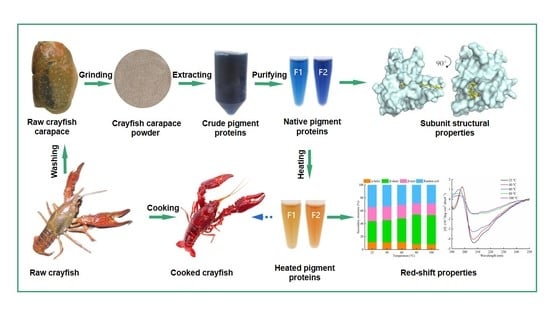
Purification and Characterisation of Two Novel Pigment Proteins from the Carapace of Red Swamp Crayfish (Procambarus clarkii)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material and Chemicals
2.2. Crude Protein Preparation
2.3. Targeted Protein Purification
2.4. Molecular Weight Determination
2.5. Colour Measurement
2.6. SDS-PAGE Analysis
2.7. LC-MS/MS Analysis
2.8. cDNA Cloning and Red-Change-Related Pigment Protein Gene Sequencing
2.9. Circular Dichroism (CD) Spectrophotometry
2.10. Bioinformatics Analysis
2.11. Statistical Analysis
3. Results and Discussion
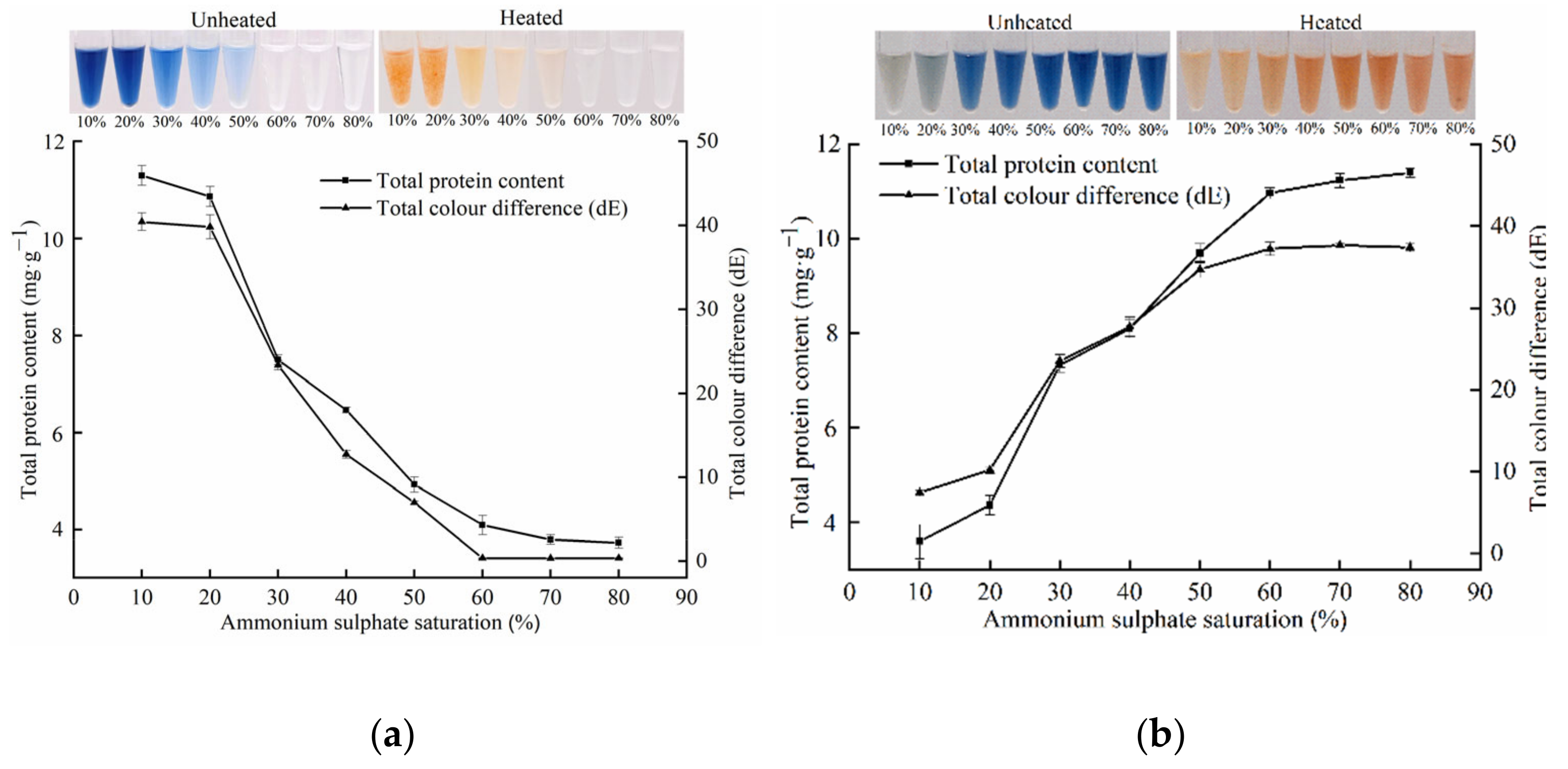
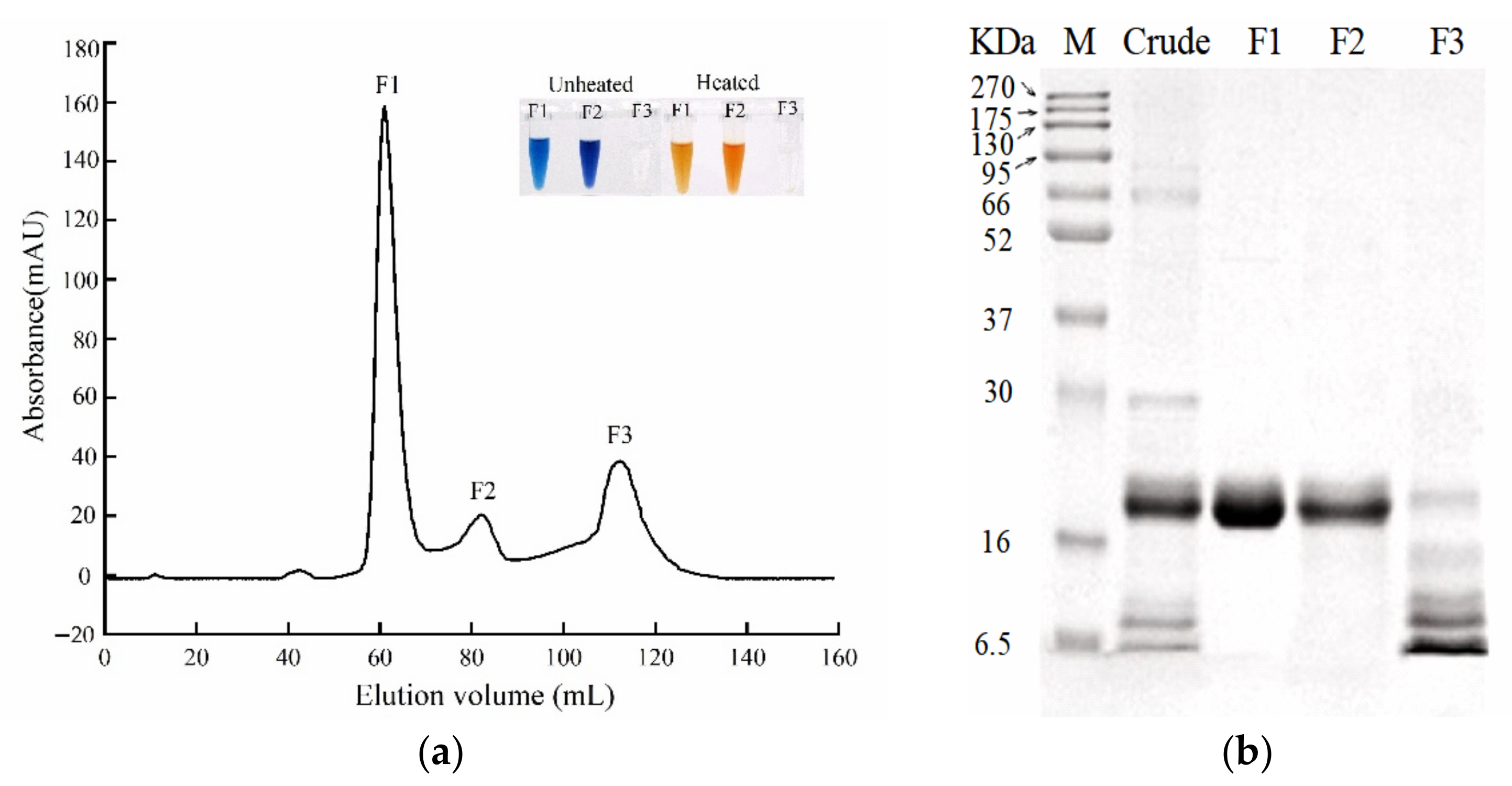
3.1. Pigment Protein Extraction and Purification
3.2. Pigment Protein Identification, cDNA Cloning and Sequence Analysis
3.3. Pigment Protein Colour Change and Secondary Structure Change
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parisenti, J.; Beirão, L.H.; Tramonte, V.L.; Ourique, F.; Brito, C.C.; Moreira, C.C. Preference ranking of colour in raw and cooked shrimps. Int. J. Food Sci. Technol. 2011, 46, 2558–2561. [Google Scholar] [CrossRef]
- Miraglia, D.; Castrica, M.; Esposto, S.; Roila, R.; Selvaggini, R.; Urbani, S.; Taticchi, A.; Sordini, B.; Veneziani, G.; Servili, M. Quality evaluation of shrimp (Parapenaeus longirostris) treated with phenolic extract from olive vegetation water during shelf-Life, before and after cooking. Foods 2021, 10, 2116. [Google Scholar] [CrossRef]
- Li, Q.Q.; Zu, L.; Cheng, Y.X.; Wade, N.M.; Liu, J.G.; Wu, X. Carapace color affects carotenoid composition and nutritional quality of the Chinese mitten crab, Eriochier sinensis. LWT Food Sci. Technol. 2020, 126, 109286. [Google Scholar] [CrossRef]
- Tume, R.K.; Sikes, A.L.; Tabrett, S.; Smith, D.M. Effect of background colour on the distribution of astaxanthin in black tiger prawn (Penaeus monodon): Effective method for improvement of cooked colour. Aquaculture 2009, 296, 129–135. [Google Scholar] [CrossRef]
- Chayen, N.E.; Cianci, M.; Grossmann, J.G.; Habash, J.; Helliwell, J.R.; Nneji, G.A.; Raftery, J.; Rizkallah, P.J.; Zagalsky, P.F. Unravelling the structural chemistry of the colouration mechanism in lobster shell. Acta Crystallogr. D 2003, 59, 2072–2082. [Google Scholar] [CrossRef]
