Novel Antioxidant Peptides from Grateloupia livida Hydrolysates: Purification and Identification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of G. Livida Protein
2.3. Preparation of G. Livida Protein Hydrolysates
2.4. Determination of Degree of Hydrolysis
2.5. Determination of Antioxidant Activity
2.5.1. Determination of Reducing Power
2.5.2. Determination of DPPH Radical Scavenging Rate
2.5.3. Determination of ABTS Radical Scavenging Rate
2.6. High-Performance Size Exclusion Chromatography
2.7. Reversed-Phase High-Performance Liquid Chromatography (RP-HPLC)
2.8. Identification of the Amino Acid Sequence by LC-MS/MS
2.9. Property Prediction of the Identified Peptides and Their Antioxidant Ability
2.10. Molecular Docking Analysis
2.11. Statistical Analysis
3. Results and Discussion
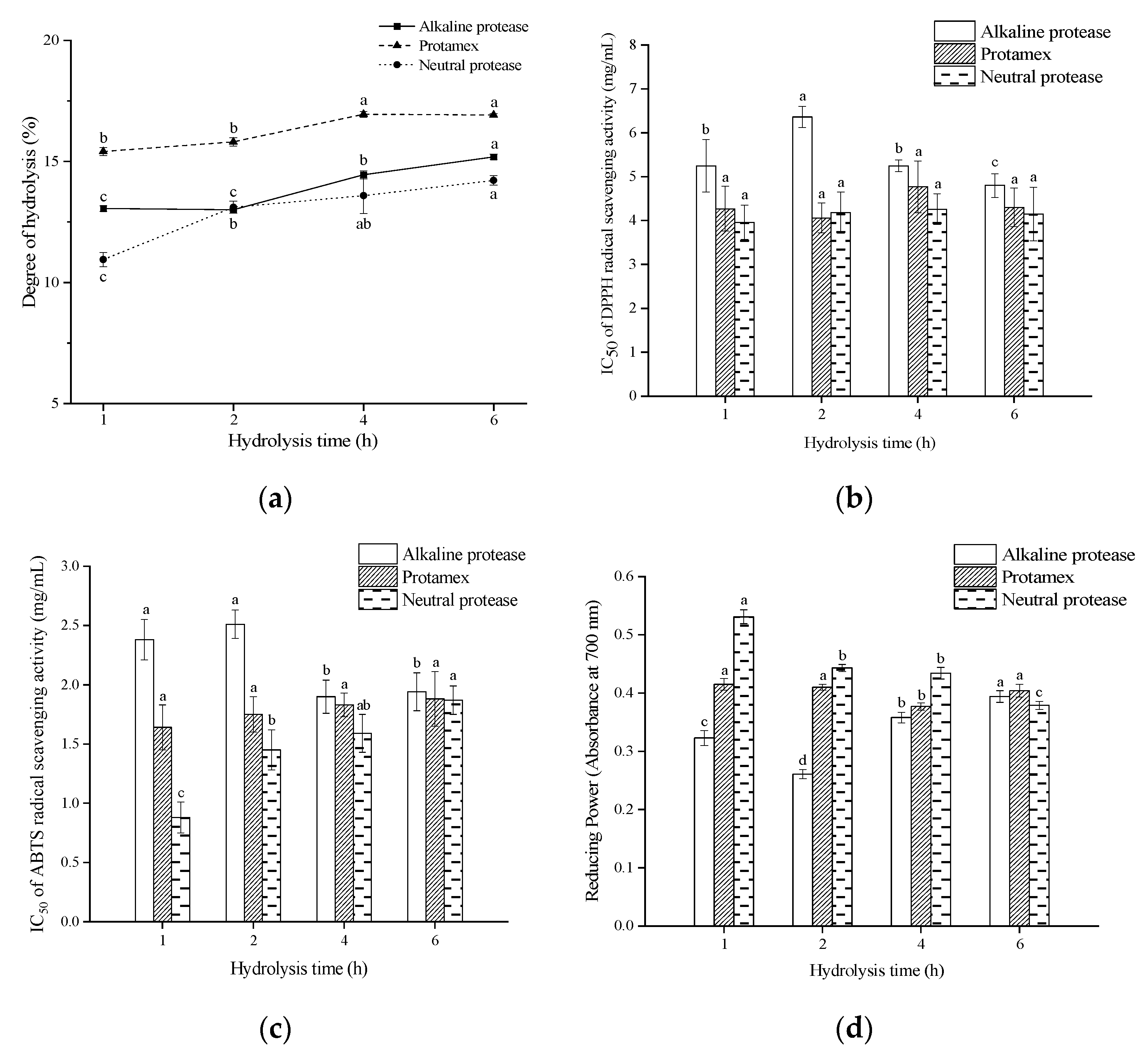
3.1. DH and Antioxidant Activity of the Protein Enzymatic Hydrolysates
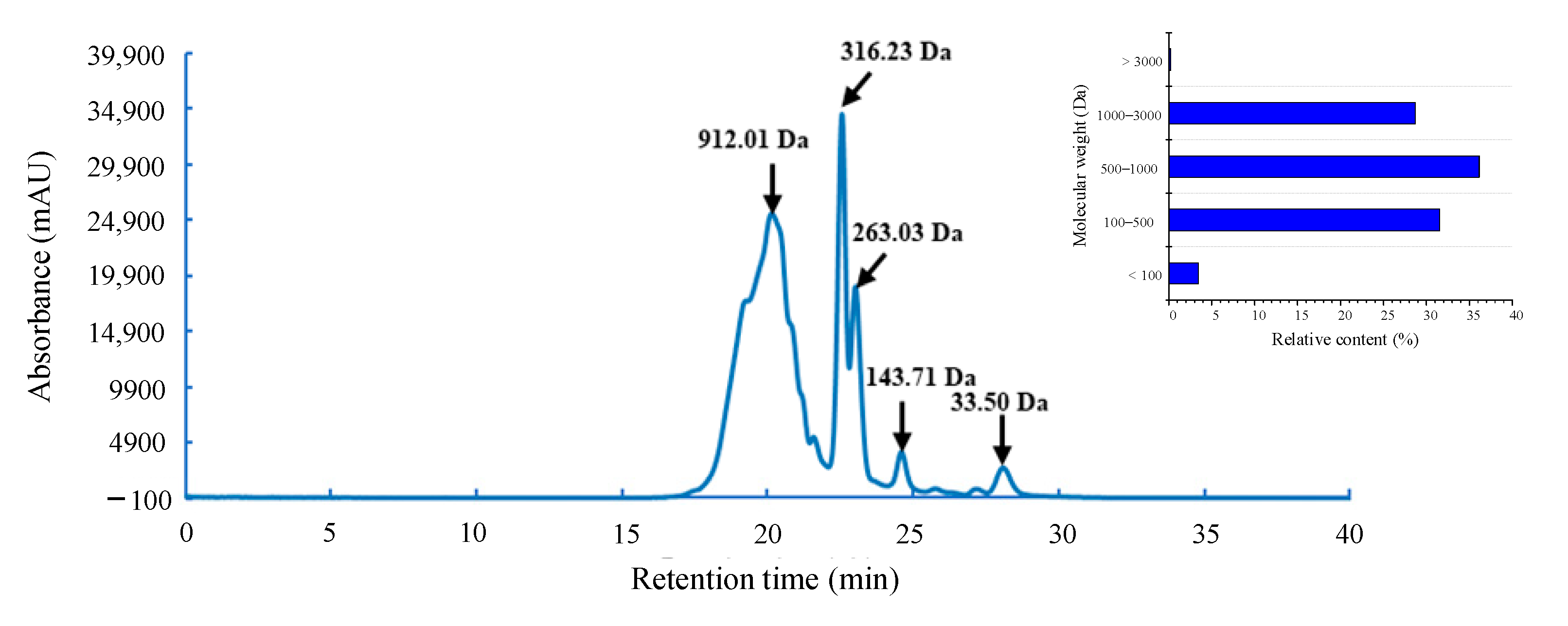
3.2. Molecular Weight Distribution of the Enzymatic Hydrolysate
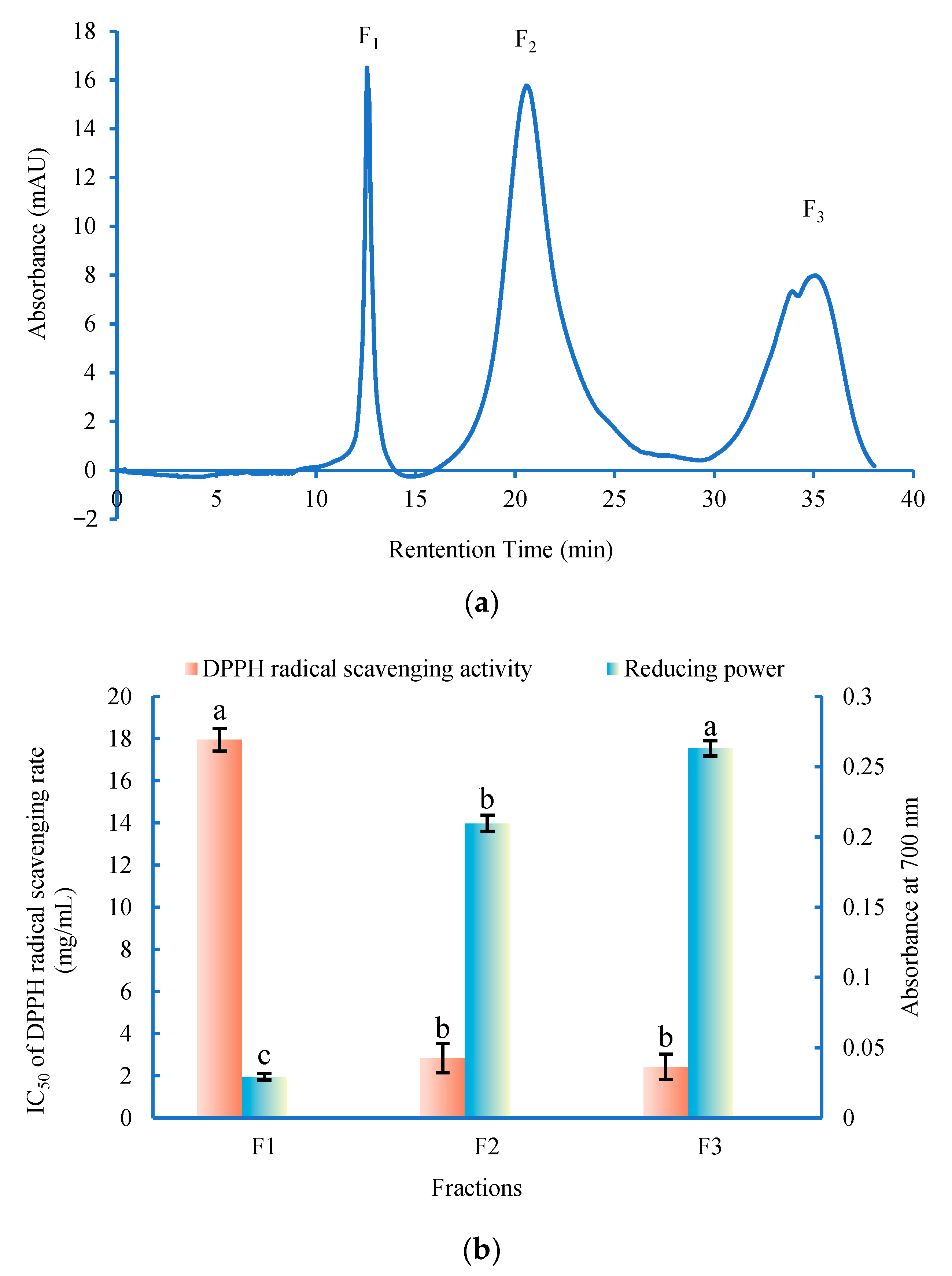
3.3. Semi-Preparative RP-HPLC Purification
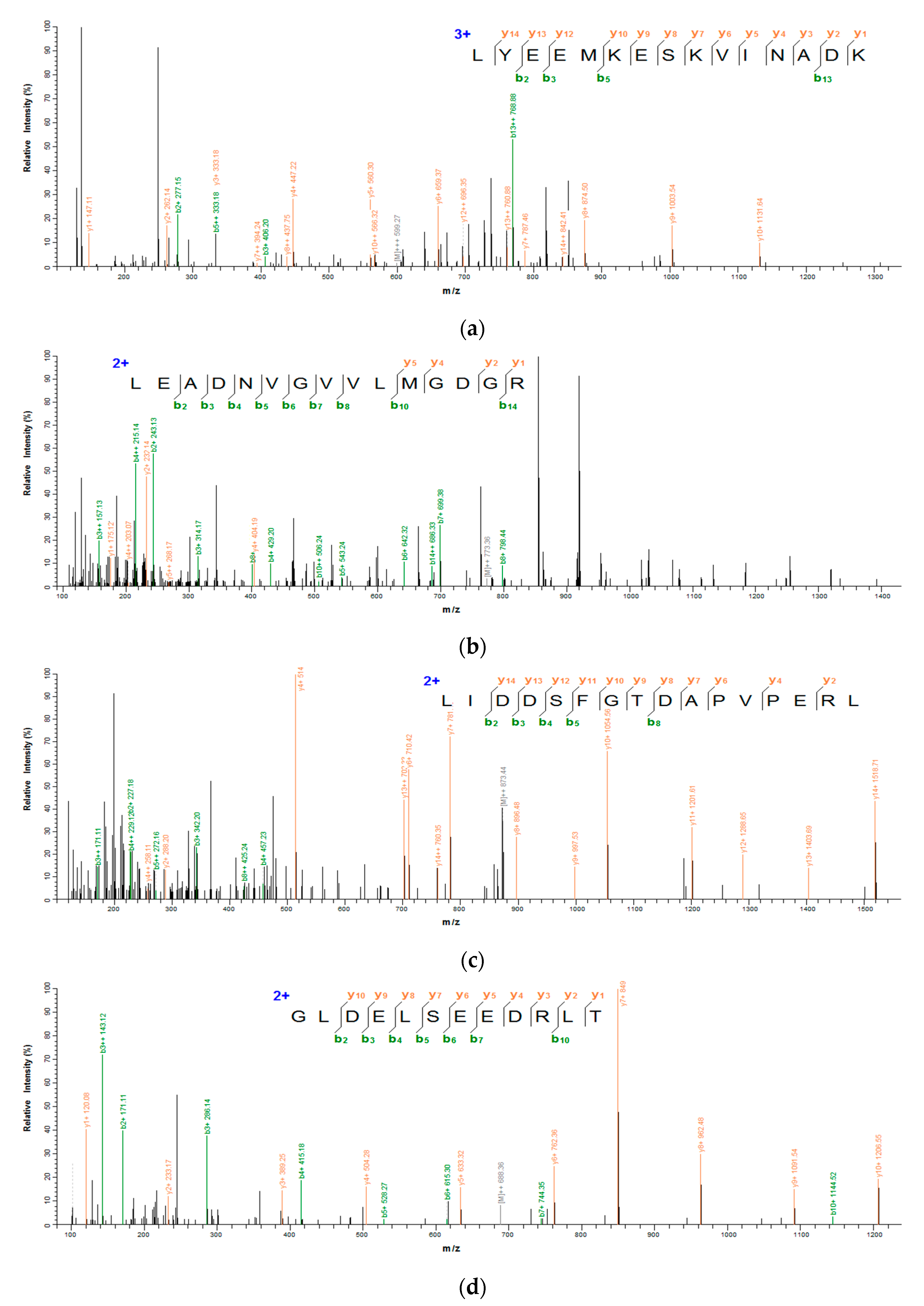
3.4. Identification of Antioxidant Peptide by LC-MS/MS
3.5. Physical and Chemical Properties of the Identified Peptides
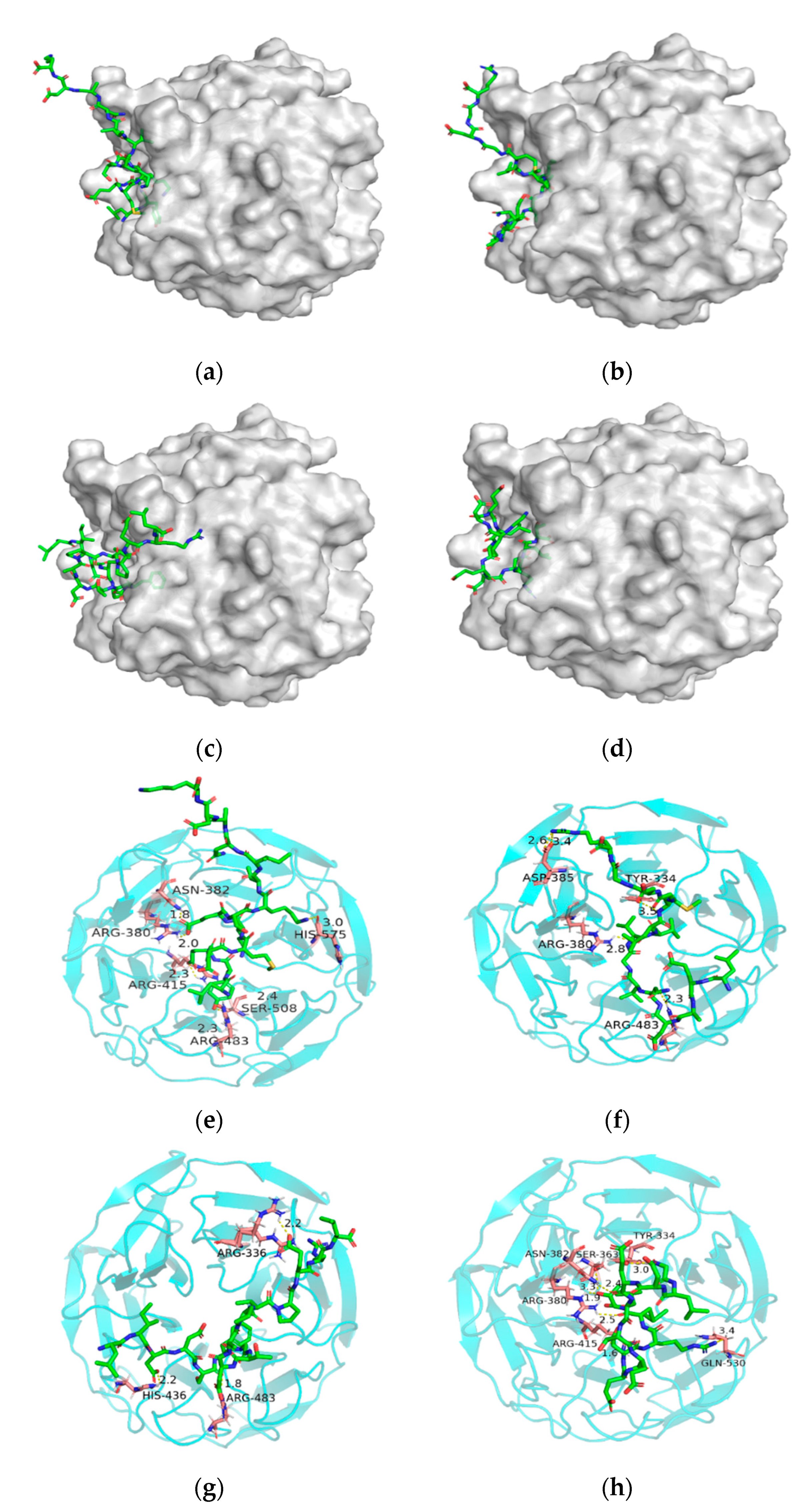
