Combining Culture-Dependent and Culture-Independent Methods: New Methodology Insight on the Vibrio Community of Ruditapes philippinarum
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Experimental Design
2.2. Microbiological Analysis
2.3. DNA Extraction and Libraries’ Preparation for Metabarcoding
2.4. Shotgun Metagenomics Libraries Preparation
2.5. Bioinformatic and Statistical Analyses for Metabarcoding
2.6. Bioinformatic and Explorative Analyses for Shotgun Metagenomics
3. Results
3.1. 16S Metabarcoding-Based Microbial Communities of Homogenate and Culture-Derived Clam Samples
3.2. recA-pyrH Metabarcoding on Homogenate and Plated Clam Samples
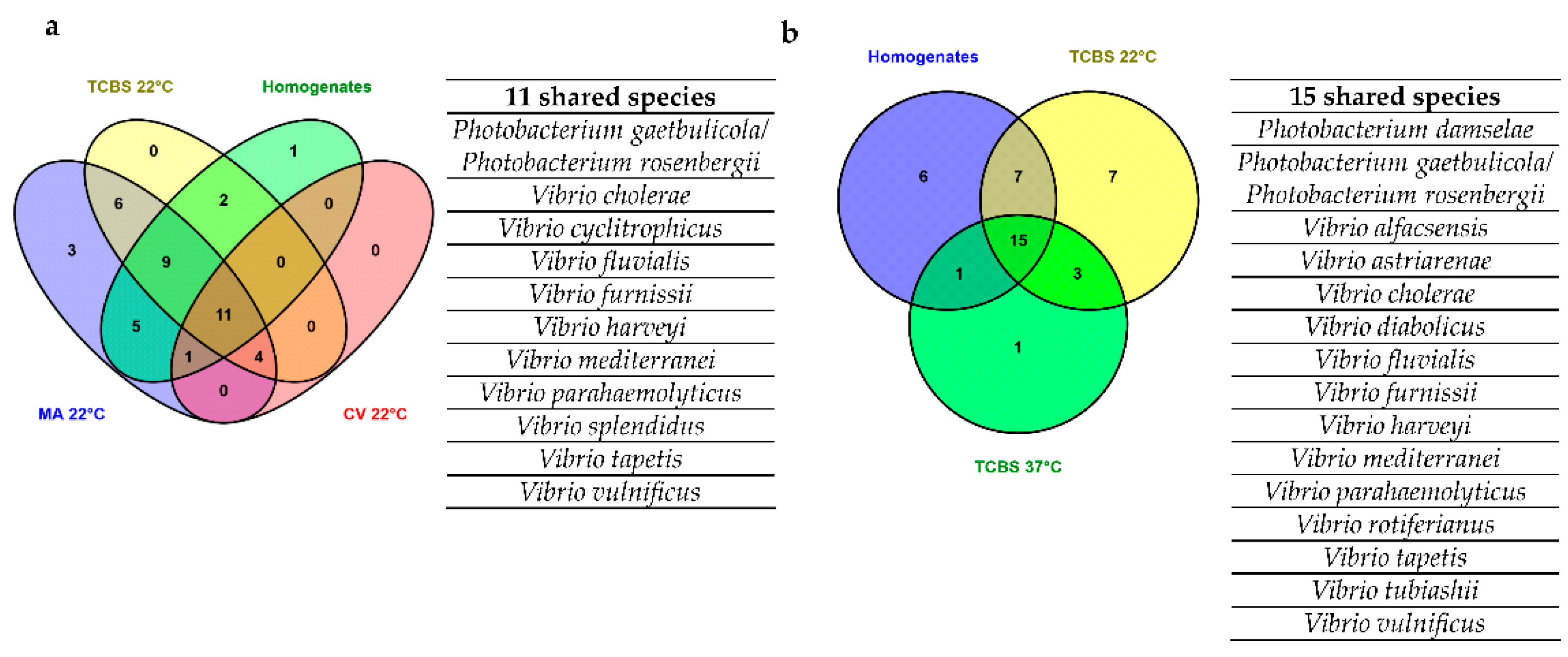
3.3. Shotgun Metagenomics of Plated Clam Samples and Comparison of Metagenomics and recA-pyrH Metabarcoding Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Romalde, J.L.; Diéguez, A.L.; Lasa, A.; Balboa, S. New Vibrio species associated to molluscan microbiota: A review. Front. Microbiol. 2014, 4, 1–11. [Google Scholar] [CrossRef]
- West, P.A. The human pathogenic vibrios—A public health update with environmental perspectives. Epidemiol. Infect. 1989, 103, 1–34. [Google Scholar] [CrossRef]
- Jiang, Y.; Chu, Y.; Xie, G.; Li, F.; Wang, L.; Huang, J.; Zhai, Y.; Yao, L. Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 2019, 290, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Passalacqua, P.L.; Zavatta, E.; Bignami, G.; Serraino, A.; Serratore, P.; Studiorum-, A.M.; Emilia, O. Occurrence of Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus in the clam Ruditapes philippinarum (Adams & Reeve, 1850) from Emilia Romagna and Sardinia, Italy. Ital. J. Food Saf. 2016, 5, 41–46. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Non-Cholera Vibrios: The Microbial Barometer of Climate Change. Trends Microbiol. 2017, 25, 76–84. [Google Scholar] [CrossRef]
- Bonnin-Jusserand, M.; Copin, S.; Le Bris, C.; Brauge, T.; Gay, M.; Brisabois, A.; Grard, T.; Midelet-Bourdin, G. Vibrio species involved in seafood-borne outbreaks (Vibrio cholerae, V. parahaemolyticus and V. vulnificus): Review of microbiological versus recent molecular detection methods in seafood products. Crit. Rev. Food Sci. Nutr. 2019, 59, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Colwell, R.R. Oceans and Health: Pathogens in the Marine Environment; Springer: New York, NY, USA, 2005; ISBN 9780387237084. [Google Scholar]
- Kriem, M.R.; Banni, B.; El Bouchtaoui, H.; Hamama, A.; El Marrakchi, A.; Chaouqy, N.; Robert-Pillot, A.; Quilici, M.L. Prevalence of Vibrio spp. in raw shrimps (Parapenaeus longirostris) and performance of a chromogenic medium for the isolation of Vibrio strains. Lett. Appl. Microbiol. 2015, 61, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Carraro, L.; Maifreni, M.; Bartolomeoli, I.; Martino, M.E.; Novelli, E.; Frigo, F.; Marino, M.; Cardazzo, B. Comparison of culture-dependent and -independent methods for bacterial community monitoring during Montasio cheese manufacturing. Res. Microbiol. 2011, 162, 231–239. [Google Scholar] [CrossRef]
