Fraud Detection in Batches of Sweet Almonds by Portable Near-Infrared Spectral Devices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. NIRS Instrumentation and Spectrum Acquisition
2.3. Study of the Population and Construction of the Training and Validation Sets
2.4. Classification Models of Almonds by Bitterness: Influence of the Composition of the Training Sets on the Detection of an Adulterated Product
3. Results and Discussion
3.1. Characteristics of the NIR Almond Spectra and Study of the Population
3.2. Development of Classification Models to Detect Adulterated Batches of Sweet Almonds
3.2.1. Strategy I
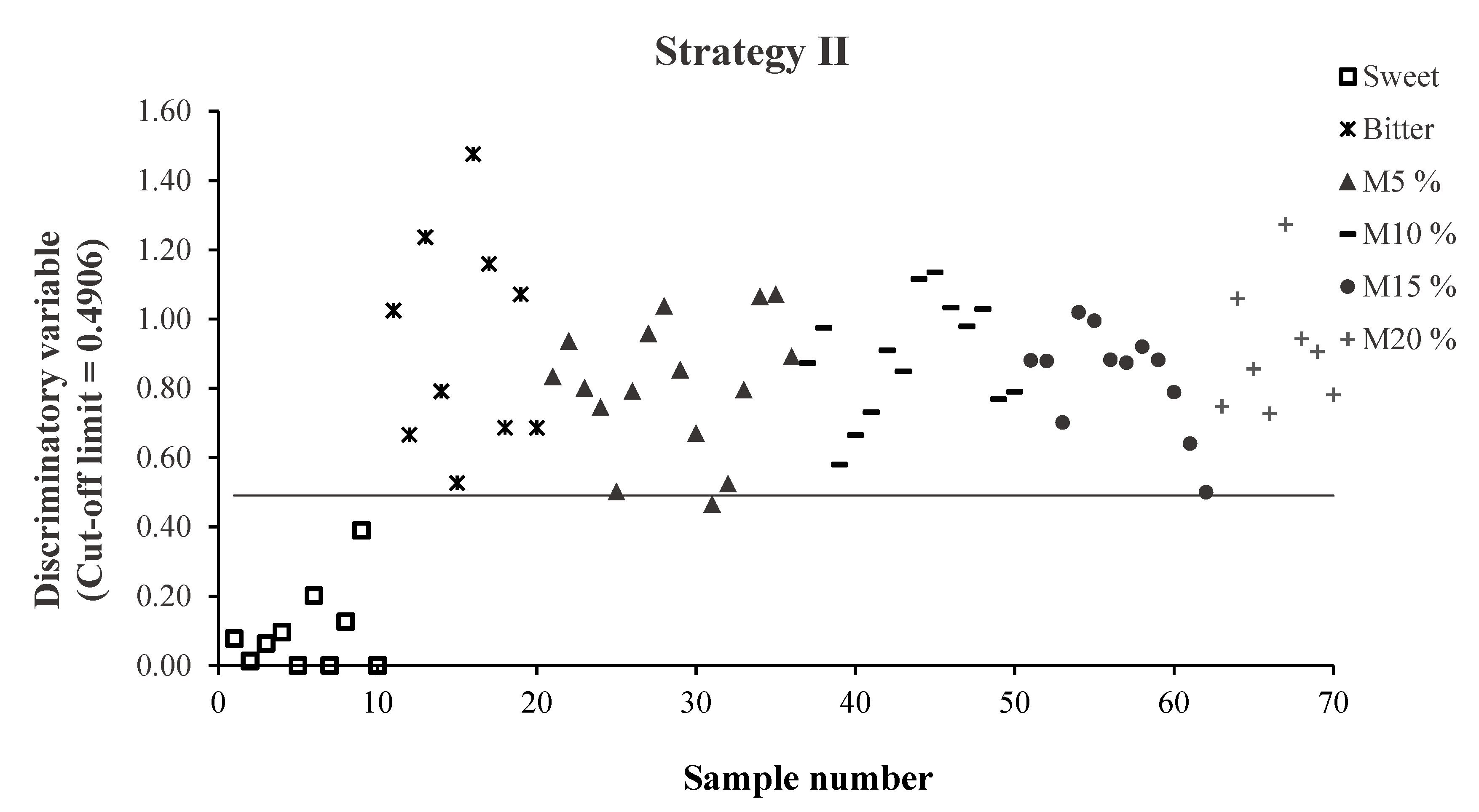
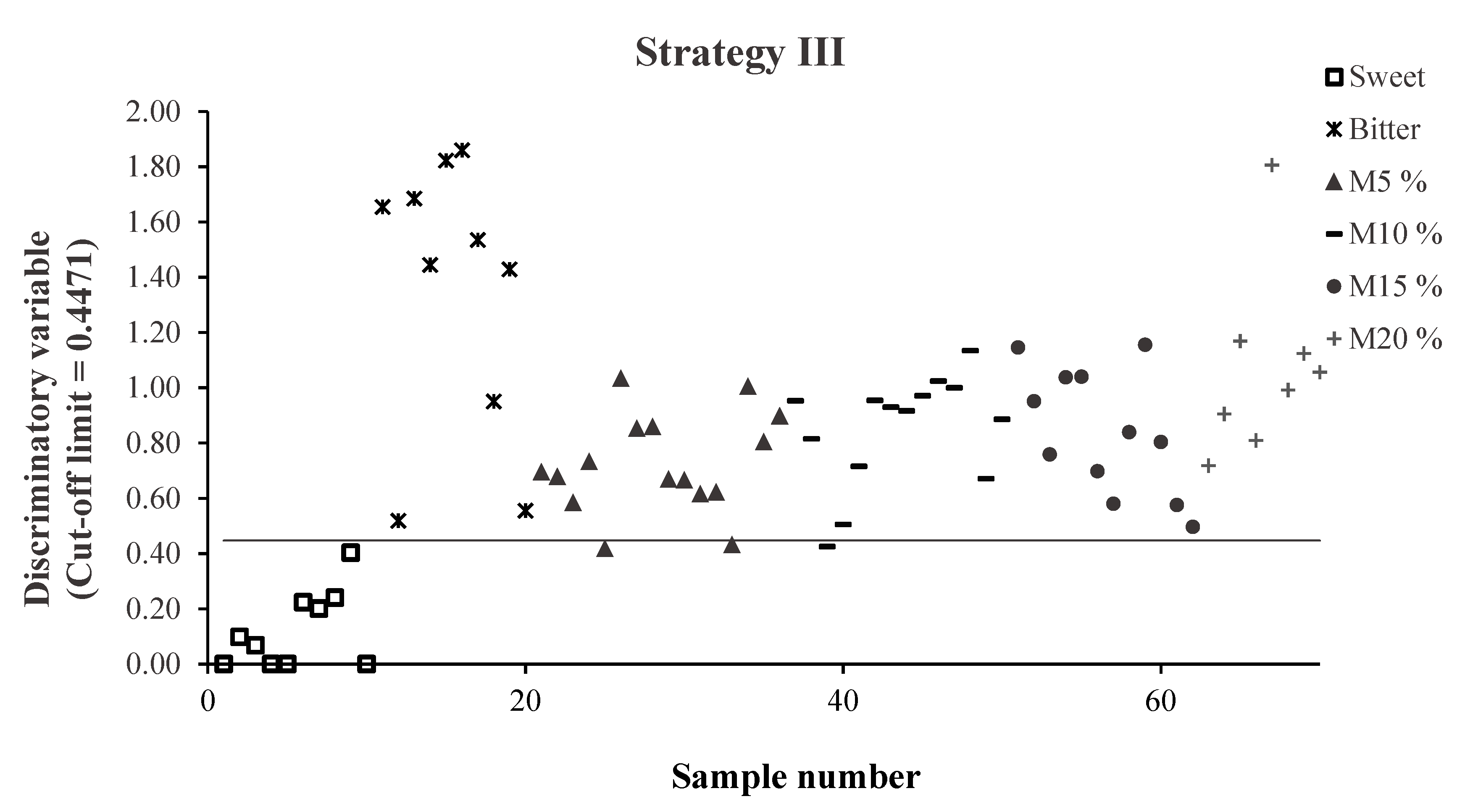
3.2.2. Strategies II and III
3.3. External Validation: Strategies II and III
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Morin, J.F.; Lees, M. Food Integrity Handbook: A Guide to Food Authenticity Issues and Analytical Solutions; Eurofins Analytics France: Nantes, France, 2018. [Google Scholar]
- Ali Meerza, S.Y.; Giannakas, K.; Yiannaka, A. Markets and welfare effects of food fraud. Aust. J. Agric. Res. Econ. 2019, 63, 759–789. [Google Scholar] [CrossRef]
- Ali Meerza, S.Y.; Gustafson, C.R. Consumers’ response to food fraud: Evidence from experimental auctions. J. Agric. Resour. Econ. 2020, 45, 219–231. [Google Scholar] [CrossRef]
- Pérez-Marín, D.; Calero, L.; Fearn, T.; Torres, I.; Garrido-Varo, A.; Sánchez, M.T. A system using in situ NIRS sensors for the detection of product failing to meet quality standards and the prediction of optimal postharvest shelf-life in the case of oranges kept in cold storage. Postharvest Biol. Tec. 2019, 147, 48–53. [Google Scholar] [CrossRef]
- Pérez-Marín, D.; Garrido, A. NIR sensors for the in-situ assessment of Iberian ham. In Comprehensive Foodomics; Cifuentes, A., Ed.; Elsevier: Oxford, UK, 2020; pp. 340–345. [Google Scholar]
- Pasquini, C. Near infrared spectroscopy: A mature analytical technique with new perspectives—A review. Anal. Chim. Acta 2018, 1026, 8–36. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Siesler, H.W. Hand-held near-infrared spectrometers: State-of-the-art instrumentation and practical applications. NIR News 2018, 29, 8–12. [Google Scholar] [CrossRef]