3.6. Molecular Docking Results of the Identified Peptides
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qian, S.; Xu, Y.; Gu, Y. Combination of spin-trapping, LC/ESR and LC/MS technique in characterization of PUFA-derived free radicals in lipid peroxidation. Acta Biophys. Sin. 2012, 28, 355–372. [Google Scholar] [CrossRef]
- Wang, J.; Hao, Y.; Li, S. Rapid determination of three synthetic antioxidants: TBHQ, BHA and BHT in vegetable oils electroanalytical method. J. Chin. Cereal Oil Assoc. 2018, 33, 126–132. [Google Scholar]
- Piu, L.D.; Tassoni, A.; Serrazanetti, D.I.; Ferri, M.; Babini, E.; Tagliazucchi, D.; Gianotti, A. Exploitation of starch industry liquid by-product to produce bioactive peptides from rice hydrolyzed proteins. Food Chem. 2014, 155, 199–206. [Google Scholar]
- Barba, F.J.; Criado, M.N.; Belda-Galbis, C.M.; Esteve, M.J.; Rodrigo, D. Stevia rebaudiana Bertoni as a natural antioxidant/antimicrobial for high pressure processed fruit extract: Processing parameter optimization. Food Chem. 2014, 148, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, C.; Gallagher, E.; Tasdemir, D.; Hayes, M. Heart health peptides from macroalgae and their potential use in functional foods. J. Agric. Food Chem. 2011, 59, 6829–6836. [Google Scholar] [CrossRef]
