A Binary Logistic Regression Model as a Tool to Predict Craft Beer Susceptibility to Microbial Spoilage
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Adaptation Process to Beer-Type Beverage
2.2. Determination of the Early Stationary Growth Phase
2.3. Data Generation for the First Group of Samples
2.3.1. Microbial and Physicochemical Analysis of Craft Beers
2.3.2. Adjusted-Beer Preparation
2.3.3. G/NG Evaluation for the First Group of Samples
2.4. Data Generation for the Second Group of Samples
2.4.1. Selection of Commercial Craft Beers
2.4.2. G/NG Evaluation for the Second Group of Samples
2.5. Model Development
2.6. Model Validation
3. Results and Discussion
3.1. Adaptation of Microorganisms to Beer-Type Beverages and Determination of the Early Stationary Phase
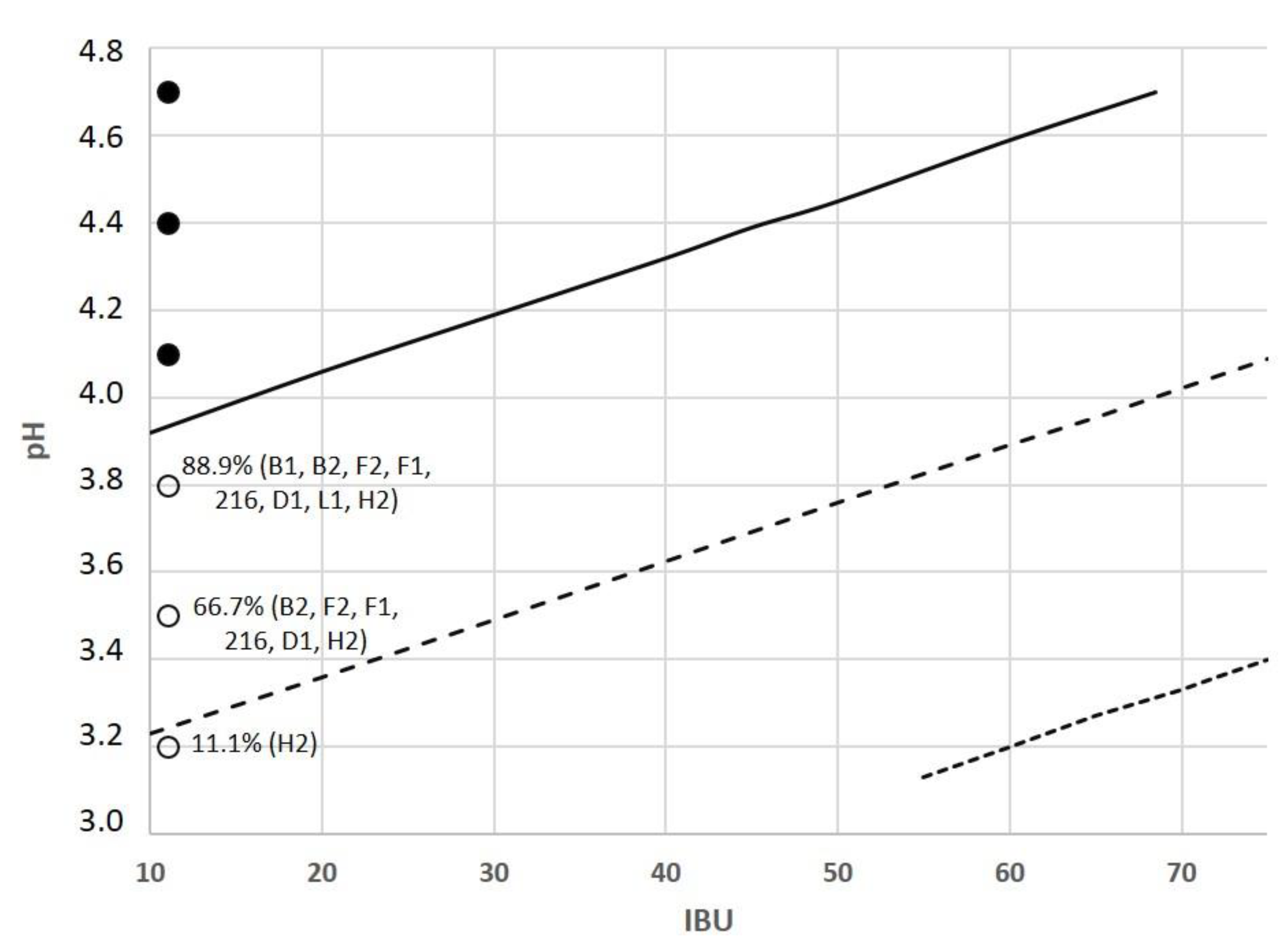
3.2. Model Data
3.2.1. First Group Data
3.2.2. Second Group Data
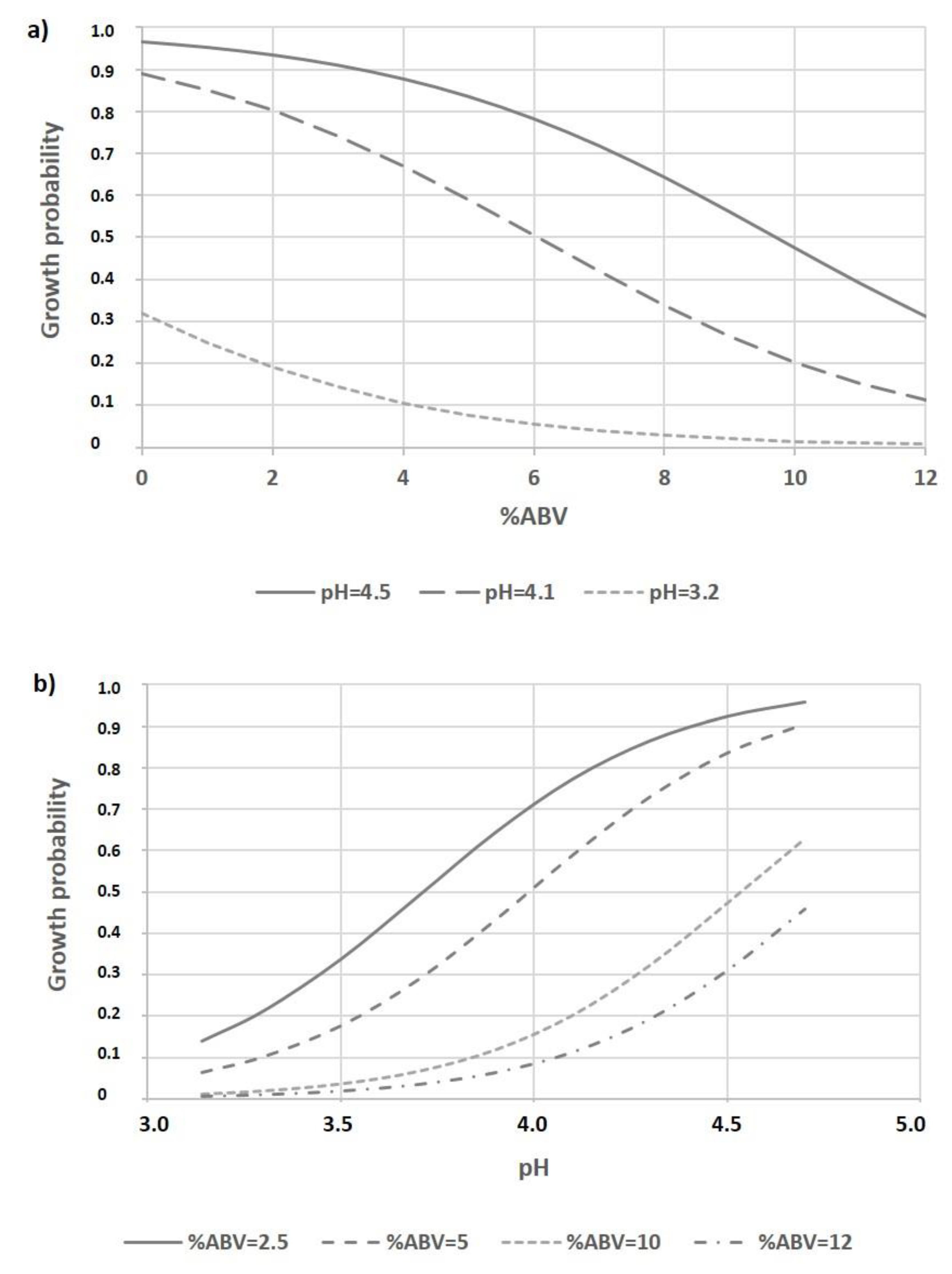
3.3. Model Development
3.3.1. Multi-Collinearity Analysis
3.3.2. Evaluation of Model Performance
3.4. Model Validation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Strong, G.; England, K. Beer Judge Certification Program 2015 Style Guidelines. Available online: http://www.bjcp.org/stylecenter.php (accessed on 19 November 2019).
- Sakamoto, K.; Konings, W.N. Beer spoilage bacteria and hop resistance. Int. J. Food Microbiol. 2003, 89, 105–124. [Google Scholar] [CrossRef]
- Suzuki, K.; Iijima, K.; Sakamoto, K.; Sami, M.; Yamashita, H. A Review of Hop Resistance in Beer Spoilage Lactic Acid Bacteria. J. Inst. Brew. 2006, 112, 173–191. [Google Scholar] [CrossRef]
- Menz, G.; Aldred, P.; Vriesekoop, F. Pathogens in Beer. In Beer in Health and Disease Prevention; Academic Press: Cambridge, MD, USA, 2009; pp. 403–413. ISBN 9780123738912. [Google Scholar]
- Suzuki, K. 125th Anniversary Review: Microbiological Instability of Beer Caused by Spoilage Bacteria. J. Inst. Brew. 2011, 117, 131–155. [Google Scholar] [CrossRef]
- Kalač, P.; Šavel, J.; Křížek, M.; Pelikánová, T.; Prokopová, M. Biogenic amine formation in bottled beer. Food Chem. 2002, 79, 431–434. [Google Scholar] [CrossRef]
- Vaughan, A.; O’Sullivan, T.; van Sinderen, D. Enhancing the Microbiological Stability of Malt and Beer—A Review. J. Inst. Brew. 2005, 111, 355–371. [Google Scholar] [CrossRef]
- Rodríguez-Saavedra, M.; González de Llano, D.; Beltran, G.; Torija, M.-J.; Moreno-Arribas, M.V. Pectinatus spp.—Unpleasant and recurrent brewing spoilage bacteria. Int. J. Food Microbiol. 2021, 336, 108900. [Google Scholar] [CrossRef]
- Thelen, K.; Beimfohr, C.; Snaidr, J. VIT-Bier: The Rapid and Easy Detection Method for Beer-Spoiling Bacteria. Tech. Q. MBAA Commun. 2004, 41, 115–119. [Google Scholar]
