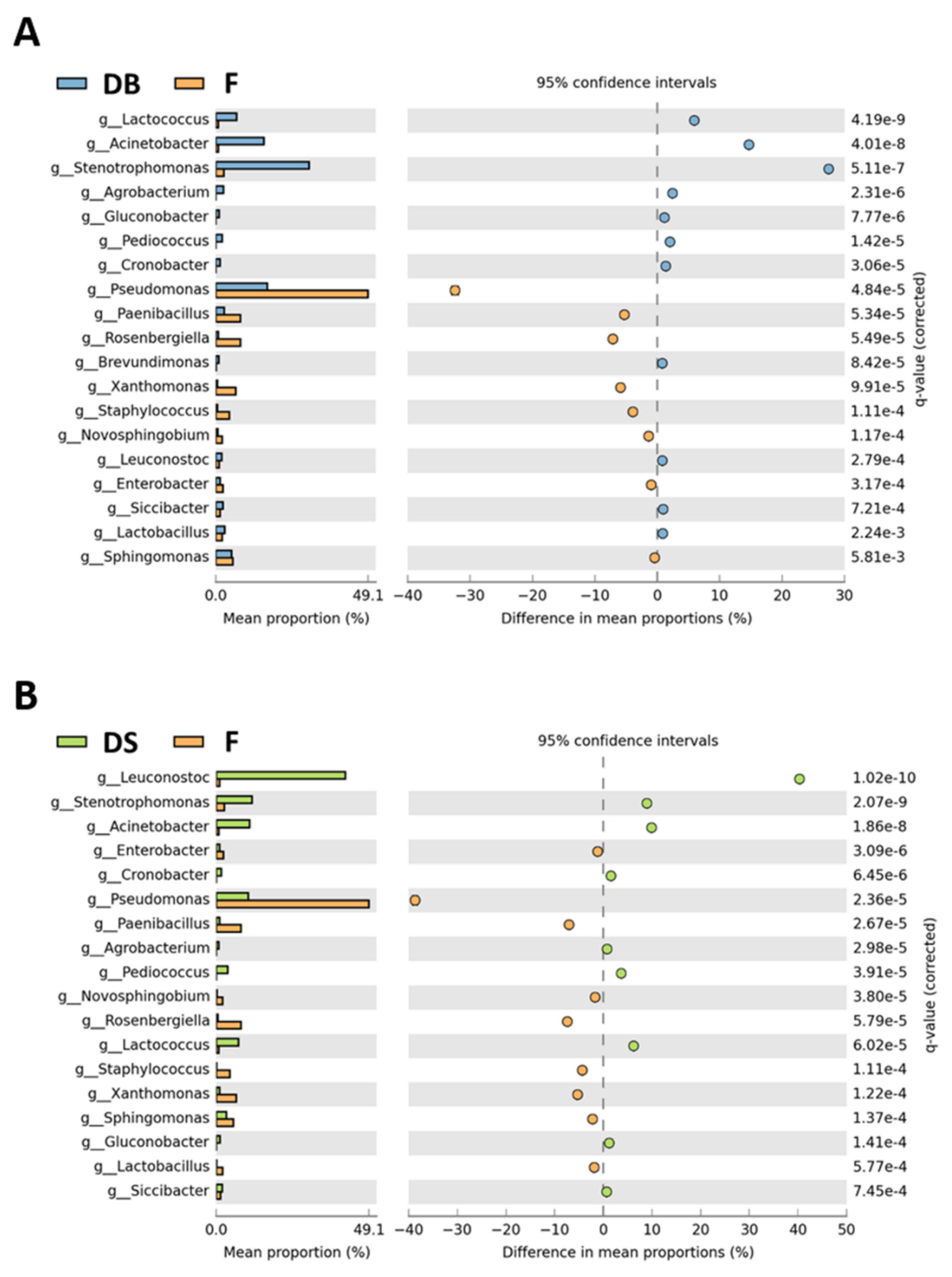
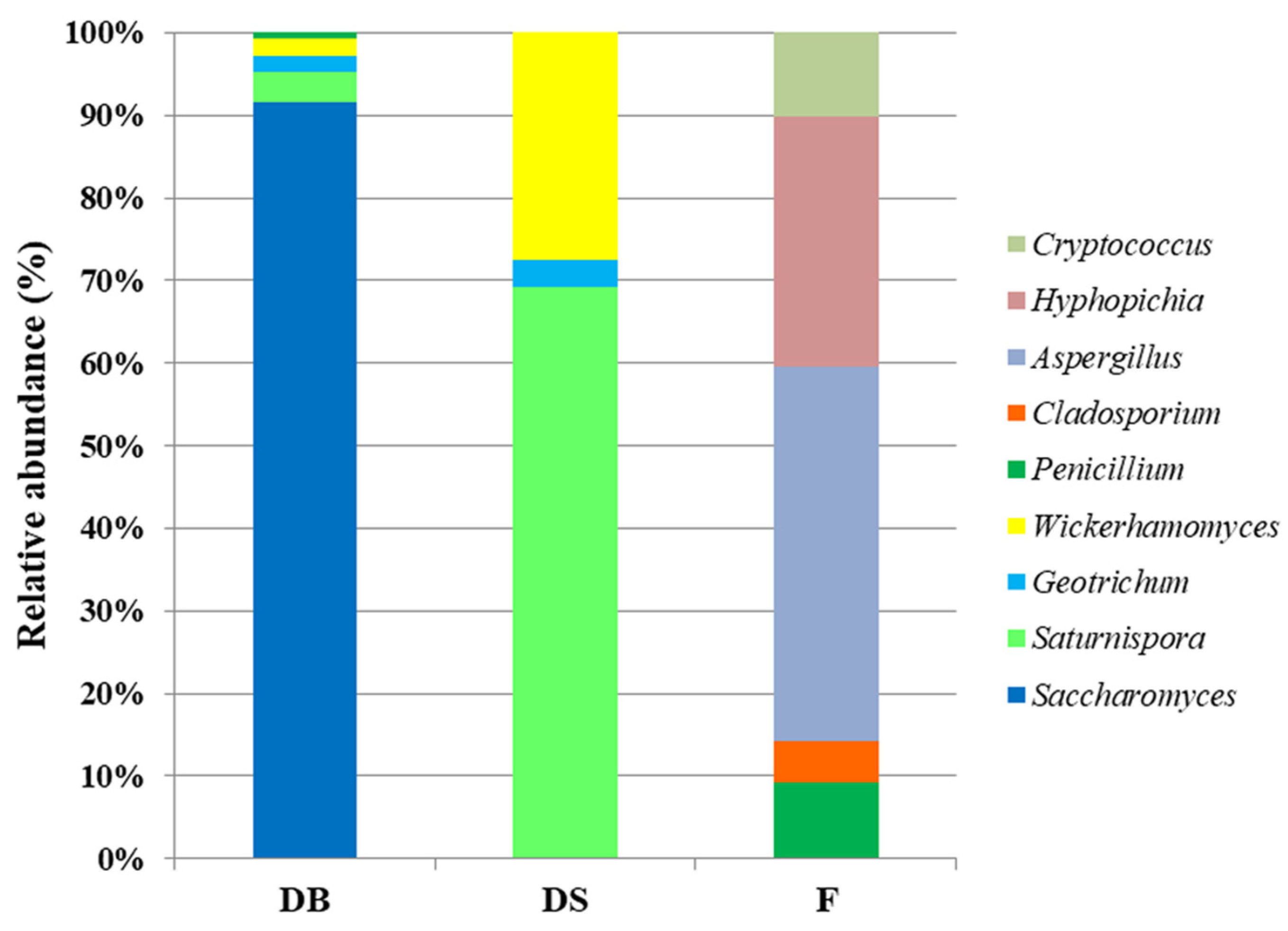
3.2. Bacterial and Fungal Microbiota in Flour and Doughs
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catzeddu, P. Sourdough Breads. In Flour and Breads and Their Fortification in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Patel, V.B., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: London, UK; Burlington, VT, USA; San Diego, CA, USA, 2011; pp. 37–46. [Google Scholar] [CrossRef]
- Liu, T.; Li, Y.; Sadiq, F.A.; Yang, H.; Gu, J.; Yuan, L.; Lee, Y.K.; He, G. Predominant yeasts in Chinese traditional sourdough and their influence on aroma formation in Chinese steamed bread. Food Chem. 2018, 242, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Suo, B.; Nie, W.; Wang, Y.; Ma, J.; Xing, X.; Huang, Z.; Xu, C.; Li, Z.; Ai, Z. Microbial diversity of fermented dough and volatile compounds in steamed bread prepared with traditional Chinese starters. LWT Food Sci. Technol. 2020, 126, 109350. [Google Scholar] [CrossRef]
- Decreto Ministeriale 7 Febbraio 2019. Aggiornamento Dell’elenco Nazionale Dei Prodotti Agroalimentari Tradizionali Ai Sensi Dell’articolo 12, Comma 1, Della Legge 12 Dicembre 2016, N. 238. Gazzetta Ufficiale N. 60 Del 12 Marzo 2019–Supplemento Ordinario N. 9. Available online: http://www.gazzettaufficiale.it/eli/gu/2019/03/12/60/so/9/sg/pdf (accessed on 3 May 2021).