- Walsh, K.B.; Blasco, J.; Zude-Sasse, M.; Sun, X. Visible-NIR ‘point’ spectroscopy in postharvest fruit and vegetable assessment: The science behind three decades of commercial use. Postharvest Biol. Technol. 2020, 168, 111246. [Google Scholar] [CrossRef]
- Vega-Castellote, M.; Pérez-Marín, D.; Torres, I.; Sánchez, M.T. Non-destructive determination of fatty acid composition of in-shell and shelled almonds using handheld NIRS sensors. Postharvest Biol. Technol. 2021, 174, 111459. [Google Scholar] [CrossRef]
- Sánchez-Pérez, R.; Jørgensen, K.; Olsen, C.E.; Dicenta, F.; Møller, B.L. Bitterness in almonds. Plant Physiol. 2008, 146, 1040–1052. [Google Scholar] [CrossRef] [Green Version]
- Cortés, V.; Talens, P.; Barat, J.M.; Lerma-García, M.J. Potential of NIR spectroscopy to predict amygdalin content established by HPLC in intact almonds and classification based on almond bitterness. Food Control 2018, 91, 68–75. [Google Scholar] [CrossRef]
- Yada, S.; Huang, G.; Lapsley, K. A review of composition studies of cultivated almonds: Macronutrients and micronutrients. J. Food Compos. Anal. 2011, 24, 469–480. [Google Scholar] [CrossRef]
- Arrázola-Paternina, G.; Dicenta-López-Higuera, F.; Grané-Teruel, N. Evolution of the amygdalin and prunasin content during the development of almond (Prunus dulcis Miller). Rev. Fac. Agron. 2015, 32, 63–81. [Google Scholar]
- Borrás, E.; Amigo, J.M.; van den Berg, F.; Boqué, R.; Busto, O. Fast and robust discrimination of almond (Prunus amygdalus) with respect to their bitterness by using near infrared and partial least squares-discriminant analysis. Food Chem. 2014, 153, 15–19. [Google Scholar] [CrossRef]
- Vega-Castellote, M.; Pérez-Marín, D.; Torres, I.; Moreno-Rojas, J.M.; Sánchez, M.T. Exploring the potential of NIRS technology for the in situ prediction of amygdalin content and classification by bitterness of in-shell and shelled intact almonds. J. Food Eng. 2021, 294, 110406. [Google Scholar] [CrossRef]