- Montone, C.M.; Chiozzi, R.Z.; Marchetti, N.; Cerrato, A.; Antonelli, M.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Laganà, A. Peptidomic approach for the identification of peptides with potential antioxidant and anti-hyperthensive effects derived from asparagus by-products. Molecules 2019, 24, 3627. [Google Scholar] [CrossRef] [Green Version]
- Jie, Y.; Hu, Y.; Xue, M.; Dun, Y.; Zhao, S. Purification and identification of antioxidant peptides from enzymatic hydrolysate of Spirulina platensis. J. Microbiol. Biotechnol. 2016, 26, 1216–1223. [Google Scholar]
- Garzon, R.; Garzón, A.; Betancur, D.; Guerrero, L.; Drago, S. Hydrolyzates from Pyropia columbina seaweed have antiplatelet aggregation, antioxidant and ACE I inhibitory peptides which maintain bioactivity after simulated gastrointestinal digestion. LWT-Food Sci. Technol. 2015, 64, 881–888. [Google Scholar]
- Hu, X.; Yang, X.; Wu, Q.; Li, L.; Wu, Y.; Chen, S.; Li, R.; Ren, J. Purification and identification of antioxidant peptides from Schizochytrium limacinum hydrolysates by consecutive chromatography and electrospray ionization-mass spectrometry. Molecules 2019, 24, 3004. [Google Scholar] [CrossRef] [Green Version]
- Larsen, R.; Eilertsen, K.; Elvevoll, E. Health benefits of marine foods and ingredients. Biotechnol. Adv. 2011, 29, 508–518. [Google Scholar] [CrossRef]