- White, C. Analysis of the First Large-Scale Testing of Craft Beer. Tech. Q. MBAA Commun. 2008, 45, 13–16. [Google Scholar] [CrossRef]
- Menz, G.; Andrighetto, C.; Lombardi, A.; Corich, V.; Aldred, P.; Vriesekoop, F. Isolation, Identification, and Characterisation of Beer-Spoilage Lactic Acid Bacteria from Microbrewed Beer from Victoria, Australia. J. Inst. Brew. 2010, 116, 14–22. [Google Scholar] [CrossRef]
- Jeon, S.H.; Kim, N.H.; Shim, M.B.; Jeon, Y.W.; Ahn, J.H.; Lee, S.H.; Hwang, I.G.; Rhee, M.S. Microbiological Diversity and Prevalence of Spoilage and Pathogenic Bacteria in Commercial Fermented Alcoholic Beverages (Beer, Fruit Wine, Refined Rice Wine, and Yakju). J. Food Prot. 2015, 78, 812–818. [Google Scholar] [CrossRef]
- Garofalo, C.; Osimani, A.; Milanović, V.; Taccari, M.; Aquilanti, L.; Clementi, F. The Occurrence of Beer Spoilage Lactic Acid Bacteria in Craft Beer Production. J. Food Sci. 2015, 80, M2845–M2852. [Google Scholar] [CrossRef]
- Ratkowsky, D.A.; Ross, T. Modelling the bacterial growth/no growth interface. Lett. Appl. Microbiol. 1995, 20, 29–33. [Google Scholar] [CrossRef]
- Debonne, E.; Vermeulen, A.; Bouboutiefski, N.; Ruyssen, T.; Van Bockstaele, F.; Eeckhout, M.; Devlieghere, F. Modelling and validation of the antifungal activity of DL-3-phenyllactic acid and acetic acid on bread spoilage moulds. Food Microbiol. 2020, 88, 103407. [Google Scholar] [CrossRef] [PubMed]
- Battey, A.S.; Schaffner, D.W. Modelling bacterial spoilage in cold-filled ready to drink beverages by Acinetobacter calcoaceticus and Gluconobacter oxydans. J. Appl. Microbiol. 2001, 91, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Battey, A.S.; Duffy, S.; Schaffner, D.W. Modeling Yeast Spoilage in Cold-Filled Ready-To-Drink Beverages with Saccharomyces cerevisiae, Zygosaccharomyces bailii, and Candida lipolytica. Appl. Environ. Microbiol. 2002, 68, 1901–1906. [Google Scholar] [CrossRef]
- Uljas, H.E.; Schaffner, D.W.; Duffy, S.; Zhao, L.; Ingham, S.C. Modeling of Combined Processing Steps for Reducing Escherichia coli O157:H7 Populations in Apple Cider. Appl. Environ. Microbiol. 2001, 67, 133–141. [Google Scholar] [CrossRef][Green Version]
- Munford, A.R.G.; Chaves, R.D.; Granato, D.; Sant’Ana, A.S. Modeling the inactivation of Lactobacillus brevis DSM 6235 and retaining the viability of brewing pitching yeast submitted to acid and chlorine washing. Appl. Microbiol. Biotechnol. 2020, 104, 4071–4080. [Google Scholar] [CrossRef]
- Vermeulen, A.; Devlieghere, F.; Bernaerts, K.; Van Impe, J.; Debevere, J. Growth/no growth models describing the influence of pH, lactic and acetic acid on lactic acid bacteria developed to determine the stability of acidified sauces. Int. J. Food Microbiol. 2007, 119, 258–269. [Google Scholar] [CrossRef]
