Identification of Maize with Different Moldy Levels Based on Catalase Activity and Data Fusion of Hyperspectral Images
Abstract
:1. Introduction
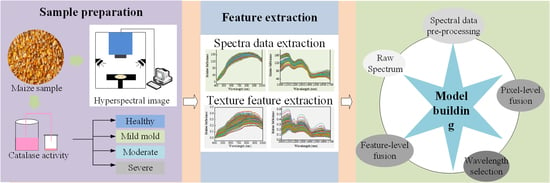
2. Materials and Methods
2.1. Maize Sample Preparation
2.2. Hyperspectral Image Acquisition System
2.3. Determination of CAT Activity
2.4. Hyperspectral Image Processing and Information Extraction
2.5. Spectral Data Preprocessing
2.6. Data Fusion
2.7. Discriminant Model and Evaluation
3. Results and Analysis
3.1. CAT Activity Analysis of Maize with Different Moldy Levels
3.2. Spectral and Texture Characterization
3.3. Comparison and Optimization of Different Classifiers and Preprocessing Methods
3.4. Pixel-Level Fusion Based on Full Wavelengths Spectra and Texture Data
3.5. Classification Model Built by Feature-Level Fusion of Spectra and Texture Data
3.6. Determination of the Optimal Feature-Level Fusion Model
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ambrose, A.; Kandpal, L.M.; Kim, M.S.; Lee, W.H.; Cho, B.K. High speed measurement of corn seed viability using hyperspectral imaging. Infrared Phys. Technol. 2016, 75, 173–179. [Google Scholar] [CrossRef]
- Hesseltine, C.W. Natural occurrence of mycotoxins in cereals. Mycopathol. Mycol. Appl. 1974, 53, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Pearson, T.C.; Wicklow, D.T.; Pasikatan, M.C. Reduction of Aflatoxin and Fumonisin Contamination in Yellow Corn by High-Speed Dual-Wavelength Sorting. Cereal Chem. J. 2004, 81, 490–498. [Google Scholar] [CrossRef] [Green Version]
- Kimuli, D.; Wang, W.; Wang, W.; Jiang, H.Z.; Zhao, X.; Chu, X. Application of SWIR hyperspectral imaging and chemometrics for identification of aflatoxin B1 contaminated maize kernels. Infrared Phys. Technol. 2018, 89, 351–362. [Google Scholar] [CrossRef]
- Wang, W.; Ni, X.Z.; Lawrence, K.C.; Yoon, S.C.; Heitschmidt, G.W.; Feldner, P. Feasibility of detecting Aflatoxin B1 in single maize kernels using hyperspectral imaging. J. Food Eng. 2015, 166, 182–192. [Google Scholar] [CrossRef]
- Campone, L.; Piccinelli, A.L.; Celano, R.; Rastrelli, L. Application of dispersive liquid-liquid microextraction for the determination of aflatoxins B1, B2, G1 and G2 in cereal products. J. Chromatogr. A 2011, 1218, 7648–7654. [Google Scholar] [CrossRef]
- Maragos, C.M.; Busman, M. Rapid and advanced tools for mycotoxin analysis: A review. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2010, 27, 688–700. [Google Scholar] [CrossRef]
- Krnjaja, V.; Stanojkovic, A.; Stankovic, S.; Lukic, M.; Bijelic, Z.; Mandic, V.; Micic, N. Fungal contamination of maize grain samples with a special focus on toxigenic genera. Biotechnol. Anim. Husb. 2017, 33, 233–241. [Google Scholar] [CrossRef]
- Guo, Z.M.; Wang, M.M.; Wu, J.Z.; Tao, F.F.; Chen, Q.S.; Wang, Q.Y.; Ouyang, Q.; Shi, J.Y.; Zou, X.B. Quantitative assessment of zearalenone in maize using multivariate algorithms coupled to Raman spectroscopy. Food Chem. 2019, 286, 282–288. [Google Scholar] [CrossRef]
- Huang, K.Y.; Chien, M.C. A Novel Method of Identifying Paddy Seed Varieties. Sensors 2017, 17, 809. [Google Scholar] [CrossRef]
- Singh, C.B.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Detection of Sprouted and Midge-Damaged Wheat Kernels Using Near-Infrared Hyperspectral Imaging. Cereal Chem. J. 2009, 86, 256–260. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Chen, W.; Wang, Z.; Wang, J.; Huo, Y. Rapid detection of Aspergillus spp. infection levels on milled rice by headspace-gas chromatography ion-mobility spectrometry (HS-GC-IMS) and E-nose. LWT 2020, 132, 109758. [Google Scholar] [CrossRef]