- Pasqualone, A.; Delcuratolo, D.; Gomes, T. Focaccia Italian flat fatty bread. In Flour and Breads and Their Fortification in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Patel, V.B., Eds.; Academic Press: Cambridge, MA, USA; Elsevier: London, UK; Burlington, VT, USA; San Diego, CA, USA, 2011; pp. 47–58. [Google Scholar] [CrossRef]
- Calorie in Focaccia Pugliese e Valori Nutrizionali. Available online: https://www.fatsecret.it/calorie-nutrizione/generico/focaccia-pugliese (accessed on 3 May 2021).
- Carnevali, P.; Ciati, R.; Leporati, A.; Paese, M. Liquid sourdough fermentation: Industrial application perspectives. Food Microbiol. 2007, 24, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Song, K.; Li, H.; Ma, R.; Cui, M. Effect of mixed Saccharomyces cerevisiae Y10 and Torulaspora delbrueckii Y22 on dough fermentation for steamed bread making. Int. J. Food Microbiol. 2019, 303, 58–64. [Google Scholar] [CrossRef]
- Lattanzi, A.; Minervini, F.; Di Cagno, R.; Diviccaro, A.; Antonielli, L.; Cardinali, G.; Cappelle, S.; De Angelis, M.; Gobbetti, M. The lactic acid bacteria and yeast microbiota of eighteen sourdoughs used for the manufacture of traditional Italian sweet leavened baked goods. Int. J. Food Microbiol. 2013, 163, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Reale, A.; Di Renzo, T.; Boscaino, F.; Nazzaro, F.; Fratianni, F.; Aponte, M. Lactic Acid Bacteria Biota and Aroma Profile of Italian Traditional Sourdoughs from the Irpinian Area in Italy. Front. Microbiol. 2019, 10, 1621. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Rizzello, C.G.; Di Cagno, R.; De Angelis, M. How the sourdough may affect the functional features of leavened baked goods. Food Microbiol. 2014, 37, 30–40. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Di Cagno, R.; Calasso, M.; Archetti, G.; Rizzello, C.G. Novel insights on the functional/nutritional features of the sourdough fermentation. Int. J. Food Microbiol. 2019, 302, 103–113. [Google Scholar] [CrossRef]
- Siepmann, F.B.; Ripari, V.; Waszczynskyj, N.; Spier, M.R. Overview of sourdough technology: From production to marketing. Food Bioprocess Technol. 2018, 11, 242–270. [Google Scholar] [CrossRef]
- Belz, M.C.E.; Axel, C.; Arendt, E.K.; Lynch, K.M.; Brosnan, B.; Sheehan, E.M.; Coffey, A.; Zannini, E. Improvement of taste and shelf life of yeasted low-salt bread containing functional sourdoughs using Lactobacillus amylovorus DSM 19280 and Weissella cibaria MG1. Int. J. Food Microbiol. 2019, 302, 69–79. [Google Scholar] [CrossRef]
- Mantzourani, I.; Plessas, S.; Odatzidou, M.; Alexopoulos, A.; Galanis, A.; Bezirtzoglou, E.; Bekatorou, A. Effect of a novel Lactobacillus paracasei starter on sourdough bread quality. Food Chem. 2019, 271, 259–265. [Google Scholar] [CrossRef]
- Minervini, F.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Ecological parameters influencing microbial diversity and stability of traditional sourdough. Int. J. Food Microbiol. 2014, 171, 136–146. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Van Kerrebroeck, S.; Harth, H.; Huys, G.; Daniel, H.M.; Weckx, S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014, 37, 11–29. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; Bavaro, A.R.; Di Biase, M.; Lonigro, S.L.; Logrieco, A.F.; Lavermicocca, P. Effect of Amaranth and Quinoa Flours on Exopolysaccharide Production and Protein Profile of Liquid Sourdough Fermented by Weissella cibaria and Lactobacillus plantarum. Front. Microbiol. 2020, 11, 967. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Georgalaki, M.; Alexandraki, V.; Kazou, M.; Anastasiou, R.; Tsakalidou, E. Sourdough Bread. In Innovations in Traditional Foods; Galanakis, C.M., Ed.; Woodhead Publishing: Cambridge, UK; Elsevier: Amsterdam, The Netherlands, 2019; pp. 127–158. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, C.; Zhao, X.; Lv, C.; He, Q.; Lei, S.; Guo, Y.; Zhi, F. Anti-Saccharomyces cerevisiae Antibodies Associate with Phenotypes and Higher Risk for Surgery in Crohn’s Disease: A Meta-Analysis. Dig. Dis. Sci. 2012, 57, 2944–2954. [Google Scholar] [CrossRef]
- Caselli, M.; Lo Cascio, N.; Rabitti, S.; Eusebi, L.H.; Zeni, E.; Soavi, C.; Cassol, F.; Zuliani, G.; Zagari, R.M. Pattern of food intolerance in patients with gastro-esophageal reflux symptoms. Minerva Med. 2017, 108, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Cannistrà, C.; Finocchi, V.; Trivisonno, A.; Tambasco, D. New perspectives in the treatment of hidradenitis suppurativa: Surgery and brewer’s yeast–exclusion diet. Surgery 2013, 154, 1126–1130. [Google Scholar] [CrossRef]
- Cordell, B.; Kanodia, A. Auto-Brewery as an Emerging Syndrome: Three Representative Case Studies. J. Clin. Med. Case Rep. 2015, 2, 5. [Google Scholar]
- De Bellis, P.; Rizzello, C.G.; Sisto, A.; Valerio, F.; Lonigro, S.L.; Conte, A.; Lorusso, V.; Lavermicocca, P. Use of a selected Leuconostoc citreum strain as a starter for making a “yeast-free” bread. Foods 2019, 8, 70. [Google Scholar] [CrossRef]
- Musatti, A.; Cappa, C.; Mapelli, C.; Alamprese, C.; Rollini, M. Zymomonas mobilis in Bread Dough: Characterization of Dough Leavening Performance in Presence of Sucrose. Foods 2020, 9, 89. [Google Scholar] [CrossRef]
- Bottani, M.; Brasca, M.; Ferraretto, A.; Cardone, G.; Casiraghi, M.C.; Lombardi, G.; De Noni, I.; Cattaneo, S.; Silvetti, T. Chemical and nutritional properties of white bread leavened by lactic acid bacteria. J. Funct. Foods 2018, 45, 330–338. [Google Scholar] [CrossRef]
- De Bellis, P.; Montemurro, M.; D’Imperio, M.; Rizzello, C.G.; Sisto, A.; Lavermicocca, P. Production of a yeast-free focaccia with reduced salt content using a selected Leuconostoc citreum strain and seawater. LWT Food Sci. Technol. 2020, 134, 109918. [Google Scholar] [CrossRef]
- De Angelis, M.; Minervini, F.; Siragusa, S.; Rizzello, C.G.; Gobbetti, M. Wholemeal wheat flours drive the microbiome and functional features of wheat sourdoughs. Int. J. Food Microbiol. 2019, 302, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Pontonio, E.; De Filippis, F.; Minervini, F.; La Storia, A.; Gobbetti, M.; Di Cagno, R. Microbial ecology dynamics during rye and wheat sourdough preparation. Appl. Environ. Microbiol. 2013, 79, 7827–7836. [Google Scholar] [CrossRef]
- Coda, R.; Kianjam, M.; Pontonio, E.; Verni, M.; Di Cagno, R.; Katina, K.; Rizzello, C.G.; Gobbetti, M. Sourdough-type propagation of faba bean flour: Dynamics of microbial consortia and biochemical implications. Int. J. Food Microbiol. 2017, 248, 10–21. [Google Scholar] [CrossRef]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Celano, G.; Gobbetti, M. House microbiotas as sources of lactic acid bacteria and yeasts in traditional Italian sourdoughs. Food Microbiol. 2015, 52, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Amaretti, A.; Rossi, M.; Fall, P.A.; Tabanelli, G.; Gardini, F.; Montanari, C. Evolution of microbial community and chemical properties of a sourdough during the production of Colomba, an Italian sweet leavened baked product. LWT Food Sci. Technol. 2017, 86, 31–39. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef]