- Nadkarni, M.A.; Martin, F.E.; Hunter, N.; Jacques, N.A. Methods for optimizing DNA extraction before quantifying oral bacterial numbers by real-time PCR. FEMS Microbiol. Lett. 2009, 296, 45–51. [Google Scholar] [CrossRef]
- Malara, D.; Mielke, C.; Oelgemöller, M.; Senge, M.O.; Heimann, K. Sustainable water treatment in aquaculture—Photolysis and photodynamic therapy for the inactivation of Vibrio species. Aquac. Res. 2017, 48, 2954–2962. [Google Scholar] [CrossRef]
- Liu, Y.; Zhong, Q.; Wang, J.; Lei, S. Enumeration of Vibrio parahaemolyticus in VBNC state by PMA-combined real-time quantitative PCR coupled with confirmation of respiratory activity. Food Control 2018, 91, 85–91. [Google Scholar] [CrossRef]
- Orruño, M.; Kaberdin, V.R.; Arana, I. Survival strategies of Escherichia coli and Vibrio spp.: Contribution of the viable but nonculturable phenotype to their stress-resistance and persistence in adverse environments. World J. Microbiol. Biotechnol. 2017, 33, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Azizah, R.N.; Kim, K. Comparative evaluation of three agar media-based methods for presumptive identi fi cation of seafood-originated Vibrio parahaemolyticus strains. Food Control 2020, 116, 107308. [Google Scholar] [CrossRef]
- Fenske, G.J.; Ghimire, S.; Antony, L.; Christopher-Hennings, J.; Scaria, J. Integration of culture-dependent and independent methods provides a more coherent picture of the pig gut microbiome. FEMS Microbiol. Ecol. 2020, 96, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kirchberger, P.C.; Orata, F.D.; Nasreen, T.; Kauffman, K.M.; Tarr, C.L.; Case, R.J.; Polz, M.F.; Boucher, Y.F. Culture-independent tracking of Vibrio cholerae lineages reveals complex spatiotemporal dynamics in a natural population. Environ. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jesser, K.J.; Rachel, T. Noble Vibrio Ecology in the Neuse River Estuary, North Carolina, Characterized by Next-Generation Amplicon Sequencing of the Gene Encoding Heat Shock Protein 60 (hsp60). Appl. Environ. Microbiol. 2018, 84, 1–21. [Google Scholar] [CrossRef] [PubMed]
- King, W.L.; Jenkins, C.; Go, J.; Siboni, N.; Seymour, J.R.; Labbate, M. Characterisation of the Pacific Oyster Microbiome During a Summer Mortality Event. Microb. Ecol. 2019, 77, 502–512. [Google Scholar] [CrossRef]
- Zampieri, A.; Carraro, L.; Cardazzo, B.; Milan, M.; Babbucci, M.; Smits, M.; Boffo, L.; Fasolato, L. Depuration processes affect the Vibrio community in the microbiota of the Manila clam, Ruditapes philippinarum. Environ. Microbiol. 2020, 22, 4456–4472. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco Baruch, M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef]
- Soejima, T.; Iida, K.I.; Qin, T.; Taniai, H.; Seki, M.; Yoshida, S.I. Method to detect only live bacteria during PCR amplification. J. Clin. Microbiol. 2008, 46, 2305–2313. [Google Scholar] [CrossRef]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS—Mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef]