- Timme, E.; Walwyn, D.; Bailey, A. Characterisation of the carotenoprotein found in carapace shells of Jasus lalandii. Comp. Biochem. Phys. B 2009, 153, 39–42. [Google Scholar] [CrossRef]
- Pan, C.; Ishizaki, S.; Nagashima, Y.; Gao, J.; Watabe, S. Isolation and cDNA cloning of a novel red colour-related pigment-binding protein derived from the shell of the shrimp, Litopenaeus vanname. Food Chem. 2018, 241, 104–112. [Google Scholar] [CrossRef]
- Begum, S.; Cianci, M.; Durbeej, B.; Falklof, O.; Hadener, A.; Helliwell, J.R.; Helliwell, M.; Regan, A.C.; Watt, C. On the origin and variation of colours in lobster carapace. Phys. Chem. Chem. Phys. 2015, 17, 16723–16732. [Google Scholar] [CrossRef] [Green Version]
- Gamiz-Hernandez, A.P.; Angelova, I.N.; Send, R.; Sundholm, D.; Kaila, V.R.I. Protein-induced colour shift of carotenoids in β-crustacyanin. Angew. Chem. Int. Ed. 2015, 54, 11564–11566. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Ishizaki, S.; Nagashima, Y.; Watabe, S. Functional and structural properties of red colour-related pigment-binding protein from the shell of litopenaeus vannamei. J. Sci. Food Agric. 2019, 99, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, J.R. The structural chemistry and structural biology of colouration in marine crustacea. Crystallogr. Rev. 2010, 16, 231–242. [Google Scholar] [CrossRef]
- Tlusty, M.F.; Metzler, A.; Huckabone, S.; Suanda, S.; Guerrier, S. Morphological colour change in the American lobster (Homarus americanus) in response to background colour and UV light. N. Z. J. Mar. Freshw. Res. 2009, 43, 247–255. [Google Scholar] [CrossRef]
- Wade, N.M.; Budd, A.; Irvin, S.; Glencross, B.D. The combined effects of diet, environment and genetics on pigmentation in the giant tiger prawn, penaeus monodon. Aquaculture 2015, 449, 78–86. [Google Scholar] [CrossRef]
- Kato, T.; Strakova, K.; García-Calvo, J.; Sakai, N.; Matile, S. Mechanosensitive fluorescent probes, changing color like lobsters during cooking: Cascade switching variations. Bull. Chem. Soc. Jpn. 2020, 93, 1401–1411. [Google Scholar] [CrossRef]