- De Filippis, F.; Laiola, M.; Blaiotta, G.; Ercolini, D. Different amplicon targets for sequencing-based studies of fungal diversity. Appl. Environ. Microbiol. 2017, 83, e00905-17. [Google Scholar] [CrossRef]
- Weckx, S.; Van Kerrebroeck, S.; De Vuyst, L. Omics approaches to understand sourdough fermentation processes. Int. J. Food Microbiol. 2019, 302, 90–102. [Google Scholar] [CrossRef]
- Valerio, F.; Favilla, M.; De Bellis, P.; Sisto, A.; De Candia, S.; Lavermicocca, P. Antifungal activity of strains of lactic acid bacteria isolated from a semolina ecosystem against Penicillium roqueforti, Aspergillus niger and Endomyces fibuliger contaminating bakery products. Syst. Appl. Microbiol. 2009, 32, 438–448. [Google Scholar] [CrossRef]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Scientific Opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA (2017–2019). EFSA J. 2020, 18, e05966. [Google Scholar] [CrossRef] [PubMed]
- Corsetti, A.; Lavermicocca, P.; Morea, M.; Baruzzi, F.; Tosti, N.; Gobbetti, M. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 2001, 64, 95–104. [Google Scholar] [CrossRef]
- De Bellis, P.; Valerio, F.; Sisto, A.; Lonigro, S.L.; Lavermicocca, P. Probiotic table olives: Microbial populations adhering on olive surface in fermentation sets inoculated with the probiotic strain Lactobacillus paracasei IMPC2.1 in an industrial plant. Int. J. Food Microbiol. 2010, 140, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Torriani, S.; Felis, G.E.; Dellaglio, F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 2001, 67, 3450–3454. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Meylan, V.; Klaenhammer, T.R.; Zink, R. Analysis, characterization, and loci of the tuf genes in Lactobacillus and Bifidobacterium species and their direct application for species identification. Appl. Environ. Microbiol. 2003, 69, 6908–6922. [Google Scholar] [CrossRef]
- Gadanho, M.; Almeida, J.M.G.C.F.; Sampaio, J.P. Assessment of yeast diversity in a marine environment in the south of Portugal by microsatellite-primed PCR. Ant. Van Leeuwenhoek 2003, 84, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Ant. Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Herbig, A.; Maixner, F.; Bos, K.I.; Zink, A.; Krause, J.; Huson, D.H. MALT: Fast alignment and analysis of metagenomic DNA sequence data applied to the Tyrolean Iceman. BioRxiv 2016. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition-Interactive Exploration and Analysis of Large-Scale Microbiome Sequencing Data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucl. Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Mengoni, A.; Bazzicalupo, M. The statistical treatment of data and the analysis of MOlecular VAriance (AMOVA) in molecular microbial ecology. Ann. Microbiol. 2002, 52, 95–101. [Google Scholar]
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Di Cagno, R.; Pontonio, E.; Buchin, S.; De Angelis, M.; Lattanzi, A.; Valerio, F.; Gobbetti, M.; Calasso, M. Diversity of the lactic acid bacterium and yeast microbiota in the switch from firm- to liquid-sourdough fermentation. Appl. Environ. Microbiol. 2014, 10, 3161–3172. [Google Scholar] [CrossRef]
- Valerio, F.; De Bellis, P.; Di Biase, M.; Lonigro, S.L.; Giussani, B.; Visconti, A.; Lavermicocca, P.; Sisto, A. Diversity of spore-forming bacteria and identification of Bacillus amyloliquefaciens as a species frequently associated with the ropy spoilage of bread. Int. J. Food Microbiol. 2012, 156, 278–285. [Google Scholar] [CrossRef]