- Vega-Castellote, M.; Sánchez, M.T.; Torres, I.; Pérez-Marín, D. An innovative non-targeted control system based on NIR spectral information for detecting non-compliant batches of sweet almonds. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 250, 119407. [Google Scholar] [CrossRef] [PubMed]
- Vichi, S.; Mayer, M.N.; León-Cárdenas, M.G.; Quintanilla-Casas, B.; Tres, A.; Guardiola, F.; Batlle, I.; Romero, A. Chemical markers to distinguish the homo- and heterozygous bitter genotype in sweet almond kernels. Foods 2020, 9, 747. [Google Scholar] [CrossRef] [PubMed]
- ISI. The Complete Software Solution Using a Single Screen for Routine Analysis, Robust Calibrations, and Networking; Manual, FOSS NIRSystems/TECATOR; Infrasoft International: Silver Spring, MD, USA, 2000. [Google Scholar]
- Shenk, J.S.; Westerhaus, M.O. Analysis of Agriculture and Food Products by Near Infrared Reflectance Spectroscopy; Monograph; NIRSystems, Inc.: Silver Spring, MD, USA, 1995. [Google Scholar]
- Barnes, R.J.; Dhanoa, M.S.; Lister, S.J. Standard Normal Variate Transformation and De-trending of near infrared diffuse reflectance spectra. Appl. Spectrosc. 1989, 43, 772–777. [Google Scholar] [CrossRef]
- Shenk, J.S.; Westerhaus, M.O. Routine Operation, Calibration, Development and Network System Management Manual; NIRSystems, Inc.: Silver Spring, MD, USA, 1995. [Google Scholar]
- Shenk, J.S.; Westerhaus, M.O. Population structuring of near infrared spectra and modified partial least squares regression. Crop Sci. 1991, 31, 1548–1555. [Google Scholar] [CrossRef]
- Naes, T.; Isaksson, T.; Fearn, T.; Davies, A. A User-Friendly Guide to Multivariate Calibration and Classification; NIR Publications: Chichester, UK, 2002. [Google Scholar]
- Shenk, J.S.; Workman, J.; Westerhaus, M. Application of NIR spectroscopy to agricultural products. In Handbook of Near Infrared Analysis, 3rd ed.; Burns, D.A., Ciurczac, E.W., Eds.; Marcel Dekker: New York, NY, USA, 2008; pp. 347–386. [Google Scholar]
- Workman, J.; Weyer, L. Practical Guide to Interpretive Near-Infrared Spectroscopy; CRC/Taylor & Francis: Boca Raton, FL, USA, 2008. [Google Scholar]
