Anti-Inflammatory Effects of Limosilactobacillus fermentum KGC1601 Isolated from Panax ginseng and Its Probiotic Characteristics
Abstract
:1. Introduction
2. Materials and Methods
2.1. L. fermentum KGC1601 and Other Bacterial Strains
2.2. L. fermentum KGC1601 Culture and Exopolysaccharide Purification
2.3. Whole-Genome Sequencing and Identification of Antibiotic Resistance and Virulence Genes
2.4. Carbohydrate Fermentation Patterns
2.5. Determination of Antibiotic Resistance
2.6. Determination of Hemolytic Activity and Biogenic Amine
2.7. Acid Resistance and Bile Salt Tolerance Tests
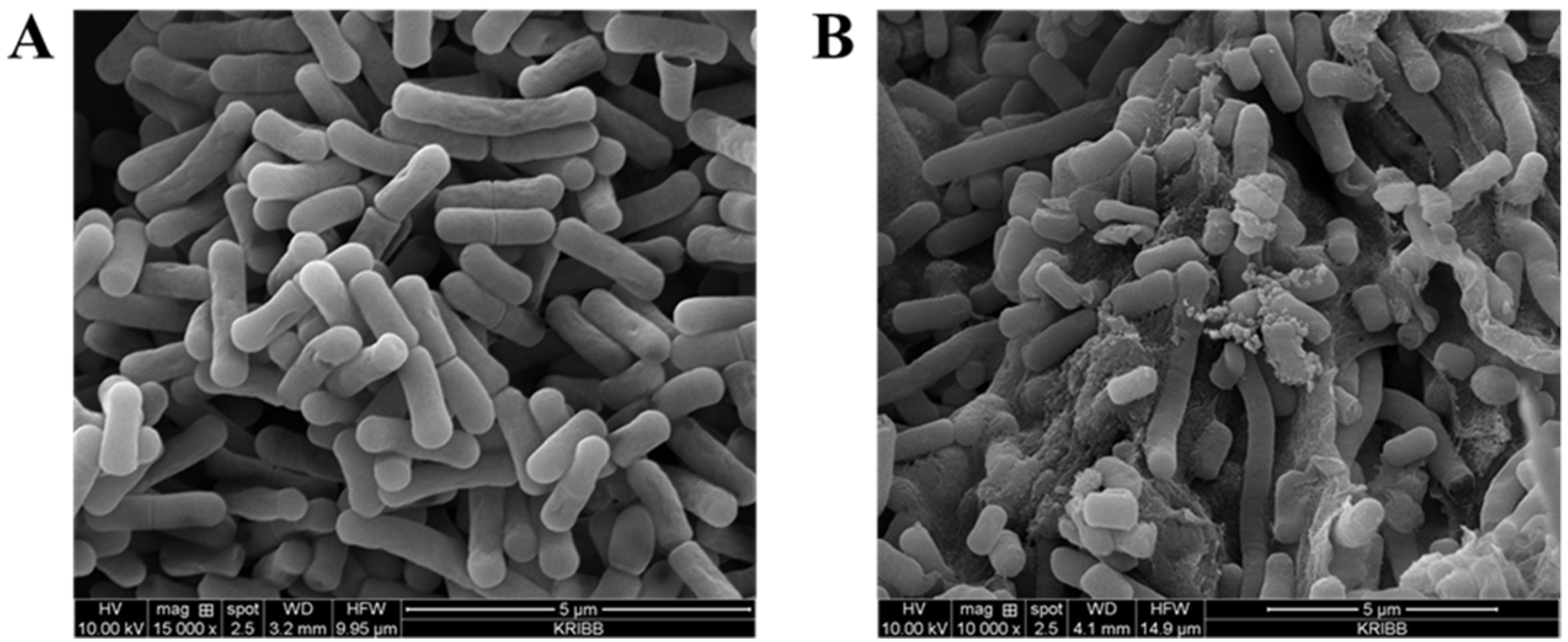
2.8. Identification of the Effect of Bile Salt Tolerance Using Scanning Electron Microscopy
2.9. Adhesion to HT-29 Cells
2.10. Evaluation of the Anti-Inflammatory Potential
2.10.1. Cell Culture
2.10.2. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.10.3. Enzyme-Linked Immunosorbent Assay (ELISA)
2.11. Statistics
3. Results and Discussion
3.1. Identification of L. fermentum Isolates
3.2. Carbohydrate Fermentation Patterns
3.3. Antibiotic Resistance
3.4. Hemolytic Activities and Biogenic Amine Production
3.5. Acid Resistance and Bile Salt Tolerance
3.6. Adhesion Abilities
3.7. Anti-Inflammatory Activity
3.7.1. Anti-Inflammatory Activity of L. fermentum KGC1601 on Raw 264.7 Cells by qRT-PCR
3.7.2. Anti-Inflammatory Activity of L. fermentum KGC1601 on Raw 264.7 Cells by ELISA
4. Conclusions
5. Patents
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gail, C.M. Use of probiotics: Benefits of a balanced microbiome in the intestinal tract. In Proceedings of the National Parent Club Canine Health Conference, St. Louis, MI, USA, 19–21 October 2007; pp. 19–21. [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO)/World Health Organization (WHO). Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation; Food and Nutrition Paper; World Health Organization: Rome, Italy, 2006; Volume 85, pp. 1–56. [Google Scholar]
- de Vrese, M.; Stegelmann, A.; Richter, B.; Fenselau, S.; Laue, C.; Schrezenmeir, J. Probiotics—Compensation for lactase insufficiency. Am. J. Clin. Nutr. 2001, 73, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Golinska, P.; Wypij, M.; Agarkar, G.; Rathod, D.; Dahm, H.; Rai, M. Endophytic actinobacteria of medicinal plants: Diversity and bioactivity. Antonie Leeuwenhoek 2015, 108, 267–289. [Google Scholar] [CrossRef] [Green Version]
- Nalini, M.S.; Prakash, H.S. Diversity and bioprospecting of actinomycete endophytes from the medicinal plants. Lett. Appl. Microbiol. 2017, 64, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, P.H. The Endosphere Microbiome of Ginseng. Plants 2022, 11, 415. [Google Scholar] [CrossRef]
- Hong, C.E.; Kim, J.U.; Lee, J.W.; Bang, K.H.; Jo, I.H. Metagenomic analysis of bacterial endophyte community structure and functions in Panax ginseng at different ages. 3 Biothech 2019, 9, 300. [Google Scholar] [CrossRef]