- Pereira-Marques, J.; Hout, A.; Ferreira, R.M.; Weber, M.; Pinto-Ribeiro, I.; Van Doorn, L.J.; Knetsch, C.W.; Figueiredo, C. Impact of host DNA and sequencing depth on the taxonomic resolution of whole metagenome sequencing for microbiome analysis. Front. Microbiol. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- McHugh, A.; Feehily, C.; Fenelon, M.; Gleeson, D.; Hill, C.; Cotter, P. Tracking the Dairy Microbiota from Farm Bulk Tank. Am. Soc. Microbiol. 2020, 5, 1–16. [Google Scholar]
- Rubiola, S.; Chiesa, F.; Dalmasso, A.; Di Ciccio, P.; Civera, T. Detection of Antimicrobial Resistance Genes in the Milk Production Environment: Impact of Host DNA and Sequencing Depth. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Anguita-Maeso, M.; Olivares-García, C.; Haro, C.; Imperial, J.; Navas-Cortés, J.A.; Landa, B.B. Culture-Dependent and Culture-Independent Characterization of the Olive Xylem Microbiota: Effect of Sap Extraction Methods. Front. Plant Sci. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Ndoye, B.; Rasolofo, E.A.; LaPointe, G.; Roy, D. A review of the molecular approaches to investigate the diversity and activity of cheese microbiota. Dairy Sci. Technol. 2011, 91, 495–524. [Google Scholar] [CrossRef]
- Udyavar, V.; Muthappa, D.M.; Venugopal, M.; Animal, K.V. Prevalence and Genomic Characterization of Vibrio parahaemolyticus isolated from Molluscan Shellfish and their Inhabiting Water of Coastal Karnataka, India. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 173–182. [Google Scholar] [CrossRef]
- Milan, M.; Carraro, L.; Fariselli, P.; Martino, M.E.; Cavalieri, D.; Vitali, F.; Bo, L.; Patarnello, T.; Bargelloni, L.; Cardazzo, B. Microbiota and environmental stress: How pollution a ff ects microbial communities in Manila clams. Aquat. Toxicol. 2018, 194, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Zakrzewski, M.; Proietti, C.; Ellis, J.J.; Hasan, S.; Brion, M.J.; Berger, B.; Krause, L. Calypso: A user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics 2017, 33, 782–783. [Google Scholar] [CrossRef]
- Feder, I.; Nietfeld, J.C.; Galland, J.; Yeary, T.; Sargeant, J.M.; Oberst, R.; Tamplin, M.L.; Luchansky, J.B. Comparison of cultivation and PCR-hybridization for detection of Salmonella in porcine fecal and water samples. J. Clin. Microbiol. 2001, 39, 2477–2484. [Google Scholar] [CrossRef]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Senderovich, Y.; Izhaki, I.; Halpern, M. Fish as reservoirs and vectors of Vibrio cholerae. PLoS ONE 2010, 5, e8607. [Google Scholar] [CrossRef]
- Serratore, P.; Ostanello, F.; Serraino, A.; Giacometti, F. First multi-year retrospective study on Vibrio parahaemolyticus and Vibrio vulnificus prevalence in Ruditapes philippinarum in Sacca di Goro, Italy. Ital. J. Food Saf. 2016, 10–12. [Google Scholar] [CrossRef][Green Version]