- Alashi, A.; Blanchard, C.; Mailer, R.; Agboola, S.; Mawson, J.; Rong, H.; Girgih, A.; Aluko, R. Antioxidant properties of Australian canola meal protein hydrolysates. Food Chem. 2014, 146, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.; Freitas, A.; Pereira, L.; Tap, R.; Duarte, A. Chemical composition of red, brown and green macroalgae from Buarcos bay in Central West Coast of Portugal. Food Chem. 2015, 183, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Park, J.; Shim, H.; Kwon, Y.; Kim, H.; Shin, H. Antioxidative effect of phycoerythrin derived from Grateloupia filicina on rat primary astrocytes. Biotechnol. Bioprocess Eng. 2016, 21, 676–682. [Google Scholar] [CrossRef]
- Oyaizu, M. Antioxidative activities of browning products of glucosamine fractionated by organic solvent and thin-layer chromatography. Nippon Shokuhin Kogyo Gakkaishi. 1988, 35, 771–775. [Google Scholar] [CrossRef]
- Boath, A.; Stewart, D.; Mcdougall, G. Berry components inhibit α-glucosidase in vitro: Synergies between acarbose and polyphenols from black currant and rowanberry. Food Chem. 2012, 135, 929–936. [Google Scholar] [CrossRef]
- Hadjadj, N.; Hazzit, M. Analysis and antioxidant activity of essential oils and methanol extracts of Origanum floribundum munby. J. Essent. Oil-Bear. Plants 2020, 23, 85–96. [Google Scholar] [CrossRef]