- Montemurro, M.; Pontonio, E.; Gobbetti, M.; Rizzello, C.G. Investigation of the nutritional, functional and technological effects of the sourdough fermentation of sprouted flours. Int. J. Food Microbiol. 2019, 302, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Minervini, F.; Pontonio, E.; Di Cagno, R.; De Angelis, M. Drivers for the establishment and composition of the sourdough lactic acid bacteria biota. Int. J. Food Microbiol. 2016, 239, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, C.G.; Cavoski, I.; Turk, J.; Ercolini, D.; Nionelli, L.; Pontonio, E.; De Angelis, M.; De Filippis, F.; Gobbetti, M.; Di Cagno, R. Organic cultivation of Triticum turgidum subsp. durum is reflected in the flour-sourdough fermentation-bread axis. Appl. Environ. Microbiol. 2015, 81, 3192–3204. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- De Vuyst, L.; Harth, H.; Kerrebroeck, S.V.; Leroy, F. Yeast diversity of sourdoughs and associated metabolic properties and functionalities. Int. J. Food Microbiol. 2016, 239, 26–34. [Google Scholar] [CrossRef]
- Vrancken, G.; De Vuyst, L.; Van der Meulen, R.; Huys, G.; Vandamme, P.; Daniel, H.M. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 2010, 10, 471–481. [Google Scholar] [CrossRef]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Influence of artisan bakery- or laboratory-propagated sourdoughs on the diversity of lactic acid bacterium and yeast microbiotas. Appl. Environ. Microbiol. 2012, 78, 5328–5340. [Google Scholar] [CrossRef] [PubMed]




| Sample | pH | ΔpH | TTA | LAB (log cfu/g) | Other LAB Species Isolates | Spore-Forming Bacteria (log cfu/g) | Yeasts (log cfu/g) | Molds (log cfu/g) | Yeast Species |
|---|---|---|---|---|---|---|---|---|---|
| F | - | - | - | 0.46 ± 0.28 | Ll. coryniformis | 0.8 ± 0.72 | 0.7 ± 1.15 | 2.59 ± 0.12 | Cr. victoriae |
| S | 3.53 ± 0.07 | 2.42 ± 0.14 | 4.15 ± 0.21 | 9.47 ± 0.13 | nd | nd | nd | 1.57 ± 0.04 | nd |
| DS | 4.23 ± 0.04 b | 1.35 ± 0.09 a | 8.17 ± 0.55 a | 9.09 ± 0.01 a | nd | 0.54 ± 0.08 | nd | 2.70 ± 0.36 | nd |
| DB | 5.38 ± 0.02 a | 0.25 ± 0.01 b | 3.55 ± 0.21 b | 4.48 ± 0.08 b | Fl. rossiae L. mesenteroides Lp. paraplantarum Lc. paracasei | nd | 8.25 ± 0.01 | nd | S. cerevisiae |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferrara, M.; Sisto, A.; Mulè, G.; Lavermicocca, P.; De Bellis, P. Metagenetic Analysis for Microbial Characterization of Focaccia Doughs Obtained by Using Two Different Starters: Traditional Baker’s Yeast and a Selected Leuconostoc citreum Strain. Foods 2021, 10, 1189. https://doi.org/10.3390/foods10061189
Ferrara M, Sisto A, Mulè G, Lavermicocca P, De Bellis P. Metagenetic Analysis for Microbial Characterization of Focaccia Doughs Obtained by Using Two Different Starters: Traditional Baker’s Yeast and a Selected Leuconostoc citreum Strain. Foods. 2021; 10(6):1189. https://doi.org/10.3390/foods10061189
Chicago/Turabian StyleFerrara, Massimo, Angelo Sisto, Giuseppina Mulè, Paola Lavermicocca, and Palmira De Bellis. 2021. "Metagenetic Analysis for Microbial Characterization of Focaccia Doughs Obtained by Using Two Different Starters: Traditional Baker’s Yeast and a Selected Leuconostoc citreum Strain" Foods 10, no. 6: 1189. https://doi.org/10.3390/foods10061189
APA StyleFerrara, M., Sisto, A., Mulè, G., Lavermicocca, P., & De Bellis, P. (2021). Metagenetic Analysis for Microbial Characterization of Focaccia Doughs Obtained by Using Two Different Starters: Traditional Baker’s Yeast and a Selected Leuconostoc citreum Strain. Foods, 10(6), 1189. https://doi.org/10.3390/foods10061189







