Modelling the Effect of Salt Concentration on the Fate of Listeria monocytogenes Isolated from Costa Rican Fresh Cheeses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of L. Monocytogenes from Cheese
2.1.1. Collection of Samples
2.1.2. Isolation of L. monocytogenes
2.1.3. Identification of L. monocytogenes
2.2. Challenge Test Study
2.2.1. Manufacturing of Fresh “Turrialba Cheese”
2.2.2. Enumeration of L. monocytogenes
2.2.3. Physico-Chemical Characterization of Fresh “Turrialba Cheese”
2.3. Modelling
2.3.1. Primary Growth Model
2.3.2. Secondary Growth Model
2.4. Determination of Growth Potential (δ) of L. monocytogenes
2.5. Model Validation
2.6. Statistics
3. Results
3.1. Listeria Monocytogenes Isolation from Fresh “Turrialba Cheese”
3.2. Challenge Test
Characterization of Fresh “Turrialba Cheese”
3.3. Modelling
3.3.1. Primary Growth Model
3.3.2. Secondary Growth Model
3.3.3. Model Validation
4. Discussion
4.1. Isolation of Listeria Monocytogenes from Fresh Cheese
4.2. Challenge Test in Fresh “Turrialba Cheese”
4.3. Modelling
4.4. Model Validation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fonnesbech, B.; Huss, H.; Ojeniyi, B.; Ahrens, P.; Gram, L. Elucidation of Listeria monocytogenes Contamination Routes in Cold-Smoked Salmon Processing Plants Detected by DNA-Based Typing Methods. Appl. Environ. Microbiol. 2001, 67, 2586–2595. [Google Scholar] [CrossRef] [Green Version]
- Keto-Timonen, R.; Tolvanen, R.; Lundén, J.; Korkeala, H. An 8-Year Surveillance of the Diversity and Persistence of Listeria monocytogenes in a Chilled Food Processing Plant Analyzed by Amplified Fragment Length Polymorphism. J. Food Prot. 2007, 70, 1866–1873. [Google Scholar] [CrossRef] [Green Version]
- Ivanek, R.; Grohn, Y.; Tauer, L.; Wiedmann, M. The Cost and Benefit of Listeria monocytogenes Food Safety Measures. Crit. Rev. Food Sci. Nutr. 2010, 44, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Goulet, V.; Jacquet, C.; Vaillant, V.; Rebière, I.; Mouret, E.; Lorente, C.; Maillot, E.; Staïner, F.; Rocourt, J. Listeriosis from consumption of raw-milk cheese. Lancet 1995, 345, 1581–1582. [Google Scholar] [CrossRef]
- Cámara Nacional de Productores de Leche. Available online: http://proleche.com/consumo-de-productos-lacteos/ (accessed on 5 May 2021).
- Ministerio de Agricultura y Ganadería de Costa Rica. Implementar el Proceso de Inocuidad y Buenas Prácticas de Manufactura en 44 Queserías Propiedad de Pequeños Productores de Queso Artesanal. Available online: http://www.mag.go.cr/bibliotecavirtual/Q02-10964.pdf (accessed on 5 May 2021).
- Pliego de Condiciones Denominación de Origen del “Queso Turrialba”. Available online: http://www.fao.org/fileadmin/templates/olq/documents/costarica/ppp/Miercoles/documentacion/PliegoCondicionesDOTurrialba.pdf (accessed on 5 May 2021).