- Leggieri, M.C.; Mazzoni, M.; Fodil, S.; Moschini, M.; Bertuzzi, T.; Prandini, A.; Battilani, P. An electronic nose supported by an artificial neural network for the rapid detection of aflatoxin B1 and fumonisins in maize. Food Control. 2021, 123, 107722. [Google Scholar] [CrossRef]
- Ng, H.F.; Wilcke, W.F.; Morey, R.V.; Lang, J.P. Machine Vision Color Calibration in Assessing Corn Kernel Damage. Trans. ASAE 1998, 41, 727–732. [Google Scholar] [CrossRef]
- Shi, Y. Research on the Visual Detection Method of Corn Seed Grain Mold and Mold Degree. Master’s Thesis, Yangzhou University, Yangzhou, China, 2021. [Google Scholar]
- Fan, Y.M.; Ma, S.C.; Wu, T.T. Individual wheat kernels vigor assessment based on NIR spectroscopy coupled with machine learning methodologies. Infrared Phys. Technol. 2020, 105, 103213. [Google Scholar] [CrossRef]
- Zhang, B.H.; Huang, W.Q.; Li, J.B.; Zhao, Z.J.; Fan, S.X.; Wu, J.T.; Liu, C.L. Principles, developments and applications of computer vision for external quality inspection of fruits and vegetables: A review. Food Res. Int. 2014, 62, 326–343. [Google Scholar] [CrossRef]
- Singh, C.B.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Identification of insect-damaged wheat kernels using short-wave near-infrared hyperspectral and digital colour imaging. Comput. Electron. Agric. 2010, 73, 118–125. [Google Scholar] [CrossRef]
- Zhou, Q.; Huang, W.Q.; Fan, S.X.; Zhao, F.; Liang, D.; Tian, X. Non-destructive discrimination of the variety of sweet maize seeds based on hyperspectral image coupled with wavelength selection algorithm. Infrared Phys. Technol. 2020, 109, 103418. [Google Scholar] [CrossRef]
- Tao, F.F.; Yao, H.B.; Hruska, Z.; Kincaid, R.; Rajasekaran, K.; Bhatnagar, D. A novel hyperspectral-based approach for identification of maize kernels infected with diverse Aspergillus flavus fungi. Biosyst. Eng. 2020, 200, 415–430. [Google Scholar] [CrossRef]
- Williams, P.J.; Geladi, P.; Britz, T.J.; Manley, M. Investigation of fungal development in maize kernels using NIR hyperspectral imaging and multivariate data analysis. J. Cereal Sci. 2012, 55, 272–278. [Google Scholar] [CrossRef]
- Dai, S.S.; Yong, Y. Fisher discriminant analysis for moldy degrees of maize samples based on the feature selection of hyperspectral data. Food Mach. 2018, 34, 68–72. [Google Scholar] [CrossRef]
- Del Fiore, A.; Reverberi, M.; Ricelli, A.; Pinzari, F.; Serranti, S.; Fabbri, A.A.; Fanelli, C. Early detection of toxigenic fungi on maize by hyperspectral imaging analysis. Int. J. Food Microbiol. 2010, 144, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.D.; Qing, L.W.; Yan, D.T.; Xia, G.; Zhang, C.; Yun, Y.H.; Zhang, W. Hyperspectral imaging in combination with data fusion for rapid evaluation of tilapia fillet freshness. Food Chem. 2021, 348, 129129. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Pu, H.; Sun, D.W.; Gao, W.; Qu, J.H.; Ma, K.Y. Application of Vis–NIR hyperspectral imaging in classification between fresh and frozen-thawed pork Longissimus Dorsi muscles. Int. J. Refrig. 2015, 50, 10–18. [Google Scholar] [CrossRef]
- Sippy, N.; Luxton, R.; Lewis, R.J.; Cowell, D.C. Rapid electrochemical detection and identification of catalase positive micro-organisms. Biosens. Bioelectron. 2003, 18, 741–749. [Google Scholar] [CrossRef]