- Ando, M.; Fukai, T.; Kawasaki, K.; Itoh, T.; Tsukamasa, Y. Improvement of red color development on the surface of kuruma prawn marsupenaeus japonicus under various conditions. J. Food Sci. 2014, 79, E202–E207. [Google Scholar] [CrossRef] [PubMed]
- Wald, G.; Nathanson, N. Crustacyanin, the blue carotenoid-protein of the lobster shell. Biol Bull. 1948, 95, 249. [Google Scholar]
- Buchwald, M.; Jencks, W.P. Properties of the crustacyanins and the yellow lobster shell pigment. Biochemistry 1968, 7, 844. [Google Scholar] [CrossRef] [PubMed]
- Budd, A.M.; Hinton, T.M.; Tonks, M.; Cheers, S.; Wad, N.M. Rapid expansion of pigmentation genes in penaeid shrimp with absolute preservation of function. J. Exp. Biol. 2017, 220, 4109–4118. [Google Scholar] [CrossRef] [Green Version]
- Pan, C.; Ishizaki, S.; Chen, S.; Hao, S.; Zhou, J.; Yang, X. Purification, characterization and antibacterial activities of red colour-related protein found in the shell of kuruma shrimp. Marsupenaeus japonicus. Food Chem. 2020, 310, 125819. [Google Scholar] [CrossRef] [PubMed]
- Bureau of Fisheries and Fishery Management. China Fisheries Statistical Yearbook in 2021; Chinese Agricultural Press: Beijing, China, 2021; pp. 24–25.
- Pan, C.; Liang, X.L.; Chen, S.J.; Tao, F.Y.; Yang, X.Q.; Cen, J.W. Red colour–related proteins from the shell of red swamp crayfish (Procambarus clarkii): Isolation, identification and bioinformatic analysis. Food Chem. 2020, 327, 127079. [Google Scholar] [CrossRef]
- Zagalsky, P.F. Invertebrate carotenoproteins. Method. Enzymol. 1985, 111, 216–247. [Google Scholar]
- Bollineni, R.C.; Guldvik, I.J.; Grnberg, H.; Wiklund, F.; Thiede, B. A differential protein solubility approach for the depletion of highly abundant proteins in plasma using ammonium sulfate. Analyst 2015, 140, 8109–8117. [Google Scholar] [CrossRef] [PubMed]
- Galetovi, A.; Seura, F.; Gallardo, V.; Graves, R.; Cortes, J.; Valdivia, C.; Nunez, J.; Tapia, C.; Neira, T.; Sanzana, S.; et al. Use of phycobiliproteins from atacama cyanobacteria as food colorants in a dairy beverage prototype. Foods 2020, 9, 244. [Google Scholar] [CrossRef] [Green Version]