- Pacheco-Zamora, M. Determinación de la Concentración Mínima de Cloruro de Sodio que se Puede Reducir en un Queso Fresco sin que Sea Perceptible Para los Consumidores Utilizando un Umbral de Diferencia y su Efecto Sobre el Desuerado y Perfil de Textura. Ph.D. Thesis, University of Costa Rica, San Pedro, Costa Rica, 2018. [Google Scholar]
- Víquez-Barrantes, D. Caracterización de la Producción Artesanal de Queso en las Empresas de la Cámara Nacional de Queseros Artesanos y Afines (CANAQUEAF), Capacitación de sus Miembros y Elaboración de una Propuesta de Plan de Acción que Permita Revalorizar los Quesos Artesanales. Ph.D. Thesis, University of Costa Rica, San Pedro, Costa Rica, 2012. [Google Scholar]
- Bolton, L.; Frank, J. Defining the Growth/No-Growth Interface for Listeria monocytogenes in Mexican-Style Cheese Based on Salt, pH, and Moisture Content. J. Food Prot. 1999, 62, 601–609. [Google Scholar] [CrossRef]
- Cadavez, V.A.P.; Campagnollo, F.B.; Rosicléia, A.S.; Duffner, C.M.; Schaffner, D.W.; Sant’Ana, A.S.; Gonzales-Barron, U. A comparison of dynamic tertiary and competition models for describing the fate of Listeria monocytogenes in Minas fresh cheese during refrigerated storage. Food Microbiol. 2019, 79, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Rodríguez, F.; Valero Díaz, A.; Carrasco Jiménez, E.; Posada-Izquierdo, G. Predictive Modelling in Food; Cambridge Scholars Publishing: Cambridge, UK, 2019; pp. 1–190. [Google Scholar]
- Ministerio de Salud de Costa Rica. Available online: https://www.ministeriodesalud.go.cr/ (accessed on 5 May 2021).
- Turck, D.; Castenmiller, J.; de Henauw, S.; Hirsch-Ernst, K.I.; Kearney, J.; Knutsen, H.K.; Maciuk, A.; Mangelsdorf, I.; McArdle, H.J.; Pelaez, C.; et al. Scientific Opinion on the dietary reference values for sodium. EFSA J. 2019, 17, 1–191. [Google Scholar] [CrossRef] [Green Version]
- Hitchins, A.D.; Jinnema, K.; Chen, Y. BAM Chapter 10: Detection of Listeria monocytogenes in Foods and Environmental Samples, and Enumeration of Listeria monocytogenes in Foods. Bacteriological Analytical Manual (BAM); 2017. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-10-detection-listeria-monocytogenes-foods-and-environmental-samples-and-enumeration (accessed on 5 May 2021).
- AOAC. Official method 981.12. pH of acidified foods. In Official Methods of Analysis, 13th ed.; AOAC International: Washington, DC, USA, 1990. [Google Scholar]
- Baranyi, J.; Roberts, T.A. A dynamic approach to predicting bacterial growth in food. Int. J. Food Microbiol. 1994, 23, 277–294. [Google Scholar] [CrossRef]
- ComBase Predictive Models (Software Tools). Available online: Browser.combase.cc/DMFit.aspx (accessed on 5 May 2021).
- Schabenberger, O.; Gotway, C.A. Statistical Methods for Spatial Data Analysis, 1st ed.; Chapman and Hall/CRC: Boca Raton, FL, USA, 2005. [Google Scholar] [CrossRef]
- González, S.C.; Possas, A.; Carrasco, E.; Valero, A.; Bolívar, A.; Posada-Izquierdo, G.D.; García-Gimeno, R.M.; Zurera, G.; Pérez-Rodríguez, F. ‘MicroHibro’: A software tool for predictive microbiology and microbial risk assessment in foods. Int. J. Food Microbiol. 2019, 2, 226–236. [Google Scholar] [CrossRef]
- Beaufort, A. The determination of ready-to-eat foods into Listeria monocytogenes growth and no growth categories by challenge tests. Food Control 2011, 22, 1498–1502. [Google Scholar] [CrossRef]
- Food Spoilage and Safety Predictor (FSSP). Available online: http://fssp.food.dtu.dk/ (accessed on 5 May 2021).
- Pathogen Modeling Program (PMP). Available online: https://pmp.errc.ars.usda.gov/PMPOnline.aspx (accessed on 5 May 2021).