- Chowdhury, E.K.; Jeon, J.; Rim, S.O.; Park, Y.-H.; Lee, S.K.; Bae, H. Composition, diversity and bioactivity of culturable bacterial endophytes in mountain-cultivated ginseng in Korea. Sci. Rep. 2017, 7, 10098. [Google Scholar] [CrossRef] [Green Version]
- Mo, S.J.; Nam, B.; Bae, C.H.; Park, S.D.; Shim, J.J.; Lee, J.L. Characterization of novel Lactobacillus paracasei HY7017 capable of improving physiological properties and immune enhancing effects using red ginseng extract. Fermentation 2021, 7, 238. [Google Scholar] [CrossRef]
- Kim, S.; Choi, S.I.; Jang, M.; Jeong, Y.; Kang, C.H.; Kim, G.H. Anti-adipogenic effect of Lactobacillus fermentum MG4231 and MG4244 through AMPK pathway in 3T3-L1 preadipocytes. Food Sci. Biotechnol. 2020, 29, 1541–1551. [Google Scholar] [CrossRef]
- Zhu, K.; Tan, F.; Mu, J.; Yi, R.; Zhou, X.; Zhao, X. Anti-obesity effects of Lactobacillus fermentum CQPC05 isolated from Sichuan pickle in high-fat diet-induced obese mice through PPAR-α signaling pathway. Microorganisms 2019, 7, 194. [Google Scholar] [CrossRef] [Green Version]
- Pan, D.D.; Zeng, X.Q.; Yan, Y.T. Characterisation of Lactobacillus fermentum SM-7 isolated from koumiss, a potential probiotic bacterium with cholesterol-lowering effects. J. Sci. Food Agric. 2011, 91, 512–518. [Google Scholar] [CrossRef]
- Zhao, Y.; Hong, K.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Lactobacillus fermentum and its potential immunomodulatory properties. J. Funct. Foods 2019, 56, 21–32. [Google Scholar] [CrossRef]
- Shih, R.H.; Wang, C.Y.; Yang, C.M. NF-kappaB signaling pathways in neurological inflammation: A mini review. Front. Mol. Neurosci. 2015, 8, 77. [Google Scholar] [CrossRef] [Green Version]
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 428–435. [Google Scholar] [CrossRef]
- Rubin, D.C.; Shaker, A.; Levin, M.S. Chronic intestinal inflammation: Inflammatory bowel disease and colitis-associated colon cancer. Front. Immunol. 2012, 3, 107. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.Y.; Chae, S.A.; Bang, W.Y.; Lee, M.; Ban, O.; Kim, S.J.; Jung, Y.H.; Yang, J. Anti-inflammatory potential of Lactiplantibacillus plantarum IDCC 3501 and its safety evaluation. Braz. J. Microbiol. 2021, 52, 2299–2306. [Google Scholar] [CrossRef]
- Kim, K.T.; Kim, J.W.; Kim, S.I.; Kim, S.; Nguyen, T.H.; Kang, C.H. Antioxidant and anti-inflammatory effect and probiotic properties of lactic acid bacteria isolated from Canine and Feline feces. Microorganisms 2021, 9, 1971. [Google Scholar] [CrossRef]
- Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.D.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); et al. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar]