- Krawczyk, S.; Britton, G. A study of protein-carotenoid interactions in the astaxanthin-protein crustacyanin by absorption and stark spectroscopy; evidence for the presence of three spectrally distinct species. BBA Biomembr. 2001, 1544, 301–310. [Google Scholar] [CrossRef]
- Garate, A.M.; Milicua, J.C.; Gomez, R.; Macarullaa, J.M.; Brittonb, G. Purification and characterization of the blue carotenoprotein from the caparace of the crayfish Procambarus clarkii (Girard). BBA Gen. Subjects. 1986, 881, 446–455. [Google Scholar] [CrossRef]
- Klomklao, S.; Benjakul, S.; Visessanguan, W.; Kishimura, H.; Simpson, B.K. Extraction of carotenoprotein from black tiger shrimp shells with the aid of bluefish trypsin. J. Food Biochem. 2009, 33, 201–217. [Google Scholar] [CrossRef]
- Wade, N.M.; Anderson, M.; Sellars, M.J.; Tume, R.K.; Preston, N.P.; Glencross, B.D. Mechanisms of colour adaptation in the prawn penaeus monodon. J. Exp. Bio. 2012, 215, 343–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garate, A.M.; Urrechaga, E.; Milicua, J.; Gomeza, R.; Brittonb, G. A blue carotenoprotein from the carapace of the crab, Carcinus maenas. Comp. Biochem. Phys. B 1984, 77, 605–608. [Google Scholar] [CrossRef]
- Czeczuga, B.; Czeczuga-Semeniuk, E.; Semeniuk, A. Carotenoids and carotenoproteins in Asellus aquaticus L. (Crustacea: Isopoda). Folia Biol. 2005, 53, 109–114. [Google Scholar] [CrossRef]
- Drozdova, P.; Saranchina, A.; Morgunova, M.; Kizenko, A.; Lubyaga, Y.; Baduev, B.; Timofeyev, M. The level of putative carotenoid-binding proteins determines the body colour in two species of endemic Lake Baikal amphipods. Peer J. 2020, 8, 9387. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, B.; Mishra, M.; Srinivasan, N.; Archunan, G. Structure-based phylogenetic analysis of the lipocalin superfamily. PLoS ONE 2015, 10, e0135507. [Google Scholar]
- Keen, J.N.; Caceres, I.; Eliopoulos, E.E.; Zagalsky, P.F.; Findlay, J.B.C. Complete sequence and model for the A2 subunit of the carotenoid pigment complex, crustacyanin. Eur. J. Biochem. 1991, 197, 407–417. [Google Scholar] [CrossRef]