- Xu, Y.G.; Sun, L.M.; Wang, Y.S.; Chen, P.P.; Liu, Z.M.; Li, Y.J.; Tang, L.J. Simultaneous detection of Vibrio cholerae, Vibrio alginolyticus, Vibrio parahaemolyticus and Vibrio vulnificus in seafood using dual priming oligonucleotide (DPO) system-based multiplex PCR assay. Food Control 2017, 71, 64–70. [Google Scholar] [CrossRef]
- Zapka, C.; Leff, J.; Henley, J.; Tittl, J.; De Nardo, E.; Butler, M.; Griggs, R.; Fierer, N.; Edmonds-Wilson, S. Comparison of standard culture-based method to culture-independent method for evaluation of hygiene effects on the hand microbiome. MBio 2017, 8, 1–17. [Google Scholar] [CrossRef]
- Di Pinto, A.; Terio, V.; Novello, L.; Tantillo, G. Comparison between thiosulphate-citrate-bile salt sucrose (TCBS) agar and CHROMagar Vibrio for isolating Vibrio parahaemolyticus. Food Control 2011, 22, 124–127. [Google Scholar] [CrossRef]
- Zhang, X.H.; Ahmad, W.; Yu, X.; Jixiang, Z.; Brian, C. Viable but nonculturable bacteria and their resuscitation: Implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 2020. [Google Scholar] [CrossRef]
- Kakizaki, E.; Takahama, K.; Seo, Y.; Kozawa, S.; Sakai, M.; Yukawa, N. Marine bacteria comprise a possible indicator of drowning in seawater. Forensic Sci. Int. 2008, 176, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Nigro, O.D.; Steward, G.F. Differential specificity of selective culture media for enumeration of pathogenic vibrios: Advantages and limitations of multi-plating methods. J. Microbiol. Methods 2015, 111, 24–30. [Google Scholar] [CrossRef]
- Kaevska, M.; Videnska, P.; Sedlar, K.; Slana, I. Seasonal changes in microbial community composition in river water studied using 454-pyrosequencing. Springerplus 2016, 5. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Wang, J.; Luo, T.; Zhang, R.; Sun, J.; Zheng, Q.; Jiao, N. Seasonal dynamics of bacterial communities in the surface seawater around subtropical Xiamen Island, China, as determined by 16S rRNA gene profiling. Mar. Pollut. Bull. 2019, 142, 135–144. [Google Scholar] [CrossRef]
- Tagliavia, M.; Salamone, M.; Bennici, C.; Quatrini, P.; Cuttitta, A. A modified culture medium for improved isolation of marine vibrios. Microbiologyopen 2019, 8, 1–9. [Google Scholar] [CrossRef]
- Pezzlo, M.; Valter, P.J.; Burns, M.J. Wound infection associated with Vibrio alginolyticus. Am. J. Clin. Pathol. 1979, 71, 476–478. [Google Scholar] [CrossRef]
- Li, R.; Lu, J.; Duan, H.; Yang, J.; Tang, C. Biofilm inhibition and mode of action of epigallocatechin gallate against Vibrio mimicus. Food Control 2020, 113, 107148. [Google Scholar] [CrossRef]
- Elhadi, N.; Radu, S.; Chen, C.H.; Nishibuchi, M. Prevalence of potentially pathogenic vibrio species in the seafood marketed in Malaysia. J. Food Prot. 2004, 67, 1469–1475. [Google Scholar] [CrossRef]
- Zarei, M.; Borujeni, M.P.; Jamnejad, A.; Khezrzadeh, M. Seasonal prevalence of Vibrio species in retail shrimps with an emphasis on Vibrio parahaemolyticus. Food Control 2012, 25, 107–109. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Balboa, S.; Romalde, J.L.; Figueras, M.J. Diversity and pathogenecity of Vibrio species in cultured bivalve molluscs. Environ. Microbiol. Rep. 2010, 2, 34–43. [Google Scholar] [CrossRef]