- Kwon, M.; Lee, J.; Park, S.; Kwon, O.H.; Seo, J.; Roh, S. Exopolysaccharide isolated from Lactobacillus plantarum L-14 has anti-inflammatory effects via the Toll-like receptor 4 pathway in LPS-induced RAW 264.7 cells. Int. J. Mol. Sci. 2020, 21, 9283. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Abriouel, H.; Muñoz, M.D.C.C.; Lerma, L.L.; Montoro, B.P.; Bockelmann, W.; Pichner, R.; Kabsch, J.; Cho, G.-S.; Franz, C.M.A.P.; Gálvez, A.; et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015, 78, 465–481. [Google Scholar] [CrossRef]
- Lee, B.S.; Ban, O.; Bang, W.Y.; Chae, S.A.; Oh, S.; Park, C.; Lee, M.; Kim, S.J.; Yang, J.; Jung, Y.H. Safety assessment of Lactobacillus reuteri IDCC 3701 based on phenotypic and genomic analysis. Ann. Microbiol. 2021, 71, 10. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.S.; Kim, Y.; Jeong, Y.; Kim, J.E.; Paek, N.S.; Kang, C.H. Antioxidant and probiotic properties of Lactobacilli and Bifidobacteria of human origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Nakajima, Y.; Ishibashi, J.; Yukuhiro, F.; Asaoka, A.; Taylor, D.; Yamakawa, M. Antibacterial activity and mechanism of action of tick defensin against Gram-positive bacteria. Biochim. Biophys. Acta Gen. Subj. 2003, 1624, 125–130. [Google Scholar] [CrossRef]
- Doeun, D.; Davaatseren, M.; Chung, M.S. Biogenic amines in foods. Food Sci. Biotechnol. 2017, 26, 1463–1474. [Google Scholar] [CrossRef]
- Ancin-Azpilicueta, C.; Gonzalez-Marco, A.; Jimenez-Moreno, N. Current knowledge about the presence of amines in wine. Crit. Rev. Food Sci. Nutr. 2008, 48, 257–275. [Google Scholar] [CrossRef]
- Shalaby, A.R. Significance of biogenic amines to food safety and human health. Food Res. Int. 1996, 29, 675–690. [Google Scholar] [CrossRef]
- Ruiz, L.; Margolles, A.; Sánchez, B. Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Immunol. 2013, 4, 396. [Google Scholar] [CrossRef] [Green Version]
- Piddock, L.J. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef] [Green Version]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [Green Version]
- Šušković, J.; Kos, B.; Matošić, S.; Besendorfer, V. The effect of bile salts on survival and morphology of a potential probiotic strain Lactobacillus acidophilus M92. World J. Microbiol. Biotechnol. 2000, 16, 673–678. [Google Scholar] [CrossRef]
- Kurdi, P.; Kawanishi, K.; Mizutani, K.; Yokota, A. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J. Bacteriol. 2006, 188, 1979–1986. [Google Scholar] [CrossRef] [Green Version]
- Gioacchini, G.; Rossi, G.; Carnevali, O. Host-probiotic interaction: New insight into the role of the endocannabinoid system by in vivo and ex vivo approaches. Sci. Rep. 2017, 7, 1261. [Google Scholar] [CrossRef] [Green Version]
- Zommiti, M.; Connil, N.; Hamida, J.B.; Ferchichi, M. Probiotic characteristics of Lactobacillus curvatus DN317, a strain isolated from chicken ceca. Probiotics Antimicrob. Proteins. 2017, 9, 415–424. [Google Scholar] [CrossRef]