- Marvig, C.L.; Kristiansen, R.M.; Nielsen, D.S. Growth/no growth models for Zygosaccharomyces rouxii associated with acidic, sweet intermediate moisture food products. Int. J. Food Microbiol. 2015, 192, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Saavedra, M.; González de Llano, D.; Moreno-Arribas, M.V. Beer spoilage lactic acid bacteria from craft brewery microbiota: Microbiological quality and food safety. Food Res. Int. 2020, 138, 109762. [Google Scholar] [CrossRef]
- McKellar, R.C.; Lu, X. Primary models. In Modeling Microbial Responses in Food; CRC Press: Boca Raton, FL, USA, 2004; pp. 21–62. [Google Scholar]
- Vermeulen, A.; Daelman, J.; Van Steenkiste, J.; Devlieghere, F. Screening of different stress factors and development of growth/no growth models for Zygosaccharomyces rouxii in modified Sabouraud medium, mimicking intermediate moisture foods (IMF). Food Microbiol. 2012, 32, 389–396. [Google Scholar] [CrossRef] [PubMed]
- American Society of Brewing Chemists. Methods of Analysis, 8th ed.; ASBC: St. Paul, MN, USA, 2009. [Google Scholar]
- Valero, A.; Pérez-Rodríguez, F.; Carrasco, E.; Fuentes-Alventosa, J.M.; García-Gimeno, R.M.; Zurera, G. Modelling the growth boundaries of Staphylococcus aureus: Effect of temperature, pH and water activity. Int. J. Food Microbiol. 2009, 133, 186–194. [Google Scholar] [CrossRef]
- Gysemans, K.P.M.; Bernaerts, K.; Vermeulen, A.; Geeraerd, A.H.; Debevere, J.; Devlieghere, F.; Van Impe, J.F. Exploring the performance of logistic regression model types on growth/no growth data of Listeria monocytogenes. Int. J. Food Microbiol. 2007, 114, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Valero, A.; Carrasco, E.; Pérez-Rodriguez, F.; García-Gimeno, R.M.; Zurera, G. Growth/no growth model of Listeria monocytogenes as a function of temperature, pH, citric acid and ascorbic acid. Eur. Food Res. Technol. 2006, 224, 91–100. [Google Scholar] [CrossRef]
- Hilbe, J.M. Logistic Regression Models; Chapman & Hall/CRC Press: London, UK, 2009; ISBN 9781420075755. [Google Scholar]
- Zhao, Y.; Knochel, S.; Siegumfeldt, H. Heterogeneity between and within Strains of Lactobacillus brevis Exposed to Beer Compounds. Front. Microbiol. 2017, 8, 239. [Google Scholar] [CrossRef]
- Asano, S.; Suzuki, K.; Iijima, K.; Motoyama, Y.; Kuriyama, H.; Kitagawa, Y. Effects of morphological changes in beer-spoilage lactic acid bacteria on membrane filtration in breweries. J. Biosci. Bioeng. 2007, 104, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Lechner, S.; Lewis, K.; Bertram, R. Staphylococcus aureus Persisters Tolerant to Bactericidal Antibiotics. J. Mol. Microbiol. Biotechnol. 2012, 22, 235–244. [Google Scholar] [CrossRef]
- Midi, H.; Sarkar, S.K.; Rana, S. Collinearity diagnostics of binary logistic regression model. J. Interdiscip. Math. 2010, 13, 253–267. [Google Scholar] [CrossRef]