- Cianci, M.; Rizkallah, P.J.; Olczak, A.; Raftery, J.; Chayen, N.E.; Zagalsky, P.F.; Helliwell, J.R. Structure of lobster apocrustacyanin A1 using softer x-rays. Acta Crystallogr. D 2001, 57, 1219–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durbeej, B.; Eriksson, L.A. On the bathochromic shift of the absorption by astaxanthin in crustacyanin: A quantum chemical study. Chem Phys. Lett. 2003, 375, 30–38. [Google Scholar] [CrossRef]
- Britton, G.; Weesie, R.J.; Askin, D.; Warburton, J.D.; Gallardo-Guerrero, L.; Jansen, F.J.H.M.; Groot, H.D.; Lugtenburg, J.; Cornard, J.P.; Merlin, J.C. Carotenoid blues: Structural studies on carotenoproteins. Pure Appl. Chem. 1997, 69, 2075–2084. [Google Scholar] [CrossRef] [Green Version]
- Lin, D.; Lin, W.; Gao, G.; Zhou, J.; Chen, T.; Ke, L.; Rao, P.; Wang, Q. Purification and characterization of the major protein isolated from semen armeniacae amarum and the properties of its thermally induced nanoparticles. Int. J. Biol. Macromol. 2020, 159, 850–858. [Google Scholar] [CrossRef]
- Trigui, I.; Zarai, Z.; Chevance, S.; Cheikh-Rouhou, S.; Attia, H.; Ayadi, M.A. Physicochemical properties, antioxidant activity and in vitro gastrointestinal digestion of purified proteins from black cumin seeds. Int. J. Biol. Macromol. 2018, 126, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.I.; Nsonzi, F.; Sedman, J.; Ismail, A.A. Enhanced unfolding of bovine β-lactoglobulin structure using microwave treatment: A multi-spectroscopic study. Food Biophys. 2016, 11, 370–379. [Google Scholar] [CrossRef]
- Ferrari, M.; Folli, C.; Pincolini, E.; McClintock, T.S.; Rossle, M.; Berni, R.; Cianci, M. Structural characterization of recombinant crustacyanin subunits from the lobster Homarus Americanus. Acta Crystallogr. F 2012, 68, 846–853. [Google Scholar] [CrossRef] [Green Version]





| Fraction | Accession | Mass | Score | Description |
|---|---|---|---|---|
| F1 | ALC79588.1 | 21,541 | 821 | Crustacyanin A (C. quadricarinatus) |
| ASY04980.1 | 20,783 | 218 | Crustacyanin A1, partial (Penaeus longistylus) | |
| pdb|1GKA|B | 19,942 | 146 | Chain B, The Molecular Basis of the Colouration Mechanism in Lobster Shell. Beta-Crustacyanin at 3.2 A Resolution | |
| ACL37121.1 | 7583 | 134 | Crustacyanin C, partial (C. quadricarinatus) | |
| ASY04979.1 | 20,871 | 131 | Crustacyanin A1, partial (Penaeus esculentus) | |
| ROT83799.1 | 21,513 | 118 | Crustacyanin subunit A (P. vannamei) | |