- Zhang, S.B.; Zhai, H.C.; Hu, Y.S.; Wang, L.; Yu, G.H.; Huang, S.X.; Cai, J.P. A rapid detection method for microbial spoilage of agro-products based on catalase activity. Food Control. 2014, 42, 220–224. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zai, H.C.; Zhang, S.B.; Cai, J.P. Early warning of AFB_1 contamination in stored maize based on monitoring of peroxidase activity. Food Mach. 2017, 33, 110–113. [Google Scholar] [CrossRef]
- Zhang, Q.B.; Li, T.; Wang, C.Q.; Cai, Y.; Yang, J.W.; Zhang, R.P.; Li, B. Microbial Agents: Effects on Activities of Urease and Catalase in Flue-cured Tobacco Rhizosphere Soil. Chin. Agric. Sci. Bull. 2016, 32, 46–50. [Google Scholar] [CrossRef]
- Li, J.B.; Huang, W.Q.; Chen, L.P.; Fan, S.X.; Zhang, B.H.; Guo, Z.M.; Zhao, C.J. Variable Selection in Visible and Near-Infrared Spectral Analysis for Noninvasive Determination of Soluble Solids Content of ‘Ya’ Pear. Food Anal. Methods 2014, 7, 1891–1902. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.S. Combined spectral and spatial processing of ERTS imagery data. Remote Sens. Environ. 1974, 3, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.M.; Wang, M.M.; Agyekum, A.A.; Wu, J.Z.; Chen, Q.S.; Zuo, M.; El-Seedi, H.R.; Tao, F.F.; Shi, J.Y.; Ouyang, Q.; et al. Quantitative detection of apple watercore and soluble solids content by near infrared transmittance spectroscopy. J. Food Eng. 2020, 279, 109955. [Google Scholar] [CrossRef]
- Wang, H.L.; Peng, J.Y.; Xie, C.Q.; Bao, Y.D.; He, Y. Fruit quality evaluation using spectroscopy technology: A review. Sensors 2015, 15, 11889–11927. [Google Scholar] [CrossRef] [Green Version]
- Ilari, J.L.; Martens, H.; Isaksson, T. Determination of Particle Size in Powders by Scatter Correction in Diffuse Near-Infrared Reflectance. Appl. Spectrosc. 1988, 42, 722–728. [Google Scholar] [CrossRef]
- Sánchez, M.T.; Haba la Haba, M.J.; Benítez-López, M.; Fernández-Novales, J.; Garrido-Varo, A.; Pérez-Marín, D. Non-destructive characterization and quality control of intact strawberries based on NIR spectral data. J. Food Eng. 2012, 110, 102–108. [Google Scholar] [CrossRef]
- Borras, E.; Ferre, J.; Boque, R.; Mestres, M.; Acena, L.; Calvo, A.; Busto, O. Olive oil sensory defects classification with data fusion of instrumental techniques and multivariate analysis (PLS-DA). Food Chem. 2016, 203, 314–322. [Google Scholar] [CrossRef]
- An, T.; Huang, W.Q.; Tian, X.; Fan, S.X.; Duan, D.D.; Dong, C.W.; Zhao, C.J.; Li, G.L. Hyperspectral imaging technology coupled with human sensory information to evaluate the fermentation degree of black tea. Sens. Actuators B. Chem. 2022, 366, 131994. [Google Scholar] [CrossRef]
- Borràs, E.; Ferré, J.; Boqué, R.; Mestres, M.; Aceña, L.; Busto, O. Data fusion methodologies for food and beverage authentication and quality assessment—A review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef]
- Huang, F.R.; Song, H.; Guo, L.; Guang, P.W.; Yang, X.H.; Li, L.Q.; Yang, M.X. Detection of adulteration in Chinese honey using NIR and ATR-FTIR spectral data fusion. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2020, 235, 118297. [Google Scholar] [CrossRef]
- Yun, Y.H.; Wang, W.T.; Deng, B.C.; Lai, G.B.; Liu, X.B.; Ren, D.B.; Liang, Y.Z.; Fan, W.; Xu, Q.S. Using variable combination population analysis for variable selection in multivariate calibration. Anal. Chim. Acta 2015, 862, 14–23. [Google Scholar] [CrossRef]
- Yun, Y.H.; Wang, W.T.; Tan, M.L.; Liang, Y.Z.; Li, H.D.; Cao, D.S.; Lu, H.M.; Xu, Q.S. A strategy that iteratively retains informative variables for selecting optimal variable subset in multivariate calibration. Anal. Chim. Acta 2014, 807, 36–43. [Google Scholar] [CrossRef]