- Ross, T. Indices for performance evaluation of predictive models in food microbiology. J. Appl. Bacteriol. 1996, 81, 501–508. [Google Scholar] [PubMed]
- Resnik, S.L.; Chirife, J. Proposed Theoretical Water Activity Values at Various Temperatures for Selected Solutions to be Used as Reference Sources in the Range of Microbial Growth. J. Food Prot. 1988, 51, 419–423. [Google Scholar] [CrossRef]
- Mejlholm, O.; Dalgaard, P. Modelling and predicting the simultaneous growth of Listeria monocytogenes and psychrotolerant lactic acid bacteria in processed seafood and mayonnaise-based seafood salads. Food Microbiol. 2015, 46, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Ratkowsky, D.A.; Wang, B.; Bowman, J.P.; Tamplin, M.L. Modelling viability of Listeria monocytogenes in paneer. Food Microbiol. 2021, 97, 103738. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Luchansky, J.B.; Tamplin, M.L.; Molina-Corral, F.J.; Anandan, S.; Porto-Fett, A.C.S. Effect of storage temperature on the growth of Listeria monocytogenes on Queso Blanco slices. J. Food Saf. 2006, 26, 202–214. [Google Scholar] [CrossRef]
- Thomas, M.; Tiwari, R.; Mishra, A. Predictive model of Listeria monocytogenes growth in queso fresco. J. Food Protect. 2019, 82, 2071–2079. [Google Scholar] [CrossRef]
- Leggett, L.; Tomasula, P.; Van Hekken, D.; Porto-fett, A.; Shoyer, B.; Renye, J.; Luchansky, J.; Farkye, N. Effect of storage at 4 and 10 °C on the growth of Listeria monocytogenes in and on Queso Fresco. J. Food Saf. 2012, 32, 236–245. [Google Scholar] [CrossRef]
- Østergaard, N.B.; Eklöw, A.; Dalgaard, P. Modelling the effect of lactic acid bacteria from starter- and aroma culture on growth of Listeria monocytogenes in cottage cheese. Int. J. Food Microbiol. 2014, 188, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Tirloni, E.; Bernardi, C.; Rosshaug, P.S.; Stella, S. Potential growth of Listeria monocytogenes in Italian mozzarella cheese as affected by microbiological and chemical-physical environment. J. Dairy Sci. 2019, 102, 4913–4924. [Google Scholar] [CrossRef] [Green Version]
- Dalzini, E.; Cosciani-Cunico, E.; Monastero, P.; Bernini, V.; Neviani, E.; Bellio, A. Decastel y of the behaviour throughout the process and growth prediction during shelf life. Int. J. Food Microbiol. 2017, 262, 71–79. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2018, 75, 1–13. [Google Scholar] [CrossRef]
- Hoelzer, K.; Pouillot, R.; Gallagher, D.; Silverman, M.B.; Kause, J.; Dennis, S. Estimation of Listeria monocytogenes transfer coefficients and efficacy of bacterial removal through cleaning and sanitation. Int. J. Food Microbiol. 2012, 157, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Sánchez, L.A.; Van Tassell, M.L.; Miller, M.J. Invited review: Hispanic-style cheeses and their association with Listeria monocytogenes. J. Dairy Sci. 2017, 100, 421–2432. [Google Scholar] [CrossRef] [PubMed]
- Mattos, R.; Staples, C.R.; Arteche, A.; Wiltbank, M.C.; Diaz, F.J.; Jenkins, T.C.; Thatcher, W. The effects of feeding fish oil on uterine secretion of PGF2alpha, milk composition, and metabolic status of periparturient Holstein cows. J. Dairy Sci. 2004, 87, 921–932. [Google Scholar] [CrossRef] [Green Version]
- Gould, L.H.; Mungai, E.; Behravesh, C.B. Outbreaks Attributed to Cheese: Differences between Outbreaks Caused by Unpasteurized and Pasteurized Dairy Products, United States, 1998–2011. Foodborne Pathog. Dis. 2014, 11, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Cháves, C.; Arias, M.L. Caracterización de Cepas de Listeria monocytogenes Realizados a Partir de Queso Fresco Proveniente de Diferentes Zonas Productoras Costarricenses. Arch. Latinoam. Nutr. 2009, 59, 66–70. Available online: http://ve.scielo.org/scielo.php?script=sci_arttext&pid=S0004-06222009000100010&lng=es&tlng=es (accessed on 5 May 2021).
- Food and Drug Administration (FDA). Contains Nonbinding Recommendations Control of Listeria monocytogenes in Ready-To-Eat Foods: Guidance for Industry Draft Guidance. Available online: https://www.fda.gov/files/food/published/Draft-Guidance-for-Industry--Control-of-Listeria-monocytogenes-in-Ready-To-Eat-Foods-%28PDF%29.pdf (accessed on 5 May 2021).