| F2 | ALC79588.1 | 21,541 | 445 | Crustacyanin A (C. quadricarinatus) |
| ASY04980.1 | 20,783 | 223 | Crustacyanin A1, partial (Penaeus longistylus) | |
| pdb|1GKA|B | 19,942 | 209 | Chain B, The Molecular Basis of the Colouration Mechanism in Lobster Shell. Beta-Crustacyanin at 3.2 Å Resolution | |
| ACL37112.1 | 21,731 | 196 | Crustacyanin-A1 precursor (Panulirus cygnus) |
| Temperature (°C) | L* | a* | b* | dE |
|---|---|---|---|---|
| 25 | 59.50 ± 0.10 d | −12.57 ± 0.12 d | −25.67 ± 0.06 d | |
| 40 | 59.90 ± 0.20 d | −12.23 ± 0.15 d | −25.37 ± 0.15 d | 0.60 |
| 60 | 61.70 ± 0.36 c | 1.13 ± 0.15 c | 7.17 ± 0.15 c | 35.65 |
| 80 | 70.77 ± 0.47 b | 23.53 ± 0.35 b | 24.13 ± 0.31 b | 62.18 |
| 100 | 74.40 ± 0.30 a | 26.03 ± 0.21 a | 25.70 ± 0.10 a | 65.50 |
| Temperature | L* | a* | b* | α-Helices | β-Sheets | β-Turns | Random Coils | |
|---|---|---|---|---|---|---|---|---|
| Temperature | 1 | 0.950 * | 0.962 ** | 0.951 * | −0.890 * | 0.950 * | −0.927 * | −0.954 * |
| L* | 1 | 0.974 ** | 0.896 * | −0.976 ** | 0.969 ** | −0.969 ** | −0.910 * | |
| a* | 1 | 0.968 ** | −0.932 * | 0.988 ** | −0.982 ** | −0.973 ** | ||
| b* | 1 | −0.815 | 0.938 * | −0.917 * | −0.980 ** | |||
| α-Helices | 1 | −0.958 * | 0.968 ** | 0.868 | ||||
| β-Sheets | 1 | −0.997 ** | −0.973 ** | |||||
| β-Turns | 1 | 0.957 * | ||||||
| Random coils | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.; Ji, H.; Pan, C.; Zhang, D.; Su, W.; Liu, S.; Deng, Y.; Huang, X. Purification and Characterisation of Two Novel Pigment Proteins from the Carapace of Red Swamp Crayfish (Procambarus clarkii). Foods 2022, 11, 35. https://doi.org/10.3390/foods11010035
Chen H, Ji H, Pan C, Zhang D, Su W, Liu S, Deng Y, Huang X. Purification and Characterisation of Two Novel Pigment Proteins from the Carapace of Red Swamp Crayfish (Procambarus clarkii). Foods. 2022; 11(1):35. https://doi.org/10.3390/foods11010035
Chicago/Turabian StyleChen, Hao, Hongwu Ji, Chuang Pan, Di Zhang, Weiming Su, Shucheng Liu, Yijia Deng, and Xiaodan Huang. 2022. "Purification and Characterisation of Two Novel Pigment Proteins from the Carapace of Red Swamp Crayfish (Procambarus clarkii)" Foods 11, no. 1: 35. https://doi.org/10.3390/foods11010035