- Gendis, M.; Marsono, Y.; Indrati, R. In vitro gastrointestinal simulation of tempe prepared from koro kratok (Phaseolus lunatus L.) as an angiotensin-converting enzyme inhibitor. J. Food Sci. Technol. 2019, 57, 1847–1855. [Google Scholar]
- Sun, Y.; Chang, R.; Li, Q.; Li, B. Isolation and characterization of an antibacterial peptide from protein hydrolysates of Spirulina platensis. Eur. Food Res. Technol. 2016, 242, 685–692. [Google Scholar] [CrossRef]
- Kimatu, B.; Zhao, L.; Biao, Y.; Ma, G.; Yang, W.; Pei, F.; Hu, Q. Antioxidant potential of edible mushroom (Agaricus bisporus) protein hydrolysates and their ultrafiltration fractions. Food Chem. 2017, 230, 58–67. [Google Scholar] [CrossRef]
- Najafian, L.; Babji, A. Isolation, purification and identification of three novel antioxidative peptides from patin (Pangasius sutchi) myofibrillar protein hydrolysates. LWT Food Sci. Technol. 2014, 60, 452–461. [Google Scholar] [CrossRef]
- Irshad, I.; Kanekanian, A.; Peters, A.; Masud, T. Antioxidant activity of bioactive peptides derived from bovine casein hydrolysate fractions. J. Food Sci. Technol. 2015, 52, 231–239. [Google Scholar] [CrossRef]
- Ma, Y.; Li, L.; Yang, X. Preparation and antioxidant activities in vitro of a designed antioxidant peptide from Pinctada fucata by Recombinant Escherichia coli. J. Microbiol. Biotechnol. 2018, 28, 1–11. [Google Scholar]