- Srisesharam, S.; Park, H.S.; Soundharrajan, I.; Kuppusamy, P.; Kim, D.H.; Jayraaj, I.A.; Lee, K.D.; Choi, K.C. Evaluation of probiotic Lactobacillus plantarum against foodborne pathogens and its fermentation potential in improving Lolium multiflorum silage quality. 3 Biotech 2018, 8, 443. [Google Scholar] [CrossRef]
- Archer, A.C.; Kurrey, N.K.; Halami, P.M. In vitro adhesion and anti-inflammatory properties of native Lactobacillus fermentum and Lactobacillus delbrueckii spp. J. Appl. Microbiol. 2018, 125, 243–256. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Jeyaram, K.; Sanni, A.I.; Banwo, K. Production of exopolysaccharide by strains of Lactobacillus plantarum YO175 and OF101 isolated from traditional fermented cereal beverage. Peer. J. 2018, 6, e5326. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.H.; Pan, T.M.; Wu, Y.J.; Chang, S.J.; Chang, M.S.; Hu, C.Y. Exopolysaccharide activities from probiotic bifidobacterium: Immunomodulatory effects (on J774A. 1 macrophages) and antimicrobial properties. Int. J. Food Microbiol. 2010, 144, 104–110. [Google Scholar] [CrossRef]
- Górska, S.; Jachymek, W.; Rybka, J.; Strus, M.; Heczko, P.B.; Gamian, A. Structural and immunochemical studies of neutral exopolysaccharide produced by Lactobacillus johnsonii 142. Carbohydr. Res. 2010, 345, 108–114. [Google Scholar] [CrossRef]
- Salazar, N.; Neyrinck, A.M.; Bindels, L.B.; Druart, C.; Ruas-Madiedo, P.; Cani, P.D.; Reyes-Gavilán, C.G.; Delzenne, N.M. Functional effects of EPS-producing bifidobacterium administration on energy metabolic alterations of diet-induced obese mice. Front. Immunol. 2019, 10, 1809. [Google Scholar] [CrossRef] [Green Version]
- Sungur, T.; Aslim, B.; Karaaslan, C.; Aktas, B. Impact of Exopolysaccharides (EPSs) of Lactobacillus gasseri strains isolated from human vagina on cervical tumor cells (HeLa). Anaerobe 2017, 47, 137–144. [Google Scholar] [CrossRef]
- Ciszek-Lenda, M.; Nowak, B.; Śróttek, M.; Gamian, A.; Marcinkiewicz, J. Immunoregulatory potential of exopolysaccharide from Lactobacillus rhamnosus KL37. Effects on the production of inflammatory mediators by mouse macrophages. Int. J. Exp. Pathol. 2011, 92, 382–391. [Google Scholar] [CrossRef]





| Gene | Primer Sequence (5′ to 3′) | Product Size (bp) | |
|---|---|---|---|
| IL-1β | Forward | AGG TCA AAG GTT TGG AAG CA | 129 |
| Reverse | TGA AGC AGC TAT GGC AAC TG | ||
| TNF-α | Forward | AGG GTC TGG GCC ATA GAA CT | 103 |
| Reverse | CCA CCA CGC TCT TCT GTC TAC | ||
| IL-6 | Forward | GTC CTT CAG AGA GAT ACA GAA ACT | 113 |
| Reverse | AGC TTA TCT GTT AGG AGA GCA TTG | ||
| GAPDH | Forward | CCA TGG AGA AGG CTG GGG | 159 |
| Reverse | CAA AGT TGT CAT GGA TGA CC |
| Title 1 | Title 3 |
|---|---|
| Identification | Limosilactobacillus fermentum |
| Genome size (bp) | 2,058,202 |
| GC contents (%) | 51.9 |
| CDS | 1998 |
| No. of rRNA genes | 11 |
| No. of tRNA genes | 54 |