- Fernández-Cuesta, A.; Fernández-Martínez, J.M.; Socias i Company, R.; Velasco, L. Near-infrared spectroscopy for analysis of oil content and fatty acid profile in almond flour. Eur. J. Lipid Sci. Technol. 2013, 115, 211–216. [Google Scholar] [CrossRef]
- Downey, G. Discriminant PLS–questions and answers from a listserver. NIR News 2000, 11, 9–12. [Google Scholar] [CrossRef]
- Pérez-Marín, D.; Garrido-Varo, A.; Guerrero, J.E. Implementation of LOCAL algorithm with near-infrared spectroscopy for compliance assurance in compound feedingstuffs. Appl. Spectrosc. 2005, 59, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, H.A.; Paoletti, C. Food and feed safety assessment: The importance of proper sampling. J. AOAC Int. 2015, 98, 252–258. [Google Scholar] [CrossRef] [PubMed]


| Genotype | Cultivar | Range | Mean | Standard Deviation | Coefficient of Variation (%) |
|---|---|---|---|---|---|
| SkSk | Antoñeta | 194.80–349.40 | 284.72 | 44.12 | 15.50 |
| Avellanera | 0.00–59.20 | 16.74 | 21.72 | 129.75 | |
| Belona | 11.64–150.84 | 62.45 | 36.61 | 58.62 | |
| Blanquilla | 25.30–229.60 | 100.96 | 75.20 | 74.48 | |
| Comuna | 37.60–230.90 | 115.00 | 70.81 | 61.57 | |
| Ferragnes | 0.00–18.80 | 10.94 | 6.86 | 62.71 | |
| Largueta | 0.00–71.90 | 40.62 | 25.13 | 61.87 | |
| Laureanne | 5.45–131.02 | 62.53 | 35.89 | 57.40 | |
| Marcona | 72.90–138.00 | 113.30 | 21.68 | 19.14 | |
| Ramillete | 0.00–56.60 | 29.28 | 23.99 | 81.93 | |
| Soleta | 77.05–165.95 | 112.37 | 25.53 | 22.72 | |
| Vairo | 26.88–125.32 | 62.59 | 27.20 | 43.46 | |
| Sksk | Garrigues | 82.20–137.90 | 104.40 | 24.10 | 23.08 |
| Guara | 0.00–551.92 | 224.06 | 148.62 | 66.33 | |
| sksk | - | 215.03–80,980.13 | 34,508.14 | 30,173.61 | 87.44 |
| Strategy I | Strategy II | Strategy III | ||||
|---|---|---|---|---|---|---|
| “Sweet” Almond Class | “Non-Sweet” Almond Class | “Sweet” Almond Class | “Non-Sweet” Almond Class | “Sweet” Almond Class | “Non-Sweet” Almond Class | |
| Training set | 100% sweet almonds (n = 125 samples) | 100% bitter almonds (n = 70 samples) | 100% sweet almonds (n = 125 samples) | 100% bitter almonds (n = 70 samples) + M5% (n = 25 samples) + M10% (n = 25 samples) + M15% (n = 25 samples) + M20% (n = 13 samples) | 100% sweet almonds (n = 125 samples) | M5% (n = 25 samples) + M10% (n = 25 samples) + M15% (n = 25 samples) + M20% (n = 13 samples) |