- Wang, M.; Li, C.; Li, H.; Wu, Z.; Shen, Y. In vitro and in silico antioxidant activity of novel peptides prepared from Paeonia ostii ‘Feng Dan’ hydrolysate. Antioxidants 2019, 8, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Hong, Y.; Jia, Z.; Huang, G. Purification and identification of antioxidant peptides from hydrolysates of large yellow croaker (Pseudosciaena crocea) scales. Trans. ASABE 2020, 63, 289–294. [Google Scholar] [CrossRef]
- Je, J.; Qian, Z.; Byun, H.; Kim, S. Purification and characterization of an antioxidant peptide obtained from tuna backbone protein by enzymatic hydrolysis. Process Biochem. 2007, 42, 840–846. [Google Scholar] [CrossRef]
- Homayouni, M.; Asoodeh, A.; Soltani, M. Cytotoxic and antioxidant capacity of camel milk peptides: Effects of isolated peptide on superoxide dismutase and catalase gene expression. J. Food Drug Anal. 2016, 25, 567–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafer, Z.; Grassian, A.; Song, L.; Jiang, Z.; Gerhart-Hines, Z.; Irie, H.; Gao, S.; Puigserver, P.; Brugge, J. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009, 461, 109–113. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Aisa, H.; Yili, A. Isolation and identification of two potential antioxidant peptides from sheep abomasum protein hydrolysates. Eur. Food Res. Technol. 2018, 244, 1615–1625. [Google Scholar] [CrossRef]