| Carbohydrates | Strains | Carbohydrates | Strains | ||
|---|---|---|---|---|---|
| KGC 1601 | KCTC 3112 | KGC 1601 | KCTC 3112 | ||
| Glycerol | − | − | Salicin | − | − |
| Erythritol | − | − | D-cellobiose | − | − |
| D-arabinose | − | − | D-maltose | + | + |
| L-arabinose | − | − | D-lactose | − | + |
| D-ribose | + | + | D-melibiose | + | + |
| D-xylose | + | − | D-saccharose(sucrose) | + | + |
| L-xylose | − | − | D-trehalose | + | − |
| D-adonitol | − | − | Inulin | − | − |
| Methyl-tolee(sucrose). o | − | − | D-melezitose | − | − |
| D-galactose | − | + | D-raffinose | + | − |
| D-glucose | + | + | Amidon (starch) | − | − |
| D-fructose | + | + | Glycogen | − | − |
| D-mannose | + | + | Xylitol | − | − |
| L-sorbose | − | − | Gentiobiose | − | − |
| L-rhamnose | − | − | D-turanose | − | − |
| Dulcitol | − | − | D-lyxose | − | − |
| Inocitol | − | − | D-tagatose | − | − |
| D-mannitol | − | − | D-fucose | − | − |
| D-sorbitol | − | − | L-fucose | − | − |
| Methylseolerch)crose). o | − | − | D-arabitol | − | − |
| Methylitolerch)crose). of | − | − | L-arabitol | − | − |
| N-acetylglucosamine | − | − | Potassium gluconate | − | − |
| Amygdalin | − | − | Potassium 2-ketogluconate | − | − |
| Arbutin | − | − | Potassium 5-ketogluconate | + | − |
| Esculin ferric citrate | + | + | |||
| Antibiotic 1 | AMP | GEN | KAN | STR | ERY | CLI | TET | CHL |
|---|---|---|---|---|---|---|---|---|
| Cut-off value (mg/L) 2 | 2 | 16 | 32 | 64 | 1 | 1 | 8 | 4 |
| Observed MICs | 0.016–0.023 | 12–16 | >256 | 96–128 | 0.19–0.25 | 0.032 | 0.25–0.38 | 4 |
| Assessment | S 3 | S | R 4 | R | S | S | S | S |
| Characteristics | Survival Rate (%) | ||
|---|---|---|---|
| KGC1601 | KCTC3112 | ||
| Acid resistance | 0 h (log CFU/mL) | 8.33 | 8.39 |
| 3 h (log CFU/mL) | 5.46 | 5.55 | |
| Survival rate (%) | 66 | 66 | |
| Bile salt tolerance | 0 h (log CFU/mL) | 8.33 | 8.39 |
| 3 h (log CFU/mL) | 6.35 | 3.44 | |
| Survival rate (%) | 76 | 41 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.; Lee, Y.-S.; Yu, H.-Y.; Kwon, M.; Kim, K.-K.; In, G.; Hong, S.-K.; Kim, S.-K. Anti-Inflammatory Effects of Limosilactobacillus fermentum KGC1601 Isolated from Panax ginseng and Its Probiotic Characteristics. Foods 2022, 11, 1707. https://doi.org/10.3390/foods11121707
Kim H, Lee Y-S, Yu H-Y, Kwon M, Kim K-K, In G, Hong S-K, Kim S-K. Anti-Inflammatory Effects of Limosilactobacillus fermentum KGC1601 Isolated from Panax ginseng and Its Probiotic Characteristics. Foods. 2022; 11(12):1707. https://doi.org/10.3390/foods11121707
Chicago/Turabian StyleKim, Heejin, Yun-Seok Lee, Hye-Young Yu, Mijin Kwon, Ki-Kwang Kim, Gyo In, Soon-Ki Hong, and Sang-Kyu Kim. 2022. "Anti-Inflammatory Effects of Limosilactobacillus fermentum KGC1601 Isolated from Panax ginseng and Its Probiotic Characteristics" Foods 11, no. 12: 1707. https://doi.org/10.3390/foods11121707