- Yun, Y.H.; Li, H.D.; Deng, B.C.; Cao, D.S. An overview of variable selection methods in multivariate analysis of near-infrared spectra. TrAC Trends Anal. Chem. 2019, 113, 102–115. [Google Scholar] [CrossRef]
- Sánchez, A.V.D. Advanced support vector machines and kernel methods. Neurocomputing 2003, 55, 5–20. [Google Scholar] [CrossRef]
- Li, J.B.; Huang, W.Q.; Zhao, C.J.; Zhang, B.H. A comparative study for the quantitative determination of soluble solids content, pH and firmness of pears by Vis/NIR spectroscopy. J. Food Eng. 2013, 116, 324–332. [Google Scholar] [CrossRef]
- Nunes Miranda, E.; Henrique Groenner Barbosa, B.; Henrique Godinho Silva, S.; Augusto Ussi Monti, C.; Yue Phin Tng, D.; Rezende Gomide, L. Variable selection for estimating individual tree height using genetic algorithm and random forest. For. Ecol. Manag. 2022, 504, 119828. [Google Scholar] [CrossRef]
- Zhao, D.D.; Hu, X.Y.; Xiong, S.W.; Tian, J.; Xiang, J.W.; Zhou, J.; Li, H.H. k-means clustering and kNN classification based on negative databases. Appl. Soft Comput. 2021, 110, 107732. [Google Scholar] [CrossRef]
- Marques, A.S.; Castro, J.N.F.; Costa, F.J.M.D.; Neto, R.M.; Lima, K.M.G. Near-infrared spectroscopy and variable selection techniques to discriminate Pseudomonas aeruginosa strains in clinical samples. Microchem. J. 2016, 124, 306–310. [Google Scholar] [CrossRef]
- Daszykowski, M.; Wrobel, M.S.; Czarnik-Matusewicz, M.; Walczak, B. Near-infrared reflectance spectroscopy and multivariate calibration techniques applied to modelling the crude protein, fibre and fat content in rapeseed meal. Analyst 2008, 133, 1523–1531. [Google Scholar] [CrossRef]
- Alhamdan, A.M.; Atia, A. Non-destructive method to predict Barhi dates quality at different stages of maturity utilising near-infrared (NIR) spectroscopy. Int. J. Food Prop. 2017, 20, 2950–2959. [Google Scholar] [CrossRef]
- Li, J.B.; Luo, W.; Wang, Z.L.; Fan, S.X. Early detection of decay on apples using hyperspectral reflectance imaging combining both principal component analysis and improved watershed segmentation method. Postharvest Biol. Technol. 2019, 149, 235–246. [Google Scholar] [CrossRef]
- Paz, P.; Sanchez, M.T.; Perez-Marin, D.; Guerrero, J.E.; Garrido-Varo, A. Nondestructive determination of total soluble solid content and firmness in plums using near-infrared reflectance spectroscopy. J. Agric. Food Chem. 2008, 56, 2565–2570. [Google Scholar] [CrossRef]
- Delwiche, S.R.; Reeves, J.B., III. The effect of spectral pre-treatments on the partial least squares modeling of agricultural products. J. Near Infrared Spectrosc. 2004, 12, 177–182. [Google Scholar] [CrossRef]
- Stasiewicz, M.J.; Falade, T.D.O.; Mutuma, M.; Mutiga, S.K.; Harvey, J.J.W.; Fox, G.; Nelson, R.J. Multi-spectral kernel sorting to reduce aflatoxins and fumonisins in Kenyan maize. Food Control 2017, 78, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Chu, X. Hyperspectral Imaging Identifications for Cereal Fungi and Detection Methods for Moldy Maize Kernels. Ph.D. Thesis, China Agriculture University, Beijing, China, 2018. [Google Scholar]
- Wang, Z.L.; Tian, X.; Fan, S.X.; Zhang, C.; Li, J.B. Maturity determination of single maize seed by using near-infrared hyperspectral imaging coupled with comparative analysis of multiple classification models. Infrared Phys. Technol. 2021, 112, 103596. [Google Scholar] [CrossRef]