- Nagelkerke, N.J.D. A note on a general definition of the coefficient of determination. Biometrika 1991, 78, 691–692. [Google Scholar] [CrossRef]
- Lemeshow, S.; Hosmer, D. A review of goodness of fit statistics for use in the development of logistic regression models. Am. J. Epidemiol. 1982, 115, 92–106. [Google Scholar] [CrossRef]
- Bernard-Morell, E. Bioestadística Básica Para Investigadores Con SPSS, 1st ed.; Bubok, Ed.; Bubok S.L.: Madrid, Spain, 2014. [Google Scholar]
- Simpson, W.J.; Hammond, J.R.M. Antibacterial action of hop resin materials. In European Brewery Convention: Proceedings of the 23rd Congress, Lisbon 1991, 1st ed.; European Brewery Convention, Ed.; IRL Press: Oxford, UK, 1991; pp. 185–193. [Google Scholar]
- Simpson, W.J.; Smith, A.R.W. Factors affecting antibacterial activity of hop compounds and their derivatives. J. Appl. Bacteriol. 1992, 72, 327–334. [Google Scholar] [CrossRef]
- Menz, G.; Aldred, P.; Vriesekoop, F. Growth and Survival of Foodborne Pathogens in Beer. J. Food Prot. 2011, 74, 1670–1675. [Google Scholar] [CrossRef] [PubMed]
- Consonni, V.; Ballabio, D.; Todeschini, R. Evaluation of model predictive ability by external validation techniques. J. Chemom. 2010, 24, 194–201. [Google Scholar] [CrossRef]
- Fernandez, J.L.; Simpson, W.J. Measurement and prediction of the susceptibility of lager beer to spoilage by lactic acid bacteria. J. Appl. Bacteriol. 1995, 78, 419–425. [Google Scholar] [CrossRef]
- Battey, A.S.; Duffy, S.; Schaffner, D.W. Modelling mould spoilage in cold-filled ready-to-drink beverages by Aspergillus niger and Penicillium spinulosum. Food Microbiol. 2001, 18, 521–529. [Google Scholar] [CrossRef][Green Version]
- Vergouwe, Y.; Steyerberg, E.W.; Eijkemans, M.J.C.; Habbema, J.D.F. Substantial effective sample sizes were required for external validation studies of predictive logistic regression models. J. Clin. Epidemiol. 2005, 58, 475–483. [Google Scholar] [CrossRef]
 A;
A;  B), Brown ale (
B), Brown ale ( A;
A;  B), Amber ale (
B), Amber ale ( A;
A;  B), Bock (
B), Bock ( A;
A;  B), Wheat beer (
B), Wheat beer ( A;
A;  B), IPA (
B), IPA ( A;
A;  B), Porter (
B), Porter ( A;
A;  B), Stout (
B), Stout ( A;
A;  B), and Non-alcoholic beer (
B), and Non-alcoholic beer ( A). Percentage of microbial growth cases with different lowercase letters (a–j) above each column are significantly different (p < 0.05), using the LSD test. A and B identify two different brands for each beer style.
A). Percentage of microbial growth cases with different lowercase letters (a–j) above each column are significantly different (p < 0.05), using the LSD test. A and B identify two different brands for each beer style.