- Calvo-Arrieta, K.; Matamoros-Montoya, K.; Arias-Echandi, M.; Huete-Soto, A.; Redondo-Solano, M. Presence of Listeria monocytogenes in Ready-to-Eat (RTE) meat products sold at retail stores in Costa Rica and analysis of contributing factors. J. Food Prot. 2021. [Google Scholar] [CrossRef]
- Campagnollo, F.B.; Gonzales-Barron, U.; Cadavez, V.A.; Sant’Ana, A.S. Quantitative risk assessment of Listeria monocytogenes in traditional minas cheeses: The cases of artisanal semi-hard and fresh soft cheeses. Food Control 2018, 92, 370–379. [Google Scholar] [CrossRef] [Green Version]
- Pouillot, R.; Klontz, K.C.; Chen, Y.; Burall, L.S.; Macarisin, D.; Doyle, M.; Bally, K.M.; Strain, E.; Datta, A.R.; Hammack, T.S.; et al. Infectious Dose of Listeria monocytogenes in Outbreak Linked to Ice Cream, United States, 2015. Emerg. Infect. Dis. 2016, 22, 2113–2119. [Google Scholar] [CrossRef] [Green Version]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Fresh Cheese Products: Principals of Manufacture and Overview of Different Varieties. In Fundamentals of Cheese Science; Springer: Boston, MA, USA, 2017. [Google Scholar]
- Lourenço, A.; Kamnetz, M.B.; Gadotti, C.; Diez-Gonzalez, F. Antimicrobial treatments to control Listeria monocytogenes in queso fresco. Food Microbiol. 2017, 64, 47–55. [Google Scholar] [CrossRef]
- Vera Pingitore, E.; Todorov, S.D.; Sesma, F.; Gombossy de Melo Franco, B.D. Application of bacteriocinogenic Enterococcus mundtii CRL35 and Enterococcus faecium ST88Ch in the control of Listeria monocytogenes in fresh Minas cheese. Food Microbiol. 2012, 32, 38–47. [Google Scholar] [CrossRef]
- Da Silva Malheiros, P.; Sant’Anna, V.; de Souza Barbosa, M.; Brandelli, A.; Gombossy de Melo Franco, B.D. Effect of liposome-encapsulated nisin and bacteriocin-like substance P34 on Listeria monocytogenes growth in Minas frescal cheese. Int. J. Food Microbiol. 2012, 156, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.R.B.; Forauer, E.C.; D’Amico, D.J. Effect of modified atmosphere packaging on the growth of spoilage microorganisms and Listeria monocytogenes on fresh cheese. J. Dairy Sci. 2018, 101, 7768–7779. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, O.M.; Christen, G.L.; Davidson, P.M. Chemical Composition of and Listeria monocytogenes Survival in White Pickled Cheese. J. Food Prot. 1993, 56, 841–846. [Google Scholar] [CrossRef]