- Shahcheraghi, S.H.; Salemi, F.; Peirovi, N.; Ayatollahi, J.; Alam, W.; Khan, H.; Saso, L. Nrf2 regulation by curcumin: Molecular aspects for therapeutic prospects. Molecules 2022, 27, 167. [Google Scholar] [CrossRef]
- Amritha, C.; Deepak Vasudevan, S.; Arjunan, K.; Divakar, S. The principal molecular mechanisms behind the activation of Keap1/Nrf2/ARE pathway leading to neuroprotective action in Parkinson’s disease. Neurochem. Int. 2022, 156, 105325. [Google Scholar]
- Adelusi, T.I.; Abdul-Hammed, M.; Idris, M.O.; Oyedele, Q.K.; Adedotun, I.O. Molecular dynamics, quantum mechanics and docking studies of some Keap1 inhibitors—An insight into the atomistic mechanisms of their antioxidant potential. Heliyon 2021, 7, e07317. [Google Scholar] [CrossRef] [PubMed]
| Pepetide Sequence | Molecular Weight (Da) | Structure Formula |
|---|---|---|
| LYEEMKESKVINADK | 1797.81 | 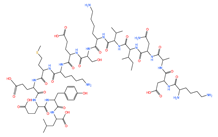 |
| LEADNVGVVLMGDGR | 1546.72 |  |
| LIDDSFGTDAPVPERL | 1746.88 |  |
| GLDELSEEDRLT | 1376.72 | 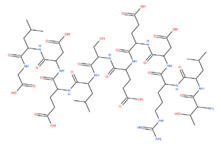 |
| Pepetide Sequence | pI | GRAVY | Water Solubility | Toxicity | Allergenicity | DPPH Radical Scavenging Activity (IC50, mg/mL) |
|---|---|---|---|---|---|---|
| LYEEMKESKVINADK | 4.87 | −1.007 | Good | non-toxin | Probable non-allergen | 0.53 ± 0.04 c |