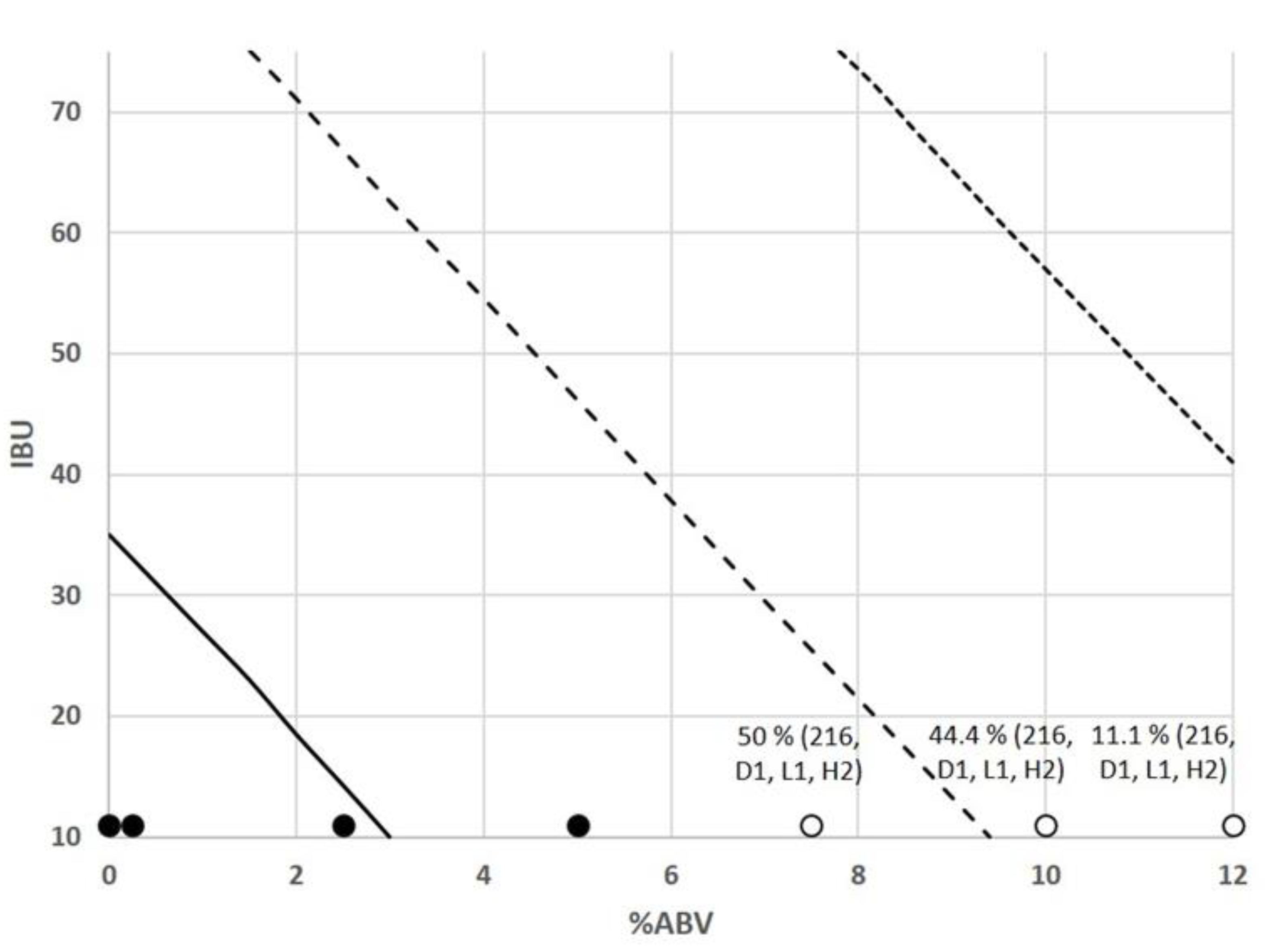
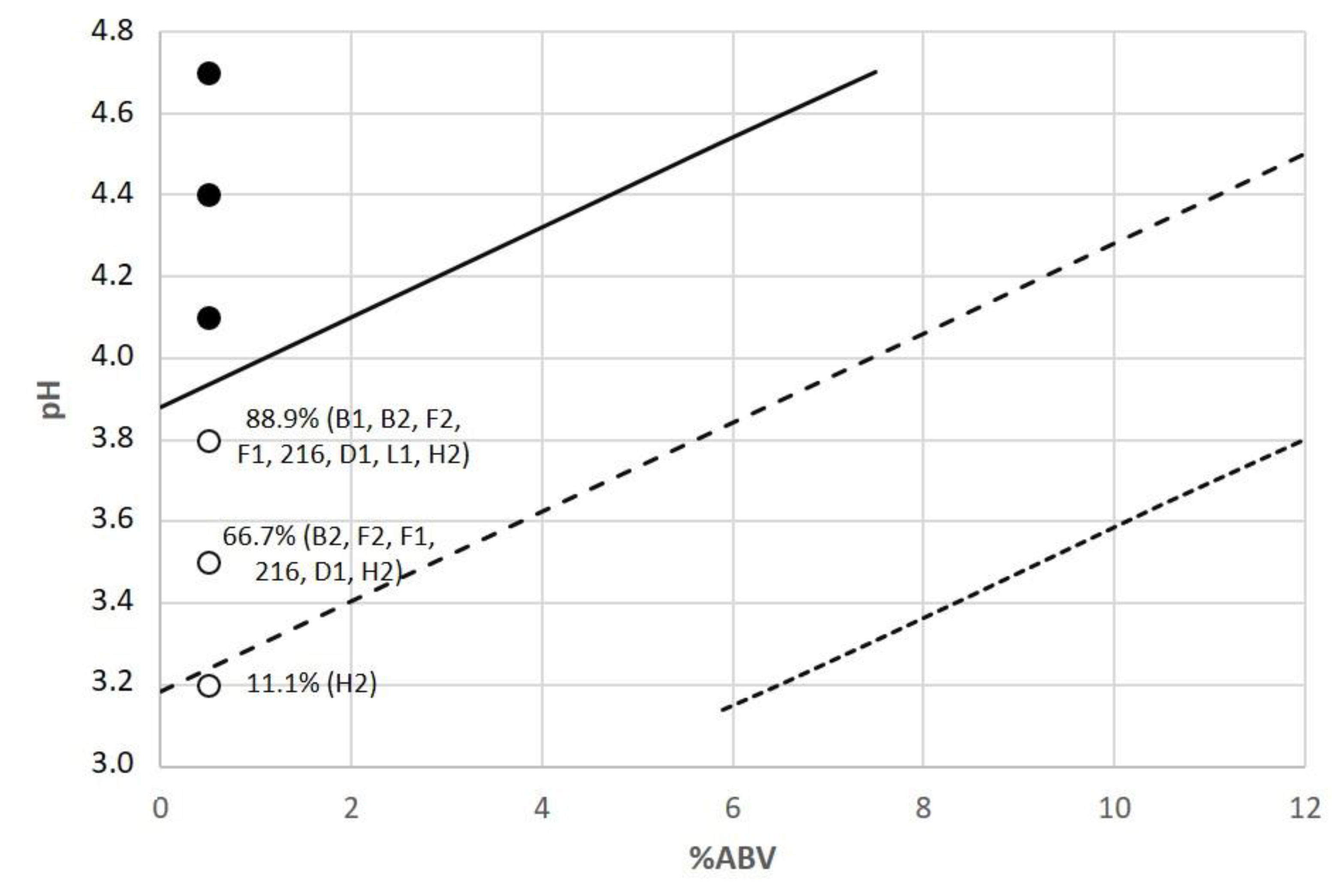
 A;
A;  B), Brown ale (
B), Brown ale ( A;
A;  B), Amber ale (
B), Amber ale ( A;
A;  B), Bock (
B), Bock ( A;
A;  B), Wheat beer (
B), Wheat beer ( A;
A;  B), IPA (
B), IPA ( A;
A;  B), Porter (
B), Porter ( A;
A;  B), Stout (
B), Stout ( A;
A;  B), and Non-alcoholic beer (
B), and Non-alcoholic beer ( A). Percentage of microbial growth cases with different lowercase letters (a–j) above each column are significantly different (p < 0.05), using the LSD test. A and B identify two different brands for each beer style.
A). Percentage of microbial growth cases with different lowercase letters (a–j) above each column are significantly different (p < 0.05), using the LSD test. A and B identify two different brands for each beer style.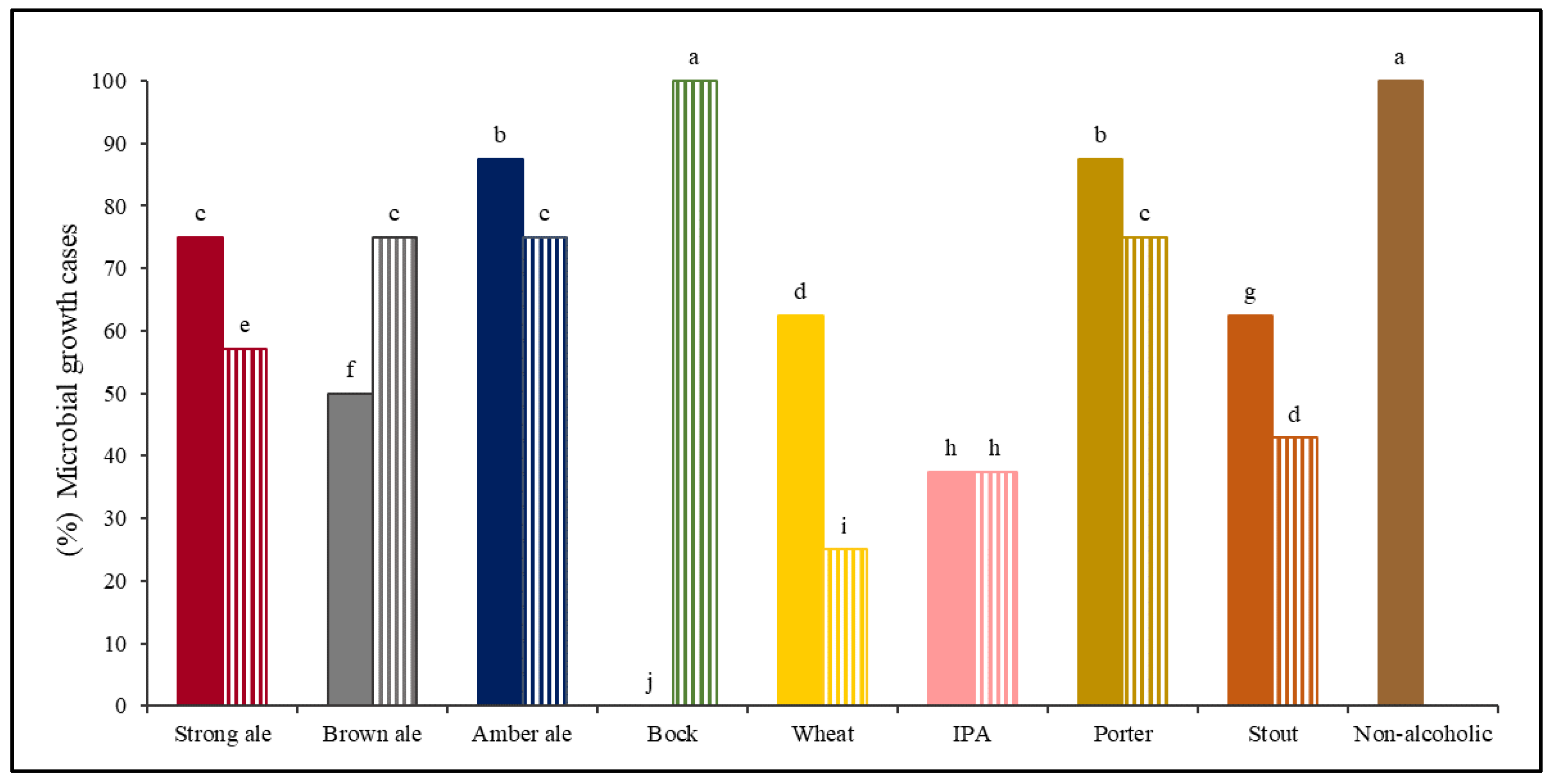
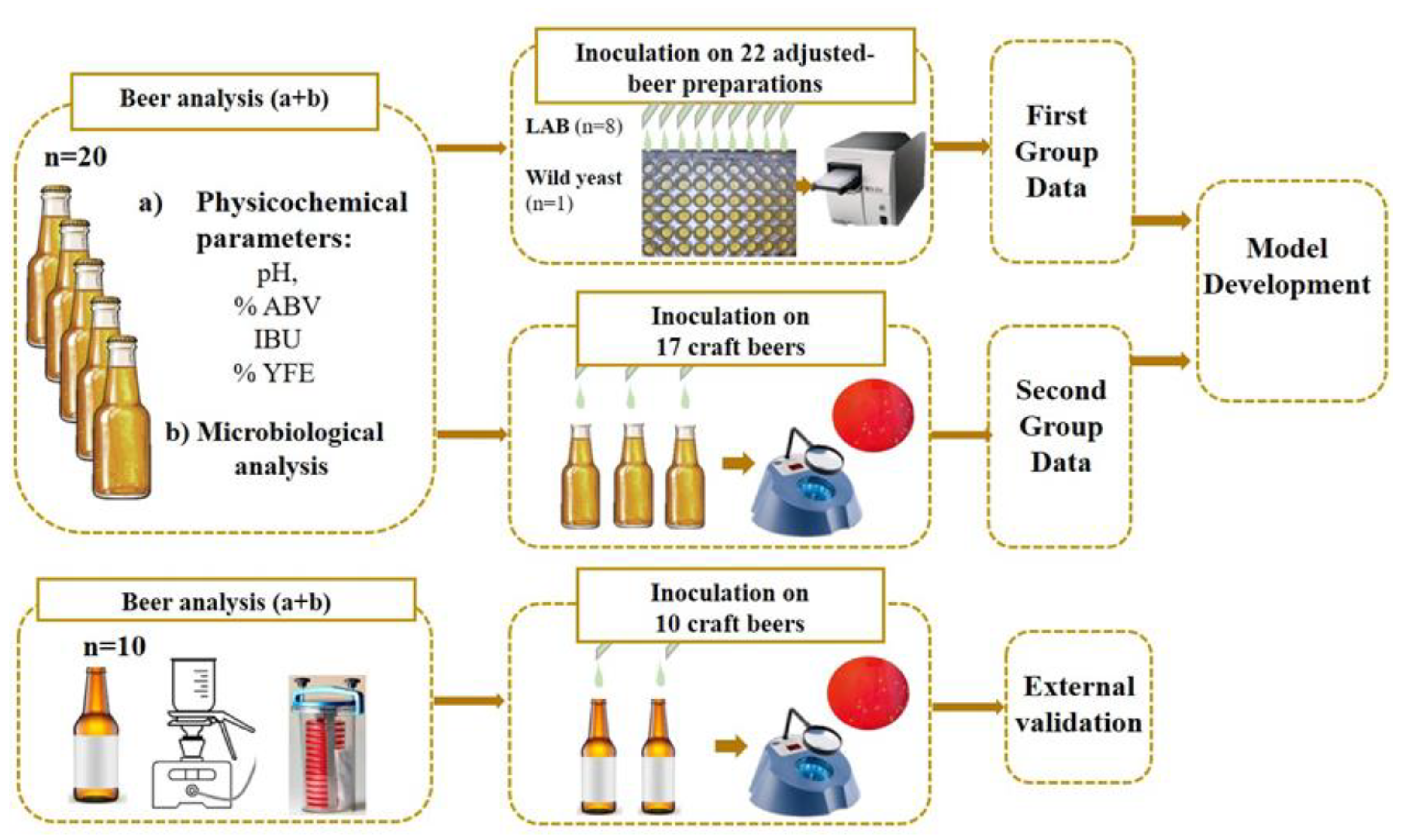
| Code | Microorganism | Original Source | Characteristics in Terms of Brewing Microbiology |
|---|---|---|---|
| L1 | Lactobacilllus brevis | Craft beer | L. brevis is the most prevalent beer spoiler causing more than a half of beer reported incidents. These three strains are hop resistant bacteria [22]. |
| D1 | Lactobacilllus brevis | Craft brewing environment | |
| 216 | Lactobacilllus brevis | Beer | |