- Leong, W.M.; Geier, R.; Engstrom, S.; Ingham, S.; Ingham, B.; Smukowski, M. Growth of Listeria monocytogenes, Salmonella spp., Escherichia coli O157:H7, and Staphylococcus aureus on cheese during extended storage at 25 °C. J. Food Prot. 2014, 77, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Millet, L.; Saubusse, M.; Didienne, R.; Tessier, L.; Montel, M.C. Control of Listeria monocytogenes in raw-milk cheeses. Int. J. Food Microbiol. 2006, 108, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, S.; Grieder, J.A.; McMahon, D.J.; Nummer, B.A. Survival of Listeria monocytogenes introduced as a post-aging contaminant during storage of low-salt Cheddar cheese at 4, 10, and 21 °C. J. Dairy Sci. 2011, 94, 4329–4335. [Google Scholar] [CrossRef] [PubMed]
- Gérard, A.; El-Hajjaji, S.; Van Coillie, E.; Bentaïb, A.; Daube, G.; Sindic, M. Determination of the growth potential of Listeria monocytogenes in various types of Belgian artisanal cheeses by challenge tests. Food Microbiol. 2020, 92, 103582. [Google Scholar] [CrossRef]
- Spanu, C.; Scarano, C.; Spanu, V.; Penna, C.; Virdis, S.; De Santis, E.P.L. Listeria monocytogenes growth potential in Ricotta salata cheese. Int. Dairy J. 2012, 24, 120–122. [Google Scholar] [CrossRef]
- Heir, E.; Møretrø, T.; Simensen, A.; Langsrud, S. Listeria monocytogenes strains show large variations in competitive growth in mixed culture biofilms and suspensions with bacteria from food processing environments. Int. J. Food Microbiol. 2018, 275, 46–55. [Google Scholar] [CrossRef]
- Sant’Ana, A.S.; Barbosa, M.S.; Destro, M.T.; Landgraf, M.; Franco, B.D.G.M. Growth potential of Salmonella spp. and Listeria monocytogenes in nine types of ready-to-eat vegetables stored at variable temperature conditions during shelf-life. Int. J. Food Microbiol. 2012, 157, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Ross, T.; Dalgaard, P.; Tienungoon, S. Predictive modeling of the growth and survival of Listeria in fishery products. Int. J. Food Microbiol. 2000, 62, 231–245. [Google Scholar] [CrossRef]