- Sharpton, T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014, 5, 1–14. [Google Scholar] [CrossRef]
- Thompson, F.L.; Gevers, D.; Thompson, C.C.; Dawyndt, P.; Naser, S.; Hoste, B.; Munn, C.B.; Swings, J. Phylogeny and Molecular Identification of Vibrios on the Basis of Multilocus Sequence Analysis. Appl. Environ. Microbiol. 2005, 71, 5107–5115. [Google Scholar] [CrossRef] [PubMed]
- Grützke, J.; Malorny, B.; Hammerl, J.A.; Busch, A.; Tausch, S.H.; Tomaso, H.; Deneke, C. Fishing in the soup—Pathogen detection in food safety using metabarcoding and metagenomic sequencing. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Srivathsan, A.; Ang, A.; Vogler, A.P.; Meier, R. Fecal metagenomics for the simultaneous assessment of diet, parasites, and population genetics of an understudied primate. Front. Zool. 2016, 13, 1–13. [Google Scholar] [CrossRef]
- Srivathsan, A.; Sha, J.C.M.; Vogler, A.P.; Meier, R. Comparing the effectiveness of metagenomics and metabarcoding for diet analysis of a leaf-feeding monkey (Pygathrix nemaeus). Mol. Ecol. Resour. 2015, 15, 250–261. [Google Scholar] [CrossRef]
- De Filippis, F.; Valentino, V.; Alvarez-Ordóñez, A.; Cotter, P.D.; Ercolini, D. Environmental microbiome mapping as a strategy to improve quality and safety in the food industry. Curr. Opin. Food Sci. 2021, 38, 168–176. [Google Scholar] [CrossRef]
- Arunkumar, M.; LewisOscar, F.; Thajuddin, N.; Pugazhendhi, A.; Nithya, C. In vitro and in vivo biofilm forming Vibrio spp: A significant threat in aquaculture. Process Biochem. 2020, 94, 213–223. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zampieri, A.; Babbucci, M.; Carraro, L.; Milan, M.; Fasolato, L.; Cardazzo, B. Combining Culture-Dependent and Culture-Independent Methods: New Methodology Insight on the Vibrio Community of Ruditapes philippinarum. Foods 2021, 10, 1271. https://doi.org/10.3390/foods10061271
Zampieri A, Babbucci M, Carraro L, Milan M, Fasolato L, Cardazzo B. Combining Culture-Dependent and Culture-Independent Methods: New Methodology Insight on the Vibrio Community of Ruditapes philippinarum. Foods. 2021; 10(6):1271. https://doi.org/10.3390/foods10061271
Chicago/Turabian StyleZampieri, Angela, Massimiliano Babbucci, Lisa Carraro, Massimo Milan, Luca Fasolato, and Barbara Cardazzo. 2021. "Combining Culture-Dependent and Culture-Independent Methods: New Methodology Insight on the Vibrio Community of Ruditapes philippinarum" Foods 10, no. 6: 1271. https://doi.org/10.3390/foods10061271
APA StyleZampieri, A., Babbucci, M., Carraro, L., Milan, M., Fasolato, L., & Cardazzo, B. (2021). Combining Culture-Dependent and Culture-Independent Methods: New Methodology Insight on the Vibrio Community of Ruditapes philippinarum. Foods, 10(6), 1271. https://doi.org/10.3390/foods10061271