| F2 | Pediococcus damnosus | Beer | The most common beer spoiler. |
| B6 | Lactobacillus paracasei | Craft brewing environment | L. paracasei and L. plantarum are species with relatively weak hop resistance [5]. |
| F1 | Lactobacilllus plantarum | Alcoholic drink | |
| B2 | Leuconostoc pseudomesenteroides | Craft brewing environment | Spoilage incidents caused by Leuconostoc sp. are rare except for beers with microbiologically weak features [5]. |
| B1 | Leuconostoc citreum | Craft brewing environment | |
| H2 | Dekkera bruxellensis | Lambic beer | Dekkera genus is a typical spoilage yeast for beer. |
| Beer | % ABV | pH | IBU | % YFE |
|---|---|---|---|---|
| Stout A | 4.2 ± 0.0 d | 4.27 ± 0.00 b | 31 ± 0 k | 1.37 ± 0.05 fg |
| Stout B | 9.0 ± 0.0 n | 4.37 ± 0.00 b | 44 ± 0 l | 1.26 ± 0.00 e |
| Pale ale A | 5.0 ± 0.0 e | 4.09 ± 0.00 b | 23 ± 0 h | 1.39 ± 0.05 fg |
| Pale ale B | 5.0 ± 0.0 e | 4.06 ± 0.00 b | 22 ± 0 g | 2.97 ± 0.05 k |
| Porter A | 5.9 ± 0.0 i | 4.08 ± 0.00 b | 22 ± 0 g | 1.31 ± 0.09 ef |
| Porter B | 5.0 ± 0.0 e | 4.14 ± 0.01 b | 29 ± 0 j | 0.18 ± 0.05 a |
| Brown ale A | 4.3 ± 0.0 q | 4.17 ± 0.01 b | 21 ± 0 f | 0.74 ± 0.05 c |
| Brown ale B | 5.2 ± 0.0 f | 4.46 ± 0.00 c | 44 ± 0 m | 1.45 ± 0.05 g |
| Amber ale A | 5.5 ± 0.0 g | 4.25 ± 0.01 b | 24 ± 0 h | 0.55 ± 0.08 b |