- Tiwari, U.; Walsh, D.; Rivas, L.; Jordan, K.; Duffy, G. Modelling the interaction of storage temperature, pH, and water activity on the growth behaviour of Listeria monocytogenes in raw and pasteurised semi-soft rind washed milk cheese during storage following ripening. Food Control 2014, 42, 248–256. [Google Scholar] [CrossRef]
- Piccinin, D.M.; Shelef, L.A. Survival of Listeria monocytogenes in Cottage Cheese. J. Food Prot. 1995, 58, 128–131. [Google Scholar] [CrossRef] [PubMed]
 .
.
 .
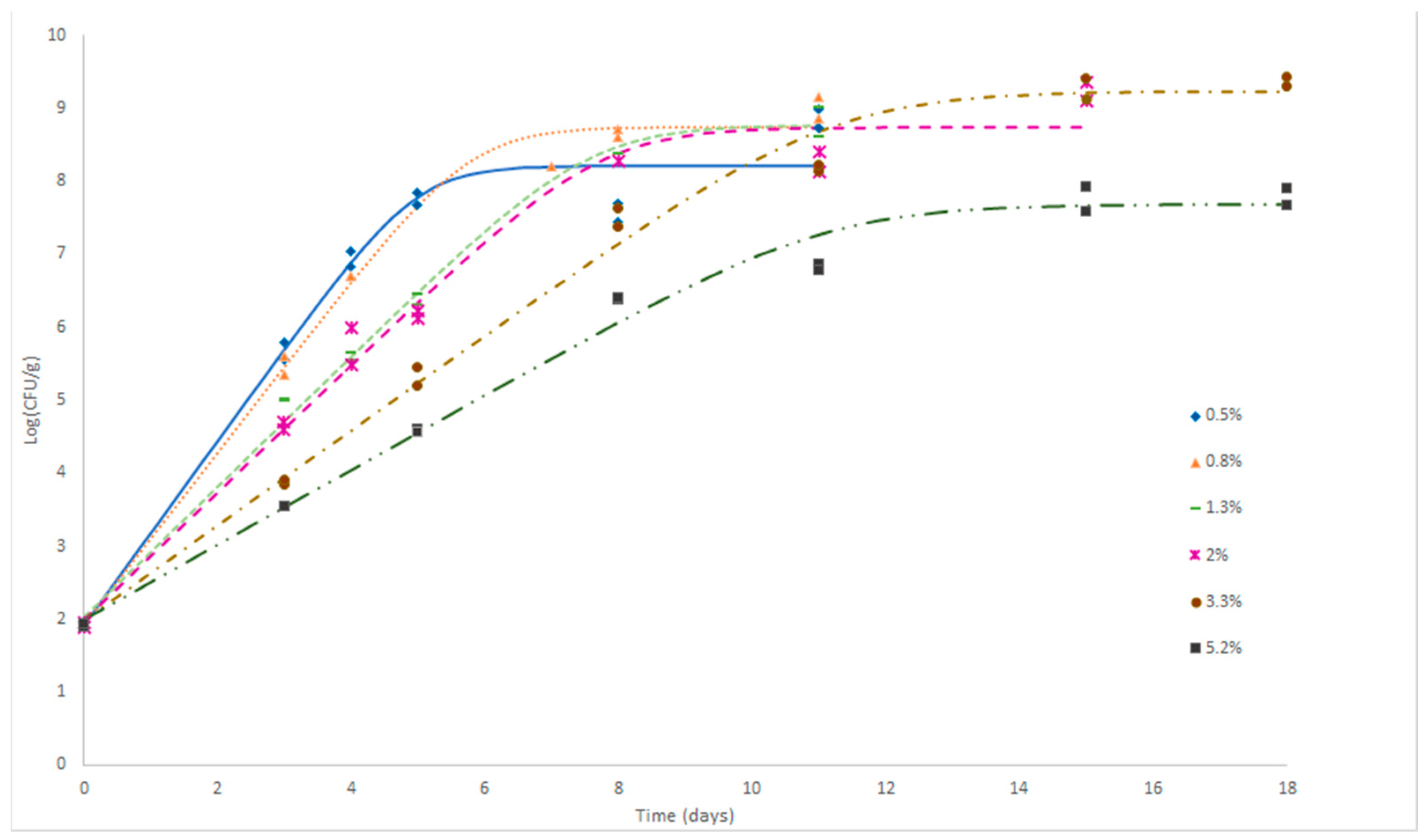
.


| Province | Location * | No. of Samples | No. of Samples Positive for L. monocytogenes |
|---|---|---|---|
| San José | 1 | 5 | 2 |
| 2 | 7 | 0 | |
| 3 | 9 | 0 | |
| 4 | 2 | 0 | |
| 5 | 5 | 0 | |
| Cartago | 6 | 2 | 0 |
| 7 | 9 | 4 | |
| Heredia | 8 | 5 | 0 |
| 9 | 8 | 3 | |
| Alajuela | 10 | 12 | 0 |
| Total | ----- | 64 | 9 |
| 3 Days | 5 Days | 7 Days | |||||||
|---|---|---|---|---|---|---|---|---|---|
| %NaCl | pH | SD | aw | pH | SD | aw | pH | SD | aw |
| 0.5 | 6.67 | 0.03 | 0.993 | 6.49 | 0.02 | 0.994 | 6.48 | 0.04 | 0.993 |
| 0.8 | 6.70 | 0.01 | 0.993 | 6.50 | 0.02 | 0.992 | 6.53 | 0.01 | 0.992 |
| 1.3 | 6.66 | 0.03 | 0.986 | 6.46 | 0.01 | 0.982 | 6.51 | 0.03 | 0.987 |
| 2.0 | 6.66 | 0.03 | 0.969 | 6.52 | 0.01 | 0.980 | 6.57 | 0.02 | 0.977 |
| 3.3 | 6.66 | 0.02 | 0.955 | 6.48 | 0.02 | 0.968 | 6.58 | 0.01 | 0.966 |
| 5.2 | 6.65 | 0.03 | 0.937 | 6.57 | 0.06 | 0.954 | 6.52 | 0.02 | 0.950 |
| %NaCl | μmax | ymax obs | δobs | y0 est | ymaxest | δest | R² | RMSE |
|---|---|---|---|---|---|---|---|---|
| 0.5 | 1.262 | 8.851 | 6.93 | 1.93 | 8.21 | 6.28 | 0.964 | 0.443 |
| 0.8 | 1.178 | 9.000 | 7.08 | 1.94 | 8.74 | 6.80 | 0.992 | 0.254 |
| 1.3 | 0.895 | 8.805 | 6.88 | 2.04 | 8.78 | 6.74 | 0.993 | 0.197 |
| 2.0 | 0.874 | 9.230 | 7.31 | 2.00 | 8.74 | 6.74 | 0.978 | 0.368 |
| 3.3 | 0.651 | 9.370 | 7.45 | 1.99 | 9.24 | 7.25 | 0.989 | 0.293 |
| 5.2 | 0.514 | 7.785 | 5.86 | 2.00 | 7.69 | 5.69 | 0.986 | 0.257 |
| References | Matrix (Broth/Type Cheese) | Factors | Bf | Af | No. |
|---|---|---|---|---|---|
| [27] | Paneer | T(°C) = 4; pH = 5.42–5.57; aw = 0.999; NaCl (%) = 0.19 | 1.82 | 1.82 | 2 |
| [28] | “Queso Blanco” slices | T(°C) = 5; pH = 6.8; aw = 0.971; NaCl (%) = 4.96 | 1.44 | 1.44 | 15 |
| [29] | “Queso fresco” | T(°C) = 4; pH = 6.40; aw = 0.96; NaCl (%) = 3 | 1.38 | 1.38 | 1 |
| [30] | “Queso fresco” | T(°C) = 4; pH = 6.75; aw = 0.983; NaCl (%) = 1.67 | 1.57 | 1.57 | 2 |