| Validation set | 100% sweet almonds (n = 10 samples) | 100% bitter almonds (n = 10 samples) + M5% (n = 41 samples) + M10% (n = 39 samples) + M15% (n = 37 samples) + M20% (n = 21 samples) | 100% sweet almonds (n = 10 samples) | 100% bitter almonds (n = 10 samples) + M5% (n = 16 samples) + M10% (n = 14 samples) + M15% (n = 12 samples) + M20% (n = 8 samples) | 100% sweet almonds (n = 10 samples) | 100% bitter almonds (n = 10 samples) + M5% (n = 16 samples) + M10% (n = 14 samples) + M15% (n = 12 samples) + M20% (n = 8 samples) |
| Instrument | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aurora | MicroNIRTM Pro 1700 | |||||||||
| Predicted Class | Predicted Class | |||||||||
| Actual Class | Sweet | Non-Sweet | Samples Correctly Classified | Actual Class | Sweet | Non-Sweet | Samples Correctly Classified | |||
| Cross-validation | Sweet | 125 | 0 | 100.00% | Sweet | 124 | 1 | 99.20% | ||
| Non-sweet | 0 | 70 | 100.00% | Non-sweet | 0 | 70 | 100.00% | |||
| Sensitivity = 1 | Specificity = 1 | NER = 100% | Sensitivity = 0.99 | Specificity = 1 | NER = 99.49% | |||||
| External validation | Predicted Class | Predicted Class | ||||||||
| Actual Class | Sweet | Non-Sweet | Samples Correctly Classified | Actual Class | Sweet | Non-Sweet | Samples Correctly Classified | |||
| Sweet | 10 | 0 | 100.00% | Sweet | 10 | 0 | 100.00% | |||
| Non-sweet | Bitter | 0 | 10 | 100.00% | Non-sweet | Bitter | 0 | 10 | 100.00% | |
| M5% | 31 | 10 | 24.39% | M5% | 35 | 6 | 14.63% | |||
| M10% | 24 | 15 | 38.46% | M10% | 30 | 9 | 23.08% | |||
| M15% | 24 | 13 | 35.14% | M15% | 28 | 9 | 24.32% | |||
| M20% | 7 | 14 | 66.67% | M20% | 18 | 3 | 14.29% | |||
| Sensitivity = 1 | Specificity = 0.42 | NER = 45.57% | Sensitivity = 1 | Specificity = 0.25 | NER = 29.75% | |||||
| Instrument | ||||||||
|---|---|---|---|---|---|---|---|---|
| Aurora | MicroNIRTM Pro 1700 | |||||||
| Predicted Class | Predicted Class | |||||||