| Amber ale B | 5.8 ± 0.0 h | 4.26 ± 0.01 b | 31 ± 0 k | 0.71 ± 0.00 c |
| IPA A | 7.2 ± 0.0 l | 4.32 ± 0.01 b | 74 ± 0 n | 1.73 ± 0.01 h |
| IPA B | 8.5 ± 0.0 m | 4.70 ± 0.02 d | 75 ± 0 o | 0.92 ± 0.05 d |
| Strong ale A | 6.5 ± 0.0 j | 4.20 ± 0.02 b | 16 ± 0 e | 2.65 ± 0.04 j |
| Strong ale B | 10.0 ± 0.0 o | 4.44 ± 0.01 c | 25 ± 0 i | 1.73 ± 0.00 h |
| No-alcohol A | 0.3 ± 0.0 a | 4.20 ± 0.02 b | 11 ± 0 b | 2.37 ± 0.00 i |
| No-alcohol B | 0.5 ± 0.0 b | 4.49 ± 0.00 c | 16 ± 0 e | 3.05 ± 0.05 k |
| Wheat beer A | 5.5±0.0 g | 4.29 ± 0.01 b | 15 ± 0 d | 1.00 ± 0.05 d |
| Wheat beer B | 3.8 ± 0.0 c | 3.14 ± 0.03 a | 12 ± 0c | 2.37 ± 0.00 i |
| Bock A | 12.0 ± 0.0 p | 4.66 ± 0.15 d | 25 ± 0i | 1.68 ± 0.12 h |
| Bock B | 7.0 ± 0.0l | 4.23 ± 0.01 b | 10 ± 0a | 0.76 ± 0.05 c |
| Nagelkerke R2 | HL Goodness-of-Fit Test | % Correctly Predicted in Classification Table | Parameter | Coefficients | Standard Error | Wald | Statistical Significance | Odds Ratio | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.41 | 0.133 | 83.4% | Constant | −9.608 | 2.030 | 22.397 | 0.000 | 0.000 | 87% | 71% |
| % ABV | −0.346 | 0.049 | 50.429 | 0.000 | 0.708 | |||||
| IBU | −0.042 | 0.010 | 16.834 | 0.000 | 0.959 | |||||
| pH | 3.161 | 0.540 | 34.306 | 0.000 | 23.58 |
| Observed | Predicted | ||
|---|---|---|---|