| LEADNVGVVLMGDGR | 4.03 | 0.28 | Good | non-toxin | Probable non-allergen | 0.69 ± 0.06 b |
| LIDDSFGTDAPVPERL | 3.84 | −0.169 | Good | non-toxin | Probable non-allergen | 0.72 ± 0.04 b |
| GLDELSEEDRLT | 3.83 | −1.042 | Good | non-toxin | Probable non-allergen | 1.07 ± 0.09 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, X.; Pan, C.; Cai, M.; Li, L.; Yang, X.; Xiang, H.; Chen, S. Novel Antioxidant Peptides from Grateloupia livida Hydrolysates: Purification and Identification. Foods 2022, 11, 1498. https://doi.org/10.3390/foods11101498
Hu X, Pan C, Cai M, Li L, Yang X, Xiang H, Chen S. Novel Antioxidant Peptides from Grateloupia livida Hydrolysates: Purification and Identification. Foods. 2022; 11(10):1498. https://doi.org/10.3390/foods11101498
Chicago/Turabian StyleHu, Xiao, Chuang Pan, Miaomiao Cai, Laihao Li, Xianqing Yang, Huan Xiang, and Shengjun Chen. 2022. "Novel Antioxidant Peptides from Grateloupia livida Hydrolysates: Purification and Identification" Foods 11, no. 10: 1498. https://doi.org/10.3390/foods11101498
APA StyleHu, X., Pan, C., Cai, M., Li, L., Yang, X., Xiang, H., & Chen, S. (2022). Novel Antioxidant Peptides from Grateloupia livida Hydrolysates: Purification and Identification. Foods, 11(10), 1498. https://doi.org/10.3390/foods11101498