| Actual Class | Sweet | Non-Sweet | Samples Correctly Classified | Actual Class | Sweet | Non-Sweet | Samples Correctly Classified | |
| Strategy II | Sweet | 121 | 4 | 96.80% | Sweet | 109 | 16 | 87.20% |
| Non-sweet | 2 | 156 | 98.73% | Non-sweet | 12 | 146 | 92.41% | |
| Sensitivity = 0.97 | Specificity = 0.99 | NER = 97.88% | Sensitivity = 0.87 | Specificity = 0.92 | NER = 90.11% | |||
| Strategy III | Sweet | 120 | 5 | 96.00% | Sweet | 109 | 16 | 87.20% |
| Non-sweet | 3 | 85 | 96.59% | Non-sweet | 9 | 79 | 89.77% | |
| Sensitivity = 0.96 | Specificity = 0.97 | NER = 96.24% | Sensitivity = 0.87 | Specificity = 0.88 | NER = 88.26% | |||
| Strategy II | Actual Category | Classified as | Correctly Classified | ||
|---|---|---|---|---|---|
| Sweet | Non-Sweet | ||||
| Sweet | 10 | 0 | 100.00% | ||
| Non-sweet | Bitter (M100%) | 0 | 10 | 100.00% | |
| M5% | 1 | 15 | 93.75% | ||
| M10% | 0 | 14 | 100.00% | ||
| M15% | 0 | 12 | 100.00% | ||
| M20% | 0 | 8 | 100.00% | ||
| Sensitivity = 1 | Specificity = 0.98 | NER = 98.57% | |||
| Strategy III | Actual Category | Classified as | Correctly Classified | ||
| Sweet | Non-Sweet | ||||
| Sweet | 10 | 0 | 100.00% | ||
| Non-sweet | Bitter (M100%) | 0 | 10 | 100.00% | |
| M5% | 2 | 14 | 87.50% | ||
| M10% | 1 | 13 | 92.86% | ||
| M15% | 0 | 12 | 100.00% | ||
| M20% | 0 | 8 | 100.00% | ||
| Sensitivity = 1 | Specificity = 0.95 | NER = 95.71% | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, I.; Sánchez, M.-T.; Vega-Castellote, M.; Pérez-Marín, D. Fraud Detection in Batches of Sweet Almonds by Portable Near-Infrared Spectral Devices. Foods 2021, 10, 1221. https://doi.org/10.3390/foods10061221
Torres I, Sánchez M-T, Vega-Castellote M, Pérez-Marín D. Fraud Detection in Batches of Sweet Almonds by Portable Near-Infrared Spectral Devices. Foods. 2021; 10(6):1221. https://doi.org/10.3390/foods10061221
Chicago/Turabian StyleTorres, Irina, María-Teresa Sánchez, Miguel Vega-Castellote, and Dolores Pérez-Marín. 2021. "Fraud Detection in Batches of Sweet Almonds by Portable Near-Infrared Spectral Devices" Foods 10, no. 6: 1221. https://doi.org/10.3390/foods10061221