| Value | % Correct | ||
| Not Easy to Spoil | Easy to Spoil | ||
| Not easy to spoil | 1 | 0 | 100 |
| Easy to spoil | 0 | 9 | 100 |
| % Global | 100 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Saavedra, M.; Pérez-Revelo, K.; Valero, A.; Moreno-Arribas, M.V.; González de Llano, D. A Binary Logistic Regression Model as a Tool to Predict Craft Beer Susceptibility to Microbial Spoilage. Foods 2021, 10, 1926. https://doi.org/10.3390/foods10081926
Rodríguez-Saavedra M, Pérez-Revelo K, Valero A, Moreno-Arribas MV, González de Llano D. A Binary Logistic Regression Model as a Tool to Predict Craft Beer Susceptibility to Microbial Spoilage. Foods. 2021; 10(8):1926. https://doi.org/10.3390/foods10081926
Chicago/Turabian StyleRodríguez-Saavedra, Magaly, Karla Pérez-Revelo, Antonio Valero, M. Victoria Moreno-Arribas, and Dolores González de Llano. 2021. "A Binary Logistic Regression Model as a Tool to Predict Craft Beer Susceptibility to Microbial Spoilage" Foods 10, no. 8: 1926. https://doi.org/10.3390/foods10081926
APA StyleRodríguez-Saavedra, M., Pérez-Revelo, K., Valero, A., Moreno-Arribas, M. V., & González de Llano, D. (2021). A Binary Logistic Regression Model as a Tool to Predict Craft Beer Susceptibility to Microbial Spoilage. Foods, 10(8), 1926. https://doi.org/10.3390/foods10081926