| [31] | Cottage | T(°C) = 5; pH = 5.18; Salt% = 1.22; 718 ppm Lactic acid | 1.72 | 1.72 | 1 |
| [32] | Mozzarella with citric acid; | T(°C) = 4; pH = 5.49–6.43; aw = 0.988; NaCl (%) = 2.18 | 0.80 | 1.25 | 2 |
| [33] | Gorgonzola | T(°C) = 8; pH = 6.80; aw = 0.978; NaCl (%) = 3.85 | 1.07 | 1.38 | 6 |
| Total | RMSE = 0.0191 | Bf = 1.35 | Af = 1.43 | n = 29 | |
| Modelling Program | Matrix (Broth/Type Cheese) | Factors | Bf | Af | No. |
| [18] | Broth media | T(°C) = 4; pH = 6.5–6.7; aw = 0.969–0.997; NaCl (%) = 0.5–5.2 | 0.67 | 1.90 | 4 |
| [22] | Cottage cheese | T(°C) = 5; pH = 5.5; aw = 0.989; NaCl (%) = 2 | 1.15 | 1.15 | 4 |
| [23] | Broth media | T(°C) = 4; pH = 6.6; aw = 0.997–0.969; NaCl (%) = 0.5–5.1 | 1.24 | 1.24 | 4 |
| Total | RMSE = 0.058 | Bf = 0.98 | Af = 2.73 | n = 12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Posada-Izquierdo, G.D.; Mazón-Villegas, B.; Redondo-Solano, M.; Huete-Soto, A.; Víquez-Barrantes, D.; Valero, A.; Fallas-Jiménez, P.; García-Gimeno, R.M. Modelling the Effect of Salt Concentration on the Fate of Listeria monocytogenes Isolated from Costa Rican Fresh Cheeses. Foods 2021, 10, 1722. https://doi.org/10.3390/foods10081722
Posada-Izquierdo GD, Mazón-Villegas B, Redondo-Solano M, Huete-Soto A, Víquez-Barrantes D, Valero A, Fallas-Jiménez P, García-Gimeno RM. Modelling the Effect of Salt Concentration on the Fate of Listeria monocytogenes Isolated from Costa Rican Fresh Cheeses. Foods. 2021; 10(8):1722. https://doi.org/10.3390/foods10081722
Chicago/Turabian StylePosada-Izquierdo, Guiomar D., Beatriz Mazón-Villegas, Mauricio Redondo-Solano, Alejandra Huete-Soto, Diana Víquez-Barrantes, Antonio Valero, Paula Fallas-Jiménez, and Rosa María García-Gimeno. 2021. "Modelling the Effect of Salt Concentration on the Fate of Listeria monocytogenes Isolated from Costa Rican Fresh Cheeses" Foods 10, no. 8: 1722. https://doi.org/10.3390/foods10081722
APA StylePosada-Izquierdo, G. D., Mazón-Villegas, B., Redondo-Solano, M., Huete-Soto, A., Víquez-Barrantes, D., Valero, A., Fallas-Jiménez, P., & García-Gimeno, R. M. (2021). Modelling the Effect of Salt Concentration on the Fate of Listeria monocytogenes Isolated from Costa Rican Fresh Cheeses. Foods, 10(8), 1722. https://doi.org/10.3390/foods10081722