- Fan, S.X.; Li, J.B.; Huang, W.Q.; Chen, L.P. Data Fusion of Two Hyperspectral Imaging Systems with Complementary Spectral Sensing Ranges for Blueberry Bruising Detection. Sensors 2018, 18, 4463. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Hruska, Z.; Kincaid, R.; Brown, R.; Cleveland, T.; Bhatnagar, D. Correlation and classification of single kernel fluorescence hyperspectral data with aflatoxin concentration in corn kernels inoculated with Aspergillus flavus spores. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2010, 27, 701–709. [Google Scholar] [CrossRef]










| Moldy Level | Mean mL/(h × g) | Standard Deviation mL/(h × g) |
|---|---|---|
| Healthy level | 0 | 0 |
| Mild level | 1.57 | 0.13 |
| Moderate level | 1.91 | 0.09 |
| Severe level | 2.24 | 0.12 |
| Moldy Level | Calibration Set | Prediction Set | ||||
|---|---|---|---|---|---|---|
| Num | Mean | Standard Deviation | Num | Mean | Standard Deviation | |
| Healthy level | 35 | 0 | 0 | 15 | 0 | 0 |
| Mild level | 35 | 1.58 | 0.17 | 15 | 1.58 | 0.14 |
| Moderate level | 35 | 1.94 | 0.18 | 15 | 1.96 | 0.20 |
| Severe level | 35 | 2.20 | 0.14 | 15 | 2.13 | 0.24 |
| Classifier | Sensor | Spectral Preprocessing Method | Calibration Set Accuracy (%) | Prediction Set Accuracy (%) |
|---|---|---|---|---|
| SVM | Vis-SWNIR | smooth-msc | 84.44 | 88.33 |
| smooth-detrend | 86.67 | 88.33 | ||
| smooth-center | 85.56 | 90.00 | ||
| LWNIR | smooth-msc | 90.56 | 86.67 | |
| smooth-detrend | 90.56 | 88.33 | ||
| smooth-center | 91.11 | 85.00 |
| Sensor | Data Source | Calibration Set Accuracy (%) | Prediction Set Accuracy (%) | |
|---|---|---|---|---|
| Spectra | Texture | |||
| Vis-SWNIR | smooth-detrend | contrast | 85.56 | 86.67 |
| correction | 85.56 | 86.67 | ||
| energy | 92.22 | 90.00 | ||
| homogeneity | 85.56 | 86.67 | ||
| LWNIR | smooth-detrend | contrast | 92.22 | 90.00 |
| correction | 92.22 | 88.33 | ||
| energy | 97.78 | 85.00 | ||
| homogeneity | 92.22 | 88.33 | ||
| Integration Method | Sensor | Data Source | Variable Selection Algorithm | Characteristic Number | Calibration Set Accuracy (%) | Prediction Set Accuracy (%) | |
|---|---|---|---|---|---|---|---|
| Spectra | Texture | ||||||
| Feature-level fusion | Vis-SWNIR | smooth-detrend energy | VCPA | 9 | 12 | 94.44 | 93.33 |
| IRIV | 21 | 28 | 97.78 | 95.00 | |||
| mVCPA-IRIV | 28 | 39 | 93.89 | 91.67 | |||
| LWNIR | smooth-detrend contrast | VCPA | 12 | 12 | 96.67 | 90.00 | |
| IRIV | 13 | 35 | 100.00 | 83.33 | |||
| mVCPA-IRIV | 17 | 41 | 99.44 | 91.97 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Huang, W.; Yu, H.; Tian, X. Identification of Maize with Different Moldy Levels Based on Catalase Activity and Data Fusion of Hyperspectral Images. Foods 2022, 11, 1727. https://doi.org/10.3390/foods11121727
Wang W, Huang W, Yu H, Tian X. Identification of Maize with Different Moldy Levels Based on Catalase Activity and Data Fusion of Hyperspectral Images. Foods. 2022; 11(12):1727. https://doi.org/10.3390/foods11121727
Chicago/Turabian StyleWang, Wenchao, Wenqian Huang, Huishan Yu, and Xi Tian. 2022. "Identification of Maize with Different Moldy Levels Based on Catalase Activity and Data Fusion of Hyperspectral Images" Foods 11, no. 12: 1727. https://doi.org/10.3390/foods11121727
APA StyleWang, W., Huang, W., Yu, H., & Tian, X. (2022). Identification of Maize with Different Moldy Levels Based on Catalase Activity and Data Fusion of Hyperspectral Images. Foods, 11(12), 1727. https://doi.org/10.3390/foods11121